Abstract
Evolution of the tandemly repeated ribosomal RNA (rRNA) genes is intriguing because in each species all units within the array are highly uniform in sequence but that sequence differs between species. In this review we summarize the origins of the current models to explain this process of concerted evolution, emphasizing early studies of recombination in yeast and more recent studies in Drosophila and mammalian systems. These studies suggest that unequal crossover is the major driving force in the evolution of the rRNA genes with sister chromatid exchange occurring more often than exchange between homologs. Gene conversion is also believed to play a role; however, direct evidence for its involvement has not been obtained. Remarkably, concerted evolution is so well orchestrated that even transposable elements that insert into a large fraction of the rRNA genes appear to have little effect on the process. Finally, we summarize data that suggest that recombination in the rDNA locus of higher eukaryotes is sufficiently frequent to monitor changes within a few generations.
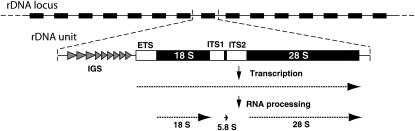
THE most conserved and most utilized genes in eukaryotes are those encoding ribosomal RNA (rRNA). All lineages organize the single rRNA of the small ribosomal subunit (18S RNA) and two of the rRNAs of the large ribosomal subunit (5.8S and 28S RNA) into 1 transcription unit (Figure 1). Because of the massive numbers of ribosomes needed during periods of rapid growth, eukaryotes typically encode hundreds of copies of this transcription unit. These rDNA units are organized in large tandem arrays, the rDNA loci, on one or a small number of chromosomes. During active synthesis these rDNA loci form the nucleoli visible in all cells. All aspects of the rRNA genes suggest that they have changed relatively little in the billion years since the separation of animals and plants.
Figure 1.—
Organization of the ribosomal RNA (rRNA) genes in eukaryotes. The genes are organized into tandemly repeated units as diagrammed at the top. A typical unit is shown in expanded detail. The positions of the three rRNA genes (18S, 5.8S, 28S) are indicated with solid boxes, while regions processed from the primary transcript are in open boxes (ETS, external transcribed spacer; ITS, internal transcribed spacer). Between the transcription units are the intergenic spacers (IGS), which in most species are composed of one or more internally repeated sequences (shaded arrowheads). The extent and direction of the transcribed region of each unit as well as the final mature rRNAs derived from that transcript are shown at the bottom as dotted arrows.
One of the most fascinating observations to arise from the study of the tandem rDNA units was their uniformity in sequence, yet that sequence could change over time. The ability of all rDNA units to change their sequence in a highly orchestrated manner is described today as concerted evolution. The mechanism by which new mutations in one gene are eliminated or spread to adjacent genes has been the subject of experimentation and speculation for nearly 35 years. However, the rDNA loci are large and few tools are available to dissect them; thus our models today remain quite general. The lessons that have been derived from studies of rRNA genes are frequently applied to other multigene families, and the lessons learned from these families have provided insights into the rRNA genes (for a recent review see Nei and Rooney 2005). This short review, however, focuses exclusively on the rRNA genes, retracing their long history of study and summarizing what we know today about their mechanism of evolution.
ORIGINAL DISCOVERY AND THE SUGGESTION OF A SIMPLE MODEL
Studies of the rRNA genes have a long history because the characterization of their sequence identity both within and between species was possible before DNA cloning and sequencing methods became available. The abundant rRNA transcripts readily available from any organism led to the development of saturation and competitive hybridization methods to estimate the number and sequence similarity of the genes (Long and Dawid 1980). Cross-hybridization of rRNA sequences from organisms as taxonomically diverse as plants and animals was observed, suggesting very high selective pressure to preserve a specific nucleotide sequence. This conservation appeared to account for the uniformity of sequence between the different copies of the genes within each organism.
An unexpected finding was obtained in the first detailed studies of the complete rDNA repeat (Brown et al. 1972). African clawed frogs (Xenopus) synthesize abundant extrachromosomal rDNA arrays during the development of their oocytes. The ability to purify these rDNA arrays allowed hybridization studies to score similarity across the entire unit. The studies revealed that like the genes themselves the regions processed from the primary transcript (the external and internal transcribed spacers, ETS and ITS in Figure 1) as well as the regions between the transcribed units (the intergenic spacer, IGS in Figure 1) were also uniform in sequence. However, these spacer regions differed significantly in sequence between two closely related species. Obviously, if the spacer regions were “free” to diverge between species, then selective pressure alone could not account for the uniformity of all units within a species. It seemed clear that a “correction” mechanism was necessary to spread new nucleotide substitutions (mutations) among all the units of the tandem array.
