Abstract
Rapid typing of genetic variation at many regions of the genome is an efficient way to survey variability in natural populations in an effort to identify segments of the genome that have experienced recent natural selection. Following such a genome scan, individual regions may be chosen for further sequencing and a more detailed analysis of patterns of variability, often to perform a parametric test for selection and to estimate the strength of a recent selective sweep. We show here that not accounting for the ascertainment of loci in such analyses leads to false inference of natural selection when the true model is selective neutrality, because the procedure of choosing unusual loci (in comparison to the rest of the genome-scan data) selects regions of the genome with genealogies similar to those expected under models of recent directional selection. We describe a simple and efficient correction for this ascertainment bias, which restores the false-positive rate to near-nominal levels. For the parameters considered here, we find that obtaining a test with the expected distribution of P-values depends on accurately accounting both for ascertainment of regions and for demography. Finally, we use simulations to explore the utility of relying on outlier loci to detect recent selective sweeps. We find that measures of diversity and of population differentiation are more effective than summaries of the site-frequency spectrum and that sequencing larger regions (2.5 kbp) in genome-scan studies leads to more power to detect recent selective sweeps.
A major goal of population genetics is to use patterns of variability in a natural population to identify regions of the genome where allele frequencies have been recently affected by the action of natural selection. Historically, studies of naturally occurring molecular variation were conducted at single loci, and uncertainties about the demographic history of natural populations frequently complicated inferences about selection. Current empirical work focuses on using either multilocus data sets (e.g., Glinka et al. 2003; Tenaillon et al. 2004; Haddrill et al. 2005b; Ometto et al. 2005; Williamson et al. 2005; Wright et al. 2005) or whole-genome polymorphism data (e.g., Carlson et al. 2005; Nielsen et al. 2005; Kelley et al. 2006) to discern the locus-specific effects of selection from the genome-wide effects of nonequilibrium demographic history. In general, this approach has been dubbed a “genome scan” for selection.
Where whole-genome variation data are unavailable, investigators will sample levels of variability from multiple regions of the genome using markers that are both relatively rapid and relatively inexpensive to type, such as microsatellites (e.g., Harr et al. 2002; Kauer et al. 2003; Bauer-DuMont and Aquadro 2005), or short fragments of nucleotide sequence to identify single-nucleotide polymorphisms (SNPs) (e.g., Glinka et al. 2003; Tenaillon et al. 2004; Ometto et al. 2005; Wright et al. 2005). From such studies, a subset of these regions may then be selected for additional sequencing, and the parameters of a model of recent positive, directional selection acting on new mutations will be estimated from the data. How such regions are chosen for additional sequencing varies from study to study, but most strategies include a comparison of individual loci to the empirical distribution of some feature of the data resulting from a genome scan. For example, genome-scan data consisting of short reads of DNA sequence may be summarized by the number of mutations in each fragment, with invariant fragments being used to identify regions for further sequencing (e.g., Glinka et al. 2003; Schlenke and Begun 2004; Beisswanger et al. 2006). Similarly, a region may be identified because variability and/or allele frequencies of microsatellite markers are extremely skewed in some regions of the genome relative to the data set as a whole (e.g., Harr et al. 2002; Bauer-DuMont and Aquadro 2005; Pool et al. 2006). The rationale for the follow-up experiment is that the statistics used to identify outlier regions (e.g., Tajima 1989; Voight et al. 2006) are not formal tests for selection, as they do not specifically reject a neutral model in favor of a model including selection. Thus, empirical distributions from genome scans are often used as a way to quickly identify regions of the genome in which to estimate the strength and target of recent positive selection. Currently, such estimates are usually obtained using the approach of Kim and Stephan (2002) and related approaches (Kim and Nielsen 2004).
The rationale for choosing such extreme loci for more detailed investigation is that models of selective sweeps (Maynard-Smith and Haigh 1974) predict both strong reductions in diversity and skews in the site-frequency spectrum, at neutral sites linked to a recent sweep (Braverman et al. 1995; Kim and Stephan 2002). However, such a procedure gives rise to at least three concerns. First, when a genome-scan study surveys a large number of (approximately) independent regions of the genome, choosing the most extreme loci imposes a multiple testing problem for subsequent analysis. Second, any empirical distribution has observations in the tails, regardless of the model that generated the data. Third, it is unclear in models of selective sweeps occurring in nonequilibrium populations the extent to which selected loci are expected to be enriched in the tails of an empirical distribution. A recent simulation study (Teshima et al. 2006) suggests that the efficacy of this approach depends on which summary statistics are used to identify outliers, as well as on the details of the underlying demographic model and the model of adaptation assumed (for example, complete sweeps vs. sweeps from standing variation).