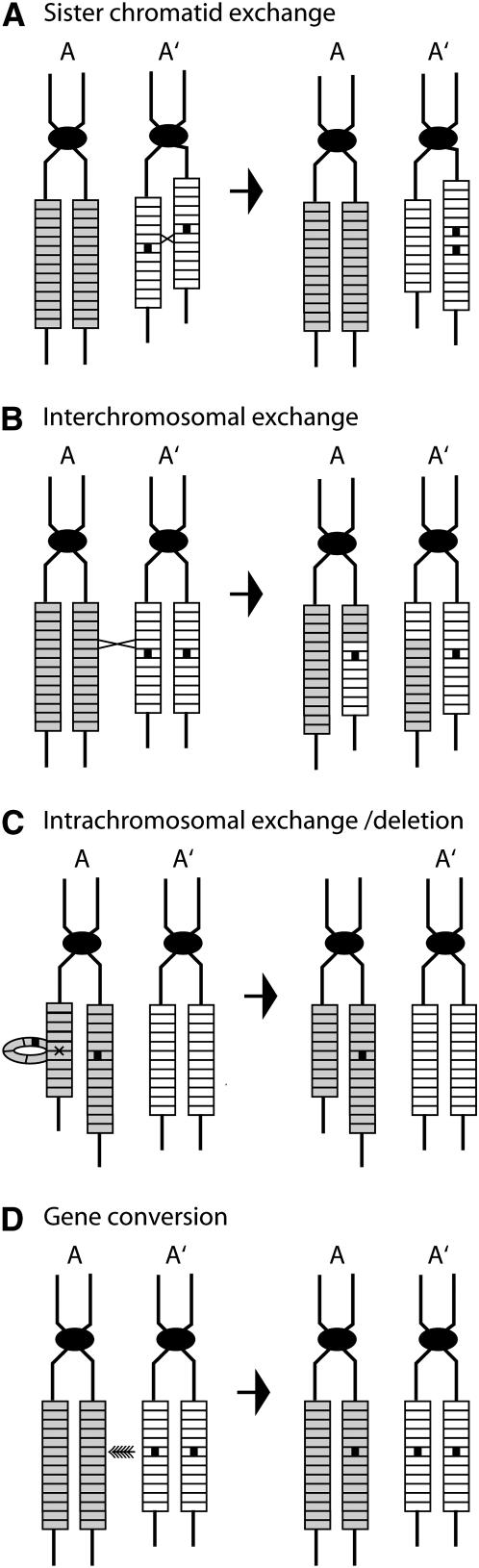
Two findings pertaining to the rRNA genes in frogs as well as fruit flies pointed to an answer. First, individuals from the same species had different numbers of rRNA genes. Second, the number of rRNA genes retained by most individuals appeared to be in excess of the number needed for survival. The variation in number of rDNA units found among individuals was hypothesized to occur by homologous recombination between rDNA units located at different positions within the loci on two chromosomes (Figure 2, A and B). Such “unequal crossover” events would generate one recombinant chromosome with more rDNA units and another chromosome with fewer units. Any chromosome with too few rDNA units would be selected against, while chromosomes with large arrays might be more susceptible to intrachromosomal crossovers (Figure 2C).
Figure 2.—
Four possible recombination mechanisms that may occur within or between rDNA loci. A and A′ represent homologous chromosomes; the rectangles, individual rDNA units; and the small solid boxes, mutations. Each chromosome is drawn after replication to show the two sister chromatids still attached by means of their centromeres (solid oval). All four recombination mechanisms can lead to the duplication or loss of a mutation on a chromosome. The three crossover events (A, B, and C) can lead to changes in the number of rDNA units on a chromosome, while gene conversion (D) will not unless a crossover also occurs.
The correction mechanism necessary to explain the evolution of the rDNA loci could be a natural outcome of the unequal crossover process. Mutations would continually arise at a slow rate in all repeats. Unequal crossovers involving units with a mutation would generate one chromosome in which the mutation was present in 2 units and another chromosome without the mutation. A random process of unequal crossover would continue to generate chromosomes with increased or decreased numbers of units with the mutation. Chromosomes containing mutations within the rRNA genes would generally be selected against, while chromosomes containing mutations in the noncoding regions of the unit would be under no adverse selective pressure. Thus substitutions in noncoding regions would increase or decrease in number of units with time in a stochastic manner. Eventually, after many crossovers, such substitutions would be either present in all the units or absent from all the units. This model readily explained how the genes would change only slowly over evolutionary time, while the noncoding region would be free to drift to new DNA sequences. Soon after these studies of the rRNA gene, Smith (1976) used computer simulations to show that high sequence identity was generated and maintained by homologous crossovers between duplicated sequences. Sequence uniformity of the rDNA units could thus be explained by the same mechanism used to explain the recently discovered uniformity in the short tandem (satellite) DNA found at many centromeres. Ohta (1976) derived mathematical models to calculate the probability and the mean time required to fix nucleotide substitutions under different parameters by the random process of unequal crossover.
This elegantly simple model for the concerted evolution of the rDNA locus could explain all the known properties of the rDNA loci and required no mechanisms other than mutation, homologous recombination, and selection. The only critical requirement of the model was that the crossover rate needed to be high relative to the mutation rate.
LESSONS FROM YEAST
Direct experimental support for the unequal crossover model of concerted evolution required the ability to “mark” individual units within the rDNA locus and follow their disappearance or duplication through recombination events. The advent of gene cloning methods made this approach possible in Saccharomyces cerevisiae. Yeast encodes ∼140 rRNA gene units on one chromosome. Using the power of yeast genetics, it was shown that LEU2 genes inserted within the rDNA locus could be used as a selectable marker to rescue an auxotroph.
Szostak and Wu (1980) followed LEU2-marked rDNA units through mitotic divisions in haploid cells. They first used fluctuation tests to show that the insertions were spontaneously lost at a measurable frequency. Determining the mechanism by which the deletion occurred required analyzing the rDNA loci from both daughters of the cell undergoing the loss. If the deletions occurred by an unequal crossover between the two copies of the chromosome after DNA synthesis (i.e., recombination between sister chromatids, Figure 2A), then each daughter cell generated without the insertion would have a sister cell with two copies of the insertion. For those crossovers that occur early in the process of yeast colony formation, one sector of the colony would contain cells with no copy of the insertion while an adjacent sector would contain cells with two copies of the insertion. Analysis of genomic DNA isolated from cells grown from the appropriate regions of sectored colonies confirmed that prediction. By digesting the genomic DNA of the cells containing two LEU2 genes with a restriction enzyme that cleaved within the LEU2 insertion but not the rDNA unit itself, Southern blots could be used to determine the distance separating the two LEU2 genes. This distance divided by the length of the rDNA unit provided a direct estimate of the number of rDNA units the two sister chromatids were displaced (offset) during the crossover. The sizes of the offsets in the seven spontaneous events analyzed varied between 6 and 8 units. Using 7 units as the average offset, 140 as the total number of rDNA units in the locus, and the loss rate from the fluctuation test, the total rate of spontaneous unequal crossovers in the rDNA locus was estimated to be 1% per mitotic division.
Petes (1980) followed LEU2 insertions in the rDNA locus through meiotic divisions by standard tetrad analysis of the four spores derived from diploid cells. An elegant aspect of the study was that the diploid cells used were generated from haploid cells that had a fixed sequence difference in their rDNA units that could be scored by restriction digestion. Thus Petes was able to follow both sister chromatid exchanges (Figure 2A) and exchanges between the two homologs (referred to as interchromosomal exchange, Figure 2B). About 10% of the tetrads analyzed contained at least one spore that had lost the marker gene from the rDNA locus. Analysis of DNA derived from all four spores indicated that these tetrads also contained a spore in which the LEU2 gene had been duplicated. Remarkably none of the unequal crossovers analyzed contained cells with rDNA units from both homologs, suggesting that all the recombination events had been between sister chromatids. This low rate of interchromosomal crossover was consistent with previous mapping experiments by Petes (1979) in which meiotic recombination in the rDNA locus had been shown to occur at rates nearly two orders of magnitude lower than that expected on the basis of the size of the locus and the average recombination rate for the yeast genome.
The combined findings of these two reports strongly suggested that unequal crossover occurred frequently in the rDNA locus and thus could serve as the basis for the concerted evolution of the locus. Furthermore because sister chromatid exchanges were more frequent than interchromosomal exchanges, the results predicted that the concerted evolution of units on individual chromosomes in a population would be faster than the concerted evolution between chromosomes. As we will see below, this prediction has been confirmed multiple times. However, concerns that the concerted evolution of the rDNA locus could not be entirely explained by unequal crossovers remained. First, the unequal crossover model worked best if the recombinations were random throughout the locus. Independently derived LEU2 marker insertions in the rDNA locus had different rates of deletion, suggesting that the location of the inserted unit in the locus influenced how often it participated in crossover events (Petes 1980). Second, applying the crossover rates determined in these studies to the mathematical formulation of Ohta (1976) suggested that it required a remarkably large number of generations for a new mutation to randomly drift to fixation on one chromosome (Szostak and Wu 1980). Finally, it was found that the sequence of the rDNA unit at one edge of the rDNA locus was identical in sequence to all other units (Zamb and Petes 1982), counter to the accumulated differences that models of unequal crossover predicted for terminal units (Smith 1976; Brutlag 1980).
The question of whether unequal crossover could entirely account for the concerted evolution of the rDNA units was eclipsed by the discovery of another type of recombination in yeast. Gene conversions are the nonreciprocal transfer of DNA sequences between two genes (Figure 2D). In yeast these events can be scored in meiotic (ratios other than 2:2 spore formation) as well as mitotic divisions. The frequency, sequence dependency, distance requirements, and possible mechanism for these events were extensively characterized (Orr-Weaver and Szostak 1985, or for a more recent review see Paques and Haber 1999). Gene conversions between two repeated sequences occur with a median frequency of ∼5% per meiosis independently of whether the DNA sequences are duplicated on the same or different (homologous or nonhomologous) chromosomes. Gene conversion usually involves short regions of DNA, and thus the large selectable gene insertions in the rDNA locus used by Szostak and Wu (1980) and Petes (1980) were unlikely to have been removed or duplicated by this process. Unfortunately no alternative method has been published to score gene conversions within the rDNA locus.
A number of analytical models of concerted evolution by gene conversion were developed (Nagylaki and Petes 1982; Nagylaki 1984; Ohta 1985; Walsh 1986). The apparent advantages of gene conversion over unequal crossovers were many. First, gene conversion enabled concerted evolution to readily occur between sequences on homologous as well as nonhomologous chromosomes, allowing for the greater homogeneity of all rDNA units in a population. Second, gene conversion could account for the sequence uniformity of the terminal repeat in a tandem array. Third, gene conversion was more broadly useful because it could also give rise to the concerted evolution of multigene families that were dispersed throughout a genome. Finally, the bias frequently encountered in gene conversion studies, even when small, greatly increased the rates of concerted evolution. It is difficult to postulate how unequal crossover events could transfer neutral information in one direction more often than in the opposite direction. All the advantages gene conversion brings to the table have led to the commonly held opinion that a combination of both unequal crossovers and gene conversions gives rise to the concerted evolution of the rDNA locus in all organisms. As is described below, evidence for the former has accumulated for many organisms, while evidence for the latter has been frustratingly difficult to obtain.
OBSERVATIONS IN HIGHER EUKARYOTES
The elegant studies conducted in yeast of recombination in the rDNA loci are inherently more difficult in higher eukaryotes. It is extremely difficult to either mark individual units in the locus to allow rapid screening for recombination or follow the daughter cells after a recombination event. However, insights into the recombinations associated with the rDNA locus are possible because of the natural sequence variation that can be found among the rDNA units in most species. While this variation can be in the rRNA genes themselves, it is most often found in the spacer regions, especially within the IGS between transcription units (see Figure 1). Greater variation is found in the IGS region because it appears to be under the lowest level of selective pressure. A second source of variation detected in the IGS region is due to its internally repeated structure. Variation in the number of these internal repeats can be scored on Southern blots even in the absence of significant nucleotide sequence differences between the units.