In this article, we use simulations to investigate the effect that choosing outlier loci has on parametric inferences of selection, when the true model is one of neutral mutations in a bottlenecked population. We study a bottleneck model to explore the properties of genome scans using parameters that may be relevant for Drosophila melanogaster and also because population bottlenecks severely confound the inference of selection (e.g., Jensen et al. 2005). We apply what is currently the state-of-the art method for “subgenomic” scans (i.e., less than whole-genome SNP data)—the composite-likelihood method of Kim and Stephan (2002) and the goodness-of-fit (GOF) test of Jensen et al. (2005). The former method estimates both the strength and target of selection, assuming the demographic null model of a large, panmictic population, and gives a composite likelihood-ratio test (CLRT) comparing the selective sweep model to the standard neutral model. Jensen et al. (2005) proposed a GOF statistic intended to be applied to data sets that reject neutrality following the procedure of Kim and Stephan (2002). They showed that the GOF procedure substantially reduces the false-positive rate under nonequilibrium demographic models and also results in a test statistic with a uniform distribution of P-values when the true model is a single selective sweep occurring in a large, constant-size, panmictic population (the model assumed by the CLRT). In Jensen et al. (2005), the calculation of the GOF test was applied to simulated data assuming that loci are random draws from a population model. In practice, however, both the method of Kim and Stephan (2002) and the GOF tests are often applied to loci that are preselected by an investigator because some feature of the region is an outlier in a multilocus genome scan (e.g., Harr et al. 2002; Bauer-DuMont and Aquadro 2005; Beisswanger et al. 2006; Pool et al. 2006).
Here, we show that the CLRT and the GOF are very sensitive to choosing outlier loci from the tails of empirical distributions, leading to false inference of selection when the true model has no selection occurring (>50% of the time for the parameters investigated). We describe a correction procedure that both is efficient and restores the false-positive rate to near nominal levels. In addition, we use a novel simulation of selective sweeps to explore the efficiency of outlier detection at identifying selected loci in models of demography-plus-selection. We find that using levels of diversity, or of population differentiation, performs better than summaries of the site-frequency spectrum, as recently found by Teshima et al. (2006). Additionally, we find that the size of the region surveyed in a genome scan (i.e., the length of each fragment sequenced) affects the efficiency of outlier detection, with a clear advantage to scanning longer fragments.
METHODS
Simulating genome-scan data:
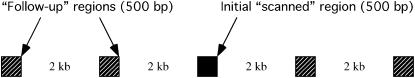
We simulated genome-scan data consisting of 100 independent loci, from a population that has undergone a recent, severe reduction in population size. Our goal here is to mimic the experimental designs that have been applied to D. melanogaster (e.g., Glinka et al. 2003; Ometto et al. 2005; Beisswanger et al. 2006). Such genome scans consist of two phases. First, short fragments of DNA are sequenced at a large number of regions of the genome (Glinka et al. 2003; Ometto et al. 2005). Second, if a fragment from the first step is identified as interesting, further sequencing will be performed in the region containing the fragment, and additional, linked fragments will be sequenced (e.g., Beisswanger et al. 2006), and a parametric test of selection will be applied, such as that of Kim and Stephan (2002). To simulate this experimental design, we simulate genealogies from 10.5-kb regions, according to the scheme shown in Figure 1. This scheme consists of five, 500-bp fragments evenly spaced over the 10.5 kb. The third, central, fragment represents the initial fragment surveyed in a genome-scan experiment. Should this fragment be chosen for further study, the simulated data from the four other linked fragments are added to the central fragment, and a parametric test of selection is performed. A genome scan data set of 100 regions is thus generated by simulating 100 of the 10.5-kb regions shown in Figure 1, and we simulated 1000 such data sets (a total of 105 10.5-kb regions).
Figure 1.—
Simulation scheme for genome-scan data. A data set consists of five linked, 500-bp fragments, with 2 kb between fragments. The central fragment (solid box) represents data obtained in a genome-scan study. The other four fragments (hatched boxes) represent the follow-up sequencing that is done if the middle fragment is chosen for further investigation of variability in the region.
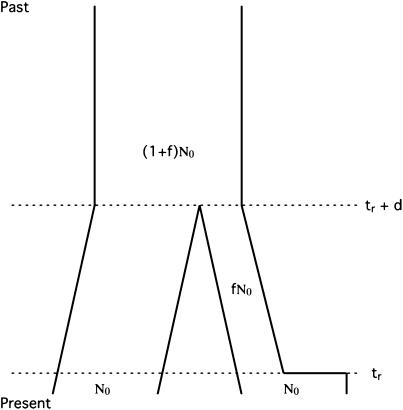
We simulated both the ancestral population and the derived, bottlenecked, population, according to the model in Figure 2. This model has five parameters: the population mutation rate (θ = 4N0μ, where N0 is the effective size of the ancestral population), the population recombination rate (ρ = 4N0r), the time at which the derived population recovered from the bottleneck (tr), the duration of the bottleneck (d), and the severity of the bottleneck (f, 0 < f ≤ 1). In this study, we use θ = 0.01/site, ρ = 0.1/site, tr = 0.004, d = 0.015, and f = 0.03, as these bottleneck parameters are compatible with data from European samples of D. melanogaster (Thornton and Andolfatto 2006). To perform these simulations, a program was written using the coalescent simulation functions in libsequence (Thornton 2003).