Many studies have appeared that examine the concerted evolution of the rDNA units or the sequence variation found within and between individuals of a species. In a few organisms either a fraction of the rDNA units have escaped the process of concerted evolution or the organism is under selective pressure to evolve multiple rDNA sequences (for examples see Fenton et al. 1998; Carranza et al. 1999; Keller et al. 2006). However, in the vast majority of organisms concerted evolution functions extremely well on all rDNA genes. Summarized here are the observations from a few of the most extensively studied species.
Probably the most studied rDNA loci in any organism are those of Drosophila melanogaster. In this organism ∼200 rRNA genes are located within heterochromatic regions on both the X and the Y chromosomes. Work on the rDNA loci of D. melanogaster also began well before the advent of DNA cloning when it was found that a short bristle phenotype known as bobbed (bb) was associated with low numbers of rRNA genes on the X chromosome (Ritossa et al. 1966). Remarkably when a bb X chromosome was maintained with a Y chromosome also deficient for many of its rDNA repeats, progeny would appear with a normal number of rDNA units (Ritossa 1968). This phenomenon, referred to as rDNA magnification, induced a series of studies focusing on both the mechanism and the possible genes involved in these dramatic changes within the rDNA locus (reviewed in Tartof 1988; Hawley and Marcus 1989). While magnification generally is assumed to involve multiple rounds of unequal crossover, the mechanism remains undefined.
Additional early studies in Drosophila compared the rDNA units between related species as well as the variation within individuals and between individuals in a population (Coen et al. 1982a,b). On the basis of studies of the rRNA genes and multigene families spread throughout the genome, Dover proposed a comprehensive model for concerted evolution (Dover 1982; Ohta and Dover 1984). The combined mechanisms of gene turnover were called “molecular drive.” A controversial suggestion in the model was that sequence homogenization occurred equally to all members of the gene family whether they were present on the same or on different chromosomes.
The phenomenon of rDNA magnification in D. melanogaster and interest in the mechanism of concerted evolution stimulated studies of meiotic recombination rates between the rDNA loci on the two X chromosomes in females and between the loci on the X and Y chromosomes in males. As in yeast, meiotic recombination between the rDNA loci on X chromosomes (∼10−4 per generation) was found to be about two orders of magnitude below that expected for a similar length of DNA elsewhere on the chromosome (Williams et al. 1989). Interestingly, these crossovers may not represent the typical meiotic recombination events found elsewhere in the genome because the rate of recombination between the rDNA loci on the X and Y chromosomes in males was found to be the same as that observed between X chromosomes, even though recombination is generally absent in D. melanogaster males (Hawley and Marcus 1989; Williams and Robins 1992).
The extremely low rate of crossover between rDNA loci on different X chromosomes or between the loci on the X and Y chromosomes suggested that, as in yeast, most of the recombinations in the rDNA loci of D. melanogaster occurred between sister chromatids. The strongest support for this suggestion was obtained by Schlotterer and Tautz (1994). These researchers identified three sequence variants in the ITS1 region of the rDNA on the X chromosome in various geographical lines. Using temperature gradient gel electrophoresis to quantitate the level of these variants, they found that the rDNA units on individual X chromosomes in a population could contain fixed differences involving these three variants. This finding suggested that unequal crossovers or gene conversion could rapidly homogenize all the units on individual X chromosomes but only slowly homogenized the units between X chromosomes in a population. Polanco et al. (1998) extended this finding to multiple populations and showed that the X and Y chromosomes in the same population also had different fixed variants in the ITS region. Polanco et al. (1998, 2000) also attempted to score IGS variants on the X and Y chromosomes in the same populations. While IGS length variants appeared to be frequently shared among the X and Y chromosomes, the internally repeated structure of the IGS and the large numbers of length variants seen on each chromosome complicate estimates of the degree to which IGS variants are exchanged.
The degree to which variants are shared within and between chromosomes has also been a major focus in the study of the rDNA genes of humans. We encode ∼400 rRNA genes that are distributed in tandem arrays on five chromosomes (nos. 13, 14, 15, 21, and 22). The presence of the rRNA genes on nonhomologous chromosomes does not prevent concerted evolution because all rDNA units in humans are more similar to each other than they are to the rDNA units of other primates (Arnheim et al. 1980). Analysis of the spread of sequence information between homologous vs. nonhomologous chromosomes utilized multiple length variants that were found within the IGS region of the repeat (Krystal et al. 1981). Individual arrays were analyzed by taking advantage of rodent–human somatic cell hybrids containing single human chromosomes. Each chromosome was found to contain several IGS length variants, and these variants were often shared among the different chromosomes, confirming that sequence information flowed between chromosomes.
The sharing of IGS length variants among human rDNA loci on different chromosomes contrasted with the results of similar studies conducted in the mouse, Mus musculus (Arnheim et al. 1982). Mouse rDNA units are also located on multiple chromosomes, but unlike humans the rDNA locus of each mouse chromosome has its own specific set of IGS length variants. The specific variants are linked to the same chromosome in unrelated strains of mice, suggesting that interchromosomal exchange between nonhomologous chromosomes was much lower in mice than in humans. The lower level of shared variants in mouse than in humans is most likely the result of the locations of the rDNA loci in each species (Figure 3). All human rDNA loci are located adjacent to the telomere on the short arm of acrocentric chromosomes. At this location a single crossover event between rDNA units on nonhomologous chromosomes would lead to an exchange of the telomeres in addition to a fraction of the rDNA loci. In mouse, however, the rDNA loci are located next to the centromere on the long arm of telocentric chromosomes. Therefore a single crossover event between two nonhomologous chromosomes would result in the exchange of not only a fraction of the rDNA loci and the telomeres but also the centromeres, an event likely to have significantly greater negative consequences to the organism.
Figure 3.—
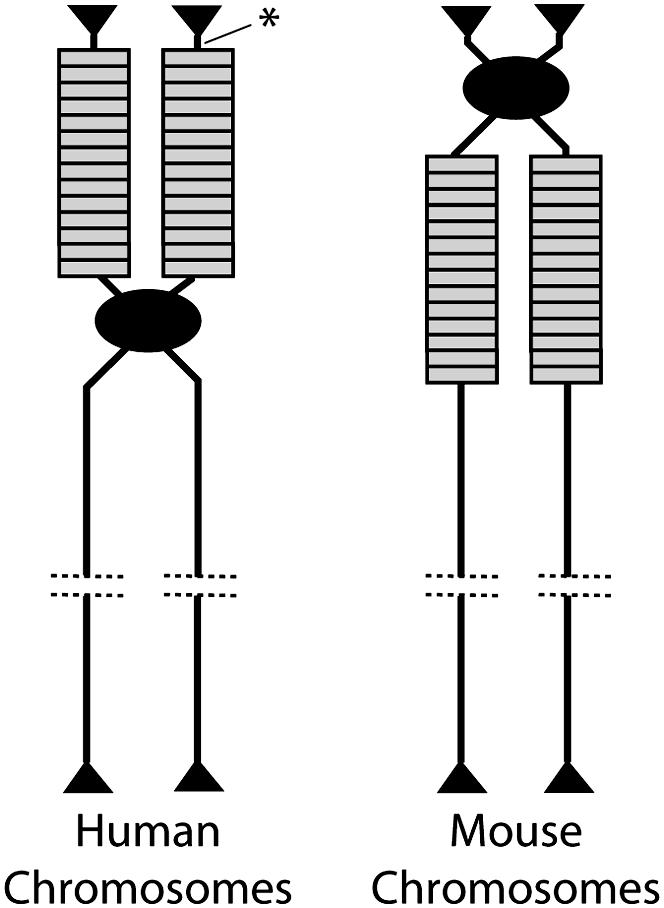
Location of the rDNA loci on the chromosomes of humans and mice. Each chromosome is drawn after replication to show the two sister chromatids still attached by means of their centromeres (solid oval). Individual rDNA units are indicated by rectangles and the telomeres by triangles. Each chromosome is representative of the multiple nonhomologous chromosomes that contain the rDNA units in each species. The noncoding region distal to the rDNA loci in humans studied by Gonzalez and Sylvester (2001) is indicated (asterisk).
In humans, as in yeast and Drosophila, sister chromatid exchange appears more frequent than exchanges between chromosomes (homologous or nonhomologous). Seperack et al. (1988) showed that there were substantial differences among individuals in the abundance of various IGS variants, suggesting that variant types were not exchanged freely among the five chromosomes. Further analysis of human–rodent cell hybrids revealed that the same chromosome from different individuals could have different IGS variants (Gonzalez and Sylvester 2001). Gonzalez and Sylvester also determined the level of sequence uniformity within and between the rDNA repeats. Sequence diversity of the rRNA genes in the short regions analyzed was extremely low whether the comparisons were made within a chromosome or between the five chromosomes. Meanwhile the IGS region of the rDNA units could be divided into multiple classes that had high sequence identity within a class but diverged by 6–8% in sequence between classes. Each chromosome had one or more of the IGS classes with all IGS classes shared to some extent between chromosomes.
Interestingly a high level of sequence identity among all five chromosomes in humans was also found for a noncoding region 6 kb distal to the last rDNA unit (Figure 3), a region presumably under little selective pressure. The high level of interchromosomal sequence identity for a noncoding region flanking the rDNA locus argues strongly for crossovers that exchange the distal (telomeric) end of the rDNA loci. While these crossovers will exchange IGS variants between chromosomes, the more rapid sister chromatid exchanges result in the differential spread of IGS variants on each chromosome. More experiments of this design to characterize the opposite (proximal) end of the human rDNA arrays as well as the rDNA arrays in mice are needed to confirm the model.
The above studies as well as many others would suggest the involvement of unequal crossovers in the concerted evolution of higher eukaryotic rDNA loci. However, one study has appeared, suggesting that gene conversion can play an important role. In a study of the Heteronotia binoei complex of lizards, Hillis et al. (1991) showed that in parthenogenetic triploid species arising by the hybridization of two sexual species, the rDNA variant from only one of the original species became fixed in many of the parthenogens. The authors suggest that this finding is most readily explained by a biased gene conversion process rather than by more neutral unequal crossovers. While a tantalizing result, the expression of the rDNA unit from only one parent in species hybrids is common (reviewed in Grummt and Pikaard 2003). Therefore, further study of this system is needed to ensure that the bias obtained is not explained by selective pressure to express the rDNA units of one parental species.