Figure 2.—
Bottleneck model in a derived population. The model considers a derived population that experiences a bottleneck upon splitting from an ancestral population. In this model, time is scaled in units of 4N0 generations, where N0 is the present size of the ancestral population. Moving backward in time, the derived population recovers from the bottleneck at time tr. The bottleneck reduced the population size of the derived population from N0 to fN0 for duration d. At time tr + d in the past, the two populations split from a common ancestor of size (1 + f)N0. For recombining regions, this model then has five parameters: the scaled mutation rate θ, the scaled recombination rate ρ = 4N0r, tr, d, and f.
Modeling selective sweeps:
We consider a contiguous fragment of M nucleotides. A beneficial mutation has swept to fixation at position X, 1 ≤ X ≤ M. We consider a coalescent process for a Wright–Fisher model with intragenic recombination (Hudson 1983) and measuring time, t, in units of 4N generations (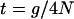 , where g is the number of generations). In this model, the selective sweep ends (i.e., the beneficial mutation fixes in the population) at time τ ≥ 0. We model the trajectory of the selected allele using the deterministic approximation given in Stephan et al. (1992), with the frequency of the beneficial allele at time t of the sweep given by
, where g is the number of generations). In this model, the selective sweep ends (i.e., the beneficial mutation fixes in the population) at time τ ≥ 0. We model the trajectory of the selected allele using the deterministic approximation given in Stephan et al. (1992), with the frequency of the beneficial allele at time t of the sweep given by
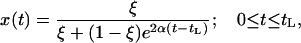 |
(1) |
where α = 2Ns and 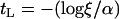 , the length of the sweep in units of 4N generations. Here, we use
, the length of the sweep in units of 4N generations. Here, we use 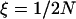 .
.
The simulation has two phases—a neutral phase and a selective phase (Braverman et al. 1995). The neutral phase is the standard coalescent model with recombination (Hudson 1983). At time τ in the past, the simulation enters the selective phase, which is modeled as a structured coalescent process (e.g., Kaplan et al. 1988; Braverman et al. 1995), and time is incremented in small units, δt, until the frequency of the beneficial allele first reaches x(t) < ξ, at which point the simulation continues in a neutral phase until the most recent common ancestor of the sample is reached. Events at time t during the sweep occur with the following probabilities. First, there are the probabilities of coalescence in the favored and unfavored classes,
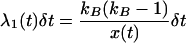 |
(2) |
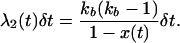 |
(3) |
In the above, at time t during the sweep, there are B lineages in the favored class and b in the unfavored. The probabilities of recombination within the same two classes are
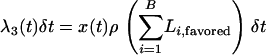 |
(4) |
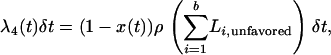 |
(5) |
where ρ = 4Nr, the population recombination rate per site, and 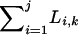 refers to the total number of positions at which recombination events may occur in the kth class. During the sweep, if the ith chromosome in class k begins at position I and ends at position J (1 ≤ I < M, 1 < J ≤ M, and I < J), then Li,k = max(X, J) − min(X, I). Finally, there are the probabilities of recombination from the favored class to the unfavored,
refers to the total number of positions at which recombination events may occur in the kth class. During the sweep, if the ith chromosome in class k begins at position I and ends at position J (1 ≤ I < M, 1 < J ≤ M, and I < J), then Li,k = max(X, J) − min(X, I). Finally, there are the probabilities of recombination from the favored class to the unfavored,
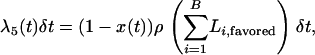 |
(6) |
and recombination from the unfavored to the favored,
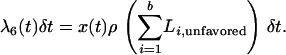 |
(7) |
For example, in Equation 6, a chromosome from the favored class is selected, and the position of the recombination event is chosen uniformly along the length of the chromosome. After the recombination event, the chromosome fragment that does not contain the selected site is placed in the unfavored class. (Recall that time is moving backward, and therefore the fragment not containing the selected site had its ancestor in the unfavored class.) A similar argument is made for Equation 7.
Our implementation of the selective phase applies the rejection algorithm of Braverman et al. (1995) to choose among the various possible events. We tested the accuracy of our simulation in two ways, using code provided by Yuseob Kim and described in Kim and Stephan (2002). First, for a given set of parameters, the distribution of several summary statistics was compared between the two implementations of the sweep process, and results were in excellent agreement (data not shown). Second, the inference machinery described in Kim and Stephan (2002), which estimates X and α on the basis of the spatial distribution of variability, was applied to the output of both programs. We checked that the distributions of 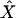 and
and 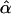 were similar when obtained from the output of both simulations, as a check that the patterns of variability surrounding the selected site were simulated accurately in our code.
were similar when obtained from the output of both simulations, as a check that the patterns of variability surrounding the selected site were simulated accurately in our code.
Sweeps in two-population models:
We extended the above model of a selective sweep to a two-population model in which one population undergoes a stepwise bottleneck (Figure 2), and no migration occurs between populations. Sweeps in the bottlenecked population occur during the period when N is reduced, constrained so that a selective sweep event does not cross a change in population size (tr < τ < tr + d and tr < τ + tL < tr + d). In this article, we do not consider the case of sweeps in the ancestral population.