rDNA LOCI SERVE AS A NICHE FOR MOBILE ELEMENTS
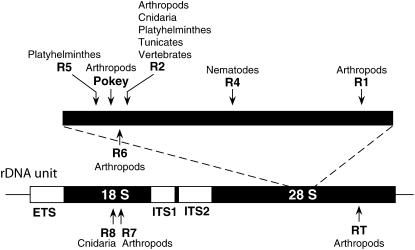
Given the remarkable efficiency of concerted evolution in rDNA loci, it is surprising that in at least five major animal taxa these loci have become the specialized niche for a number of mobile elements (Figure 4) (reviewed in Eickbush 2002). The R elements are non-long terminal repeat (non-LTR) retrotransposons, while Pokey is a DNA-mediated element (Penton and Crease 2004). R2, R4, R5, and R8 are members of a common lineage found in many animal taxa (Kojima et al. 2006), while R1, R6, R7, and RT represent an independent lineage of elements in arthropods whose members have moved to different positions within and sometimes outside the rDNA locus (Kojima and Fujiwara 2003). R elements have been shown to encode endonucleases highly specific for their insertion sites (Xiong and Eickbush 1988). The endonuclease of the R2 family of elements is related to type IIS restriction enzymes (Yang et al. 1999), while the endonuclease for the R1 family is related to apurinic endonucleases (Feng et al. 1998).
Figure 4.—
Location of mobile element insertions in the rDNA unit. Abbreviations within the rDNA transcription unit are as described in Figure 1. A small region of the 28S gene, which contains many insertion classes, is magnified above this repeat. Arrows indicate the insertion site of the various elements based on their 3′ junction with the gene. The current known distribution of each element is also shown. For more detailed descriptions of these elements see Eickbush (2002), Kojima and Fujiwara (2003), and Kojima et al. (2006).
The R1 and R2 elements of arthropods have been extensively studied since early in the characterization of the rRNA genes of various Drosophila species (Long and Dawid 1980). R1 and R2 elements appear completely adapted to life in the rDNA locus (reviewed in Eickbush 2002). They are widely distributed in all lineages of arthropods where they frequently insert into 10–30% of the units, but insertion percentages well above 50% have been observed. Because the insertion of either element gives rise to a 28S gene that can no longer make functional rRNA, it was initially assumed that selection against defective genes and concerted evolution would frequently eliminate R1 and R2 elements from the rDNA locus. Thus attempts were made to show that horizontal transfers were responsible for the broad distribution of these elements. However, phylogenetic analyses have provided no evidence for such horizontal transfers (Malik et al. 1999). We are left with the remarkable conclusion that these two transposable elements are highly stable even though they insert into a locus that is highly adapted to rid itself of variation. This paradox has led to speculation that the elements are involved in the regulation of rRNA synthesis or provide mechanisms to initiate recombination. However, the multiple lineages of R1 or R2 found in many taxa (Gentile et al. 2001) are not consistent with the elements providing a host function. Thus the model that best explains the wide distribution of R1 and R2 elements remains that they are simply highly successful parasites.
Are R1 and R2 elements affecting the concerted evolution of the rDNA locus? The sequence of the rDNA units with the insertions is identical to that of the units without the insertions, suggesting that the simple answer is no. The presence of the mobile elements within the rDNA locus provides a series of “variants” that can be used to study the concerted evolution of the loci. R1 and R2, like many other non-LTR retrotransposons, insert by a mechanism that often generates copies truncated (deleted) at their 5′ end. These truncations, which can extend the full length of the elements and thus are readily scored by simple PCR assays, serve to mark individual rDNA units within the locus. Studies of the R1 and R2 elements in Drosophila have shown that these truncated copies are present in only one or a few copies per chromosome and that these copies differ between individuals from the same population (Perez-Gonzalez and Eickbush 2001). These results indicate that new insertions are rapidly eliminated from the rDNA locus. Because gene conversions are typically inhibited by large insertions (R1/R2 elements are 3.5–5.5 kb in length) most of the eliminations presumably occur by unequal crossover between sister chromatids.
The model for turnover of the R1 and R2 insertions is thus merely an extension of the original unequal crossover models of concerted evolution. New variants (insertions) are subject to random crossovers with the strong selective pressure against inactive rDNA units eliminating them from the loci. The remarkable efficiency by which the concerted evolution process removes these elements is, however, somewhat of a double-edged sword. It allows the loci to function smoothly in the face of a continuous onslaught of mobile element insertions, but by doing so continually provides these elements with new target sites, allowing them to remain active.
FOLLOWING CHANGES IN THE rDNA LOCUS OVER TIME
The studies in higher organisms described to this point measured differences in the rDNA loci between individuals in the same or different populations. The studies suggested rapid change in evolutionary terms. Addressing the issue of the rate of recombination is best accomplished by monitoring the changes that accumulate in specific rDNA loci over short time frames.
Averbeck and Eickbush (2005) evaluated the short-term dynamics of an rDNA locus by measuring differences in the locus among 15 replicate D. melanogaster lines after 400 generations. All lines were initially started from the progeny of one pair of flies from a highly inbred stock. Changes in the rDNA locus were dramatic. The total number of rDNA units on the X chromosome across the lines varied from 140 to 310, similar to the range found within or between natural populations of D. melanogaster. These lines had several unique as well as 14 common IGS length variants. The common IGS variants varied significantly in abundance: from near zero to 30 copies in most cases and from 25 to 70 copies for the two most abundant variants. The observed changes in number of rDNA units suggested frequent unequal sister chromatid exchanges, given the slow rate of interchromosomal exchange (Williams et al. 1989). The dramatic change in abundance of all IGS variant types further suggested that these crossover events occurred at many regions throughout the locus. Additional support for the rapid changes in the rDNA loci came from the study of the R1 and R2 elements. Two hundred new R1 and R2 insertions as well as 100 independent losses of insertions present in the parental stock were scored in these lines (Perez-Gonzalez et al. 2003).
An equally dramatic demonstration of the dynamic nature of the rDNA locus came from McTaggart et al. (2007) and their study of the rDNA loci of the freshwater crustacean, Daphnia obtusa. An advantage of this organism is that females can produce diploid eggs via parthenogenesis that develop directly into female adults. Daughters from a single stem mother were used to establish multiple lines, and every generation a single daughter was selected at random to produce the next generation. Recombination in the locus was scored at 5-generation time intervals in two ways: variation in the total number of genes and variation in the ratio of six nucleotide variants within the 18S gene. The changes were again remarkable. Over the 90 generations of this study, each line (i.e., chromosome) underwent from one to six measurable shifts in the ratio of sequence variants. These changes appear to involve unequal crossovers with large offsets because the number of rRNA genes varied from 50 to 230 units and the shifts observed in variant ratios between time points were as large as 33%. On the basis of the number of scored changes the recombination rate was estimated to be between 2 and 6% per generation. This is clearly an underestimate because unequal crossovers involving small offsets or sequential events with canceling effects would not have been scored.
These two studies clearly indicate that rDNA loci of both Drosophila and Daphnia are in a continual, dynamic state of flux. Therefore monitoring changes in the loci in real time is possible and does not require selection schemes.
CONCLUDING COMMENTS
The many studies summarized in this review suggest that unequal crossover is undoubtedly involved in the process of concerted evolution. Further, both direct demonstration in yeast and the greater intrachromosomal than interchromosomal homogeneity observed in Drosophila and in humans indicate that these crossovers are more frequent between sister chromatids than between chromosomes (homologous and nonhomologous). While gene conversion is clearly an enticing mechanism to help explain the sequence homogeneity, direct evidence for its involvement remains elusive. As our genetic and molecular tools increase, the involvement of gene conversion as well as the rules governing crossovers can be further addressed. These tools may also help address the many unresolved questions regarding the expression of the locus (reviewed in Grummt and Pikaard 2003). Like attending a good symphony, one could simply sit back and marvel at the intricate movements and synchronous effect of the many players. However, to truly understand its complexity, one needs to know the role played by each instrument.
Acknowledgments
We thank Alan Spradling, William Burke, and Deb Stage for comments on the manuscript. We thank Steve Henikoff and members of his laboratory for providing a stimulating environment during our sabbatical stay at the Fred Hutchison Cancer Research Center. Our work on the rDNA locus and its insertion elements was conducted with support from the National Institutes of Health (GM42790) and the National Science Foundation (MCB-0544071).
References
- Arnheim, N., M. Krystal, R. Schmickel, G. Wilson, O. Ryder et al., 1980. Molecular evidence for genetic exchange among ribosomal genes on non-homologous chromosomes. Proc. Natl. Acad. Sci. USA 77: 7323–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim, N., D. Treco, B. Taylor and E. M. Eicher, 1982. Distribution of ribosomal gene length variants among mouse chromosomes. Proc. Natl. Acad. Sci. USA 79: 4677–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck, K. T., and T. H. Eickbush, 2005. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics 171: 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. D., P. C. Wensink and E. Jordan, 1972. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 63: 57–73. [DOI] [PubMed] [Google Scholar]
- Brutlag, D. L., 1980. Molecular arrangement and evolution of heterochromatic DNA. Annu. Rev. Genet. 14: 121–144. [DOI] [PubMed] [Google Scholar]
- Carranza, S., J. Baguna and M. Riutort, 1999. Origin and evolution of paralogous rRNA gene clusters within the flatworm family Dugesiidae (Platyhelminthes, Tricladida). J. Mol. Evol. 49: 250–259. [DOI] [PubMed] [Google Scholar]
- Coen, E., T. Strachan and G. Dover, 1982. a Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J. Mol. Biol. 158: 17–35. [DOI] [PubMed] [Google Scholar]
- Coen, E., J. M. Thoday and G. Dover, 1982. b Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature 295: 564–568. [DOI] [PubMed] [Google Scholar]
- Dover, G., 1982. Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. [DOI] [PubMed] [Google Scholar]
- Eickbush, T. H., 2002. R2 and related site-specific non-long terminal repeat retrotransposons, pp. 813–835 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellart and A. M. Lambowitz. American Society of Microbiology, Washington, DC.