We calculate the trajectory of the favored allele using 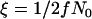 and α = 2fN0s in Equation 1. This calculation results in tL, the length of the sweep, being in units of 4fN0 generations; i.e.,
and α = 2fN0s in Equation 1. This calculation results in tL, the length of the sweep, being in units of 4fN0 generations; i.e., 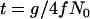 , where g is the length of the sweep in generations. However, we measure time in the simulation in units of 4N0 generations, and therefore events during the selective phase occur on different timescales in the two populations, which is accounted for as described below.
, where g is the length of the sweep in generations. However, we measure time in the simulation in units of 4N0 generations, and therefore events during the selective phase occur on different timescales in the two populations, which is accounted for as described below.
During the selective phase of a two-population model, there are three demes that must be considered: the favored class, the unfavored class, and the population not undergoing a sweep. Events in the derived population occur according to Equations 2–7, with 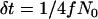 , and we simulate along the trajectory of the beneficial allele from
, and we simulate along the trajectory of the beneficial allele from 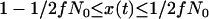 . Events in the unswept deme occur with probabilities
. Events in the unswept deme occur with probabilities
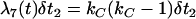 |
(8) |
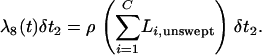 |
(9) |
In the above, there are C lineages in the population not experiencing a sweep, and 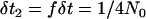 , representing that scaled time moves f-fold slower in the larger, ancestral population. Note that Li,unswept = J − I because the position of the selected site is not relevant for the population not undergoing the sweep (for the case of no migration between populations considered here). If j generations pass between events, the time in the simulation is incremented from t to t + jδt2, ensuring that the total time on the genealogy is in units of 4N0 generations.
, representing that scaled time moves f-fold slower in the larger, ancestral population. Note that Li,unswept = J − I because the position of the selected site is not relevant for the population not undergoing the sweep (for the case of no migration between populations considered here). If j generations pass between events, the time in the simulation is incremented from t to t + jδt2, ensuring that the total time on the genealogy is in units of 4N0 generations.
We simulated genealogies for an equilibrium, ancestral population and for a derived population under demography-and-selection, as described in methods and Figure 2. For each population the sample size was n = 24 chromosomes, using the model parameters described above. We chose selection parameters to maximize the effect of a sweep on the genealogy. We simulated 10.5-kb regions, and X, the position of the selected site, was assigned uniformly from 1 ≤ X ≤ 10, 500 for each replicate. We considered two different sampling schemes, sampling either 500 bp in the center of the 10.5 kb or 2500 bp. The beneficial mutation fixed in the recent past at τ = 0.0041 or 0.015, and we examined two strengths of selection—α = 2fN0s = 100 or α = 1500. We assume N0 = 2.4 million (Thornton and Andolfatto 2006), and therefore our values of α correspond to s ≈ 7 × 10−4 and ≈ 0.01, respectively. We are therefore studying the effect of a recent and relatively strong (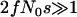 ) sweep occurring at all loci in the history of the derived population.
) sweep occurring at all loci in the history of the derived population.
From these simulations, we explore three summary statistics. First, 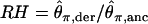 , where
, where 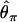 is Tajima's (1983) estimator of θ in the derived and ancestral populations, respectively. A natural-log transformation of these distributions would be analagous to the ln RH statistic for microsatellite data (Kauer et al. 2002). Second, we explore the FST-statistic of Hudson et al. (1992). Finally, we study the distribution of
is Tajima's (1983) estimator of θ in the derived and ancestral populations, respectively. A natural-log transformation of these distributions would be analagous to the ln RH statistic for microsatellite data (Kauer et al. 2002). Second, we explore the FST-statistic of Hudson et al. (1992). Finally, we study the distribution of 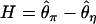 , where
, where 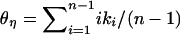 , which is a sum over the ki occurrences of derived mutations at frequency i in a sample of size n (J. Shapiro and C.-I Wu, personal communication; see also Thornton and Andolfatto 2006).
, which is a sum over the ki occurrences of derived mutations at frequency i in a sample of size n (J. Shapiro and C.-I Wu, personal communication; see also Thornton and Andolfatto 2006).
Parametric tests of selection:
We apply two parametric methods to simulated data to test for a recent selective sweep. The first method is the CLRT of Kim and Stephan (2002), and the second is the GOF test of Jensen et al. (2005). Both tests require the simulation of a null distribution, which is simulated under a neutral model for the former method and under a model of selection for the latter. For the CLRT, all null distributions consist of 104 simulated samples, and for the GOF test, we used 103 simulations to generate null distributions.
Following previous work, null distributions of the CLRT were simulated using the observed 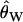 (Watterson 1975) as the mutation rate (Kim and Stephan 2002; Jensen et al. 2005), and the null distributions for the GOF test were simulated using S, the observed number of mutations in the data (Jensen et al. 2005), as simulating with
(Watterson 1975) as the mutation rate (Kim and Stephan 2002; Jensen et al. 2005), and the null distributions for the GOF test were simulated using S, the observed number of mutations in the data (Jensen et al. 2005), as simulating with 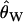 does not result in a uniform distribution of P-values when the true null model is a recent selective sweep (J. Jensen, unpublished results).
does not result in a uniform distribution of P-values when the true null model is a recent selective sweep (J. Jensen, unpublished results).
RESULTS
The “goodness-of-fit” test applied to neutral genome-scan data:
We simulated 1000 100-locus genome-scan data sets that mimic the sample sizes and locus lengths of the largest studies to date in D. melanogaster (Glinka et al. 2003; Ometto et al. 2005; see methods), using the model from Figure 2 (see methods for parameters).
We apply two methods to choose outlier loci for follow-up studies in the derived population. First, we choose a locus if the fragment surveyed is invariant in the derived population. Second, we choose a locus if the value of Tajima's (1989) D statistic in the derived population is less than or equal to the value of D at the lower 2.5th percentile of the empirical distribution (D ≤ D0.025). These two ascertainment schemes identify nonoverlapping sets of loci for further analysis, as D is undefined for invariant regions.
From the 1000 100-locus, neutral data sets, we obtained 10,827 regions chosen on the basis of having no variation and 3300 on the basis of D ≤ D0.025. Note that D is a discrete statistic, and therefore the value of D may be identical at different percentiles of the empirical distribution, which is why we obtained more than the expected 2500 outliers.
We then applied the CLRT of Kim and Stephan (2002) and, for those loci rejecting neutrality at P < 0.05, calculated P-values for the GOF method as previously described (Jensen et al. 2005). For the GOF test, there is a range of P-values (P ≤ ∼0.05–0.2) where it is unclear if selection can be distinguished from demography (Jensen et al. 2005). For our purposes, we apply a strict cutoff at P = 0.05, such that P ≤ 0.05 implies that a recent selective sweep with the parameters estimated from the composite-likelihood method is not the best fit to the data. Likewise, P > 0.05 implies that the rejection of neutrality by the CLRT is more likely due to a sweep than due to demography alone.
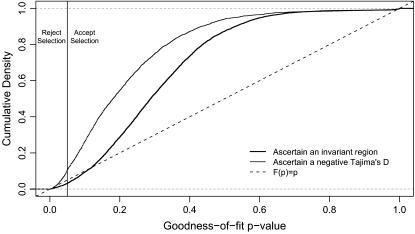
When regions were chosen because the scanned fragment was invariant, 7048 (65%) of the simulated data sets rejected neutrality according to the CLRT, and 6822 (96.7%) of those had GOF P-values >0.05, indicating that the selection model fit the data better than a demographic scenario (despite the data being simulated under a strictly neutral model). If outliers are chosen on the basis of D ≤ D0.025, 2931 (88.8%) reject neutrality using the CLRT, and 2614 (89.1%) of those had GOF P-values >0.05. If the false-positive rate were properly controlled, the empirical cumulative density function (ECDF) of P-values for the GOF test would have a cumulative density of 0.95 at P = 0.05 when applied to neutral data. This is not the case when regions are ascertained from a genome scan—very little of the cumulative density is <0.05, indicating false acceptance of the selection model for the majority of data sets (Figure 3). This leads to total type I errors of 63 and 79.2% when choosing regions because they are invariant or have an unusual Tajima's D, respectively. We should note, however, that this is a substantial improvement over relying solely on the tails of the empirical distribution. If we had simply assumed that our outliers were subject to selection, our type I error would have been 100%, but applying the GOF method reduces the error rate by 20–40%.
Figure 3.—
Empirical cumulative density functions of GOF P-values when applied to outliers in genome scans. One thousand 100-locus data sets were simulated under a bottleneck model with no selection (Figure 2, see methods for parameters), using the sampling scheme from Figure 1, and outlier loci were chosen for follow-up sequencing and application of the CLRT (Kim and Stephan 2002) and GOF (Jensen et al. 2005) methods (see methods). The vertical line at P = 0.05 is the significance threshold for the GOF test. P ≤ 0.05 leads to the inference that the selection model is rejected in favor of a demographic explanation, while P > 0.05 is taken to mean that selection is inferred to be a better explanation than demography. In this case, any P > 0.05 is therefore a false positive. If the false-positive rate were nominal (i.e., 5%), then the cumulative density of P-values should reach 0.95 by P = 0.05 (vertical line), which is not the case for either of the ascertainment schemes plotted.
Controlling the false-positive rate:
In this section, we explore controlling the false-positive rate when loci are not randomly sampled from the genome. In practice, follow-ups to genome-scan experiments have to deal with the issue of nonequilibrium demography and of how loci are selected for further analysis, and it is not clear which issue has a greater impact on downstream analysis. For example, is it necessary to correct for both demography and ascertainment, or is it sufficient to correct for either demography or ascertainment? From a statistical point of view, the appropriate quantity to keep track of is the distribution of P-values for each of these procedures and then to choose the procedure that results in a uniform distribution of P-values when the null model is correct. Although we discuss the problem in terms of genome scans that survey single-nucleotide polymorphisms and follow up with the CLRT/GOF tests, the statistical issues addressed here are quite general. All statistical tests of neutrality that we are aware of assume a null distribution where loci are random draws from the model, but the ascertainment of a region and its use in a subsequent hypothesis test samples from a different null distribution. The general issue here is how to sample from the correct null distribution, illustrated with specific examples using the Kim and Stephan (2002) framework. Further, as the GOF test is applied only to regions that reject the null model with the CLRT, it is sufficient to control the false-positive rate of the CLRT. In other words, if a null distribution for the CLRT gives a 5% false-positive rate, then the total false-positive rate of the entire procedure (the CLRT + GOF tests) is necessarily ≤5%, and therefore we can identify the maximum false-positive rate.