- Feng, Q., G. Schumann and J. D. Boeke, 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95: 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton, B., G. Malloch and F. Germa, 1998. A study of variation in rDNA ITS regions shows that two haplotypes coexist within a single aphid genome. Genome 41: 337–345. [PubMed] [Google Scholar]
- Gentile, K. L., W. D. Burke and T. H. Eickbush, 2001. Multiple lineages of R1 retrotransposable elements can co-exist in the rDNA loci of Drosophila. Mol. Biol. Evol. 18: 235–245. [DOI] [PubMed] [Google Scholar]
- Gonzalez, I. L., and J. E. Sylvester, 2001. Human rDNA: evolutionary patterns within genes and tandem arrays derived from multiple chromosomes. Genomics 73: 255–263. [DOI] [PubMed] [Google Scholar]
- Grummt, I., and C. S. Pikaard, 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell. Biol. 4: 641–649. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., and C. H. Marcus, 1989. Recombinational controls of rDNA redundancy in Drosophila. Annu. Rev. Genet. 23: 87–120. [DOI] [PubMed] [Google Scholar]
- Hillis, D. M., C. Moritz, C. A. Porter and R. J. Baker, 1991. Evidence for biased gene conversion in the concerted evolution of ribosomal DNA. Science 251: 308–310. [DOI] [PubMed] [Google Scholar]
- Keller, I., I. C. Chintauan-Marquier, P. Veltsos and R. A. Nichols, 2006. Ribosomal DNA in the grasshopper Podisma pedestris: escape from concerted evolution. Genetics 174: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. K., and H. Fujiwara, 2003. Evolution of target specificity in R1 clade non-LTR retrotransposons. Mol. Biol. Evol. 20: 351–361. [DOI] [PubMed] [Google Scholar]
- Kojima, K. K., K. Kuma, H. Toh and H. Fujiwara, 2006. Identification of rDNA-specific non-LTR retrotransposons in Cnidaria. Mol. Biol. Evol. 23: 1984–1993. [DOI] [PubMed] [Google Scholar]
- Krystal, M., P. D'Eustachio, F. H. Ruddle and N. Arnheim, 1981. Human nucleolus organizers on non-homologous chromosomes can share the same ribosomal gene variants. Proc. Natl. Aad. Sci. USA 78: 5744–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, E. O., and I. B. Dawid, 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke and T. H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805. [DOI] [PubMed] [Google Scholar]
- McTaggart, S. J., J. L. Dudycha, A. Omilian and T. J. Crease, 2007. Rates of recombination in the ribosomal DNA of apomictically propagated Daphnia obtusa lines. Genetics 175: 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T., 1984. Evolution of multigene families under interchromosomal gene conversion. Proc. Natl. Acad. Sci. USA 81: 3796–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T., and T. D. Petes, 1982. Intrachromosomal gene conversion and the maintenance of sequence homogeneity among repeated genes. Genetics 100: 315–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and A. P. Rooney, 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39: 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1976. Simple model for treating evolution of multigene families. Nature 262: 74–76. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1985. A model of duplicative transposition and gene conversion for repetitive DNA families. Genetics 110: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., and G. A. Dover, 1984. The cohesive population genetics of molecular drive. Genetics 108: 501–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., and J. W. Szostak, 1985. Fungal recombination. Microbiol. Rev. 49: 33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton, E. H., and T. J. Crease, 2004. Evolution of the transposable element Pokey in the ribosomal DNA of species in the subgenus Daphnia (Crustacea: Cladocera). Mol. Biol. Evol. 21: 1727–1739. [DOI] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., and T. H. Eickbush, 2001. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., W. D. Burke and T. H. Eickbush, 2003. R1 and R2 retrotransposition and elimination from the rDNA loci of the X and Y chromosomes of Drosophila melanogaster. Genetics 165: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 1979. Meiotic mapping of yeast ribosomal DNA on chromosome XII. J. Bacteriol. 138: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 1980. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell 19: 765–774. [DOI] [PubMed] [Google Scholar]
- Polanco, C., A. I. Gonzalez, A. de la Fuente and G. A. Dover, 1998. Multigene family of ribosomal DNA in Drosophila melanogaster reveals contrasting patterns of homogenization for IGS and ITS spacer regions: a possible mechanism to resolve this paradox. Genetics 149: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco, C., A. I. Gonzalez and G. A. Dover, 2000. Patterns of variation in the intergenic spacers of ribosomal DNA in Drosophila melanogaster support a model for genetic exchanges during X-Y pairing. Genetics 155: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa, F. M., 1968. Unstable redundancy of genes for ribosomal RNA. Proc. Natl. Acad. Sci. USA 60: 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa, F. M., K. C. Atwood, D. L. Lindsley and S. Spiegelman, 1966. On the chromosomal distribution of DNA complementary to ribosomal and soluble RNA. Natl. Cancer Inst. Monogr. 23: 449–472. [PubMed] [Google Scholar]
- Schlotterer, C., and D. Tautz, 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Smith, G. P., 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528–535. [DOI] [PubMed] [Google Scholar]
- Seperack, P., M. Slatkin and N. Arnheim, 1988. Linkage disequilibrium in human ribosomal genes: implications for multigene family evolution. Genetics 119: 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J. W., and R. Wu, 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284: 426–430. [DOI] [PubMed] [Google Scholar]
- Tartof, K. D., 1988. Unequal crossing over then and now. Genetics 120: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J. B., 1986. Selection and biased gene conversion in a multigene family: consequences of interallelic bias and threshold selection. Genetics 112: 699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S. M., and L. G. Robins, 1992. Molecular genetic analysis of Drosophila rDNA arrays. Trends Genet. 8: 335–340. [DOI] [PubMed] [Google Scholar]
- Williams, S. M., J. A. Kennison, L. G. Robbins and C. Strobeck, 1989. Reciprocal recombination and the evolution of the ribosomal gene family of Drosophila melanogaster. Genetics 122: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. E., and T. H. Eickbush, 1988. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell 55: 235–246. [DOI] [PubMed] [Google Scholar]
- Yang, J., H. S. Malik and T. H. Eickbush, 1999. The identification of the endonuclease domain encoded by R2 and other site-specific non-LTR retrotransposons. Proc. Natl. Acad. Sci. USA 96: 7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamb, T. J., and T. D. Petes, 1982. Analysis of the junction between ribosomal RNA genes and single-copy chromosomal sequences in the yeast Saccharomyces cerevisiae. Cell 28: 355–364. [DOI] [PubMed] [Google Scholar]