For the results described above, there are two factors that contribute to a high false-positive rate. First, in practice, one obtains P-values for the CLRT by simulating a null distribution under the standard neutral model (Harr et al. 2002; Kim and Stephan 2002; Bauer-DuMont and Aquadro 2005; Beisswanger et al. 2006; Pool et al. 2006), which is problematic when the demographic assumptions of that model are violated (Jensen et al. 2005). Second, the CLRT (and the subsequent GOF) are not applied to randomly chosen loci in practice, but to outlier loci identified by a genome scan (e.g., Harr et al. 2002; Bauer-DuMont and Aquadro 2005; Beisswanger et al. 2006; Pool et al. 2006). Such ascertainment procedures choose loci with very unusual underlying genealogies, resulting in a pattern of spatial variability that may mimic that of a recent selective sweep, such as an excess of high-frequency, derived mutations surrounding a region of reduced diversity (Figure 4). This pattern is observed because lineages in the invariant region reach common ancestors in the relatively recent past during the bottleneck, whereas lineages in the flanking regions have different genealogies due to recombination and reach common ancestors further back in the past (either later during the bottleneck or they coalesce at a time more ancient than the bottleneck). This effect is also predicted by Barton (1998), who showed that many of the properties of genealogies are very similar between bottlenecks and selective sweeps. In this section, we show that the false-positive rate of genome scans can be controlled if the demographic model is known and the ascertainment procedure accounted for when simulating the null distribution. We explore the case of ascertaining a region of the genome on the basis of the original scanned fragment having no variation, but the principles apply to any ascertainment scheme.
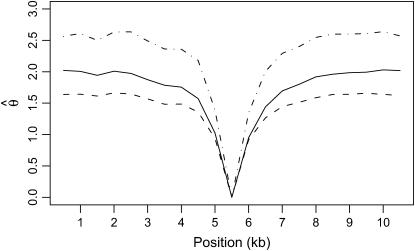
Figure 4.—
The expected spatial pattern of variability for a 10.5-kb region is plotted, given that the region has been ascertained because it contains a small invariant fragment. Estimates of the expectation of three estimators of θ = 4Neμ are plotted for nonoverlapping 500-bp windows—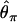 (solid line, Tajima 1983),
(solid line, Tajima 1983), 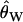 (dashed line, Watterson 1975), and
(dashed line, Watterson 1975), and 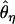 (dotted/dashed line, Equation 1 of Thornton and Andolfatto 2006). These data were simulated under the bottleneck model in Figure 2 with parameters θ = 0.01/site, ρ = 0.1/site, tr = 0.004, d = 0.015, and f = 0.03. The expectations are for n = 20 and are based on 1000 simulated replicates, analyzing only the derived, bottlenecked population. One-hundred-kilobase regions were simulated, a single window of 500 bp with no variation was identified, and the flanking 5 kb on either side were then analyzed.
(dotted/dashed line, Equation 1 of Thornton and Andolfatto 2006). These data were simulated under the bottleneck model in Figure 2 with parameters θ = 0.01/site, ρ = 0.1/site, tr = 0.004, d = 0.015, and f = 0.03. The expectations are for n = 20 and are based on 1000 simulated replicates, analyzing only the derived, bottlenecked population. One-hundred-kilobase regions were simulated, a single window of 500 bp with no variation was identified, and the flanking 5 kb on either side were then analyzed.
The statistic of interest is λ, the composite likelihood-ratio statistic from the CLRT of Kim and Stephan (2002). We wish to obtain a null distribution of λ that is both generated from the correct demographic model and conditional on the ascertainment scheme (asc). In other words, for a specified demographic model, we wish to sample λ from the conditional distribution Pr(λ|asc) = Pr(λ ∩ asc)/Pr(asc). An estimate of Pr(asc) is used as a weight on the observed statistic λobs, and P-values are estimated using n draws from the conditional null distribution as
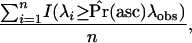 |
(10) |
where I(x) = 1 if the condition x is true and 0 otherwise. These corrected P-values can be calculated using a rejection algorithm (described below) and available software (e.g., Hudson 2002). In practice, one can simply run a simulation until n replicates satisfying the ascertainment scheme are recorded, keeping track of the k trials required, allowing Pr(asc) to be estimated as n/k. The steps of this algorithm are detailed in the appendix.
To mimic the ascertainment scheme, we accept only simulation runs where the middle of the five fragments is invariant. In practice, the null distribution of the CLRT is obtained using 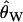 (Watterson 1975) as the mutation rate in the simulations. This poses a practical problem when simulating under demographic models that reduce diversity—if fragments of a region are sampled, the probability that all fragments are invariant can be relatively high for small
(Watterson 1975) as the mutation rate in the simulations. This poses a practical problem when simulating under demographic models that reduce diversity—if fragments of a region are sampled, the probability that all fragments are invariant can be relatively high for small 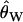 . We therefore accept simulation runs only if the middle fragment is invariant and there is at least one segregating site in the data. Therefore, Pr(asc) = Pr(middle region invariant ∩ S > 0). Conditioning on the data set being variable is also appropriate as an investigator would not perform the CLRT on a region completely devoid of variation.
. We therefore accept simulation runs only if the middle fragment is invariant and there is at least one segregating site in the data. Therefore, Pr(asc) = Pr(middle region invariant ∩ S > 0). Conditioning on the data set being variable is also appropriate as an investigator would not perform the CLRT on a region completely devoid of variation.
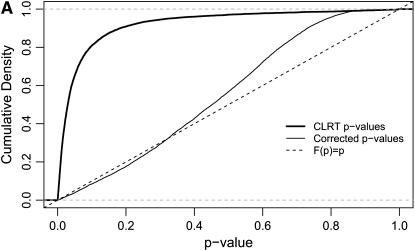
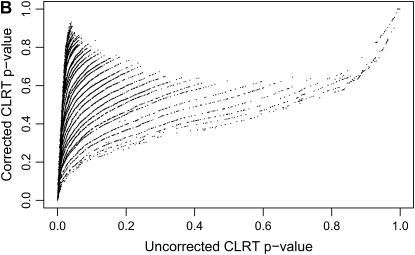
We applied this procedure to the 10,827 data sets that were ascertained from our simulated neutral data on the basis of having an invariant region in the scanned fragment. As described above, 65% of these data sets falsely reject the equilibrium neutral model in favor of selection. When the correct demographic null model is used (a stepwise bottleneck with tr = 0.004, d = 0.015, and f = 0.03), and ascertainment is accounted for, 3.8% of loci reject neutrality, making the test slightly conservative. The ECDFs of P-values for these two cases are shown in Figure 5A. If the P-values are truly drawn from the null distribution, then the cumulative density function (CDF) of P-values should be a linear function F(P) = P. When ascertainment and demography are not accounted for, ∼65% of P-values are <0.05 (thick solid line in Figure 5A). Accounting for demography and ascertainment leads to an ECDF that grows approximately as expected (thin line in Figure 5A). Further, accounting for ascertainment and demography affects the rank order of P-values (Figure 5B). The change in rank order shows that the standard CLRT P-values are not an appropriate metric to compare the evidence in favor of selection at different loci, when loci are ascertained from a genome-scan experiment. For each value of 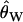 , the null distribution consisted of 50,000 replicates matching the ascertainment criteria under the bottleneck model. Results were nearly identical using only 1000 replicates (data not shown). The acceptance rates in the simulations [i.e.,
, the null distribution consisted of 50,000 replicates matching the ascertainment criteria under the bottleneck model. Results were nearly identical using only 1000 replicates (data not shown). The acceptance rates in the simulations [i.e., 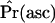 ] ranged from 0.12 to 0.52, depending on the value of
] ranged from 0.12 to 0.52, depending on the value of 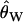 . This procedure is therefore efficient enough to be performed using available software to simulate from the neutral coalescent (e.g., Hudson 2002).
. This procedure is therefore efficient enough to be performed using available software to simulate from the neutral coalescent (e.g., Hudson 2002).
Figure 5.—
The effect of region ascertainment on the P-values for the CLRT of Kim and Stephan (2002). These plots are generated from our simulated data for the case where neutral regions are ascertained on the basis of an invariant fragment in a population that has undergone a recent, severe bottleneck (see methods) and are calculated from the same data used in Figure 3. (A) Cumulative densities of P-values for the CLRT of Kim and Stephan (2002) for ascertained genome-scan data. When the null model is the standard neutral model and ascertainment is not accounted for, the cumulative density grows quickly; i.e., there is a large false-positive rate (thick solid line). When both the demographic model and ascertainment are accounted for, the cumulative density grows approximately as expected, with a 3.8% false-positive rate (thin solid line). (B) The P-values in A are plotted against each other. The P-values on the x-axis do not account for ascertainment or demography and correspond to the thick solid line in A. The corrected P-values are on the y-axis.
Application to data:
Beisswanger et al. (2006) recently analyzed levels of nucleotide variability in a Netherlands and a Zimbabwe sample of D. melanogaster. They sequenced in this region because a previous study (Glinka et al. 2003) had identified a small (∼500 bp) fragment without variation near the wapl locus on the X chromosome in the Netherlands sample. Beisswanger et al. (2006) sequenced 12 short (again, ∼500 bp) fragments distributed along a 110-kb region surrounding the wapl locus. The data therefore consist of 13 fragments in total, including the fragment originally discovered in Glinka et al. (2003). In this section, we explore the effects that ascertainment and demography have on the P-value of the CLRT applied to the Netherlands data from this region. Specifically, we estimate the CLRT P-value under four scenarios:
Using the standard neutral model as the null model and not accounting for ascertainment: This is the standard application of the CLRT.
Using the standard neutral model as the null, but accounting for ascertainment.
Using the point estimates for a bottleneck model for the Netherlands population (Thornton and Andolfatto 2006) as the demographic null model and not accounting for ascertainment. We use point estimates here, rather than simulate from the full posterior distribution on the parameter space, to keep the procedure as practical as possible using available tools, such as ms (Hudson 2002).
Using the point estimates for a bottleneck model for the Netherlands population (Thornton and Andolfatto 2006) as the demographic null model and accounting for ascertainment.
To improve simulation efficiency when generating a null distribution for an ascertained region under the standard neutral model, we applied a slightly different scheme from that described in the previous section. For each simulated replicate, we calculated T, the total length of the genealogy in the ascertained fragment (the invariant fragment identified in Glinka et al. 2003), and placed no mutations on that fragment. For the ith replicate, 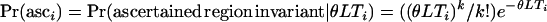 , where θ is the mutation rate per site, L is the length of the fragment, and k = 0. The P-values are then estimated as
, where θ is the mutation rate per site, L is the length of the fragment, and k = 0. The P-values are then estimated as
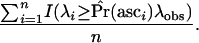 |
(11) |
This approach is appropriate for sparsely sampled fragments (Figure 1) and has the advantage that all simulation replicates can be used, rather than relying on rejection sampling, which would be inefficient as the probability of an invariant fragment is low under the standard neutral model.
The CLRT P-values estimated under these four schemes are shown in Table 1. In all calculations, we used estimates of θ and ρ from Beisswanger et al. (2006) and an input file for the CLRT kindly provided by Steffen Beisswanger. When the standard CLRT is applied to the data, the P-value is nearly significant (0.054). When either ascertainment or demography is accounted for individually, the P-value is much larger (0.99 in both cases). Finally, when both demography and ascertainment are accounted for in the null distribution, P = 0.81. Clearly, the impacts both of demography and of how regions are selected for analysis may have a large influence on the strength of evidence in favor of selection. Although correcting either for ascertainment alone or for demography resulted in a nonsignificant CLRT for this example, it is not guaranteed that either procedure adequately controls the false-positive rate. We explore these issues below.
TABLE 1.
The effect of different null distributions on CLRT P-values, applied to the Netherlands data in Beisswanger et al. (2006)
| Null model | Ascertainment | 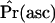 |
CLRT P-value |
|---|---|---|---|
| SNMa | Ignored | NA | 0.054 |
| SNM | Accounted forb | 0.025 | 0.99 |
| SNM | Accounted forc | 0.024d | 0.99 |
| Bottlenecke | Ignored | 0.998f | 0.27 |
| Bottlenecke | Accounted for | 0.414g | 0.81 |
SNM, standard neutral model. When ascertainment of regions is ignored, this is the standard CLRT (Kim and Stephan 2002), and 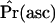 is not relevant.
is not relevant.
Calculated using rejection sampling and Equation 10.
Calculated using Equation 11, see text for details.
Calculated as the mean of 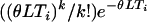 , from 105 simulated replicates.
, from 105 simulated replicates.
Using the parameters from Thornton and Andolfatto (2006), tr = 0.004, d = 0.015, f = 0.03 (see Figure 2), and the mutation and recombination rates used in Beisswanger et al. (2006).
Calculated using Equation 10, with 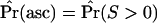 .
.
Calculated using Equation 10, with 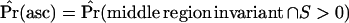 .
.
Correcting for ascertainment under the standard neutral model:
The results in Table 1 suggest that accounting for ascertainment of regions alone, and assuming the standard neutral model, may have a strong effect on CLRT P-values. Given that there is considerable uncertainty concerning the appropriate demographic model to use for the null distribution, we explore here the effect of accounting for ascertainment, but assuming the standard neutral model as the demographic null model.
We generated null distributions for the CLRT for 10,827 data sets ascertained from our genome-scan simulations based on having an invariant region. The simulation scheme is as described above for the Beisswanger et al. (2006) data, calculating P-values according to Equation 11 from 1000 simulated data sets. Accounting for ascertainment alone resulted in an overly conservative distribution of P-values (100% of the data sets had P ≥ 0.422). The reason that this procedure is so conservative is due to the demographic assumptions. In an equilibrium population, the probability of ascertaining an invariant, 500-bp region is high for small 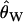 , in which case there is little information in the data and therefore little power to reject the null model. At the other extreme, it is very unlikely to ascertain a 500-bp invariant region for high
, in which case there is little information in the data and therefore little power to reject the null model. At the other extreme, it is very unlikely to ascertain a 500-bp invariant region for high 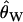 , and therefore the weight placed on the observed λ is very small, such that
, and therefore the weight placed on the observed λ is very small, such that 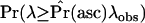 is high when estimated using the standard neutral model as the null.
is high when estimated using the standard neutral model as the null.
Correcting only for demographic effects:
In the analysis of the Beisswanger et al. (2006) data, correcting for demographic effects alone resulted in a nonsignificant CLRT. We explore here whether or not correcting for demography, while ignoring ascertainment of regions, is sufficient to control the false-positive rate. To do this, we generated a null distribution for the same 10,827 data sets under the correct demographic model, but ignoring ascertainment. Because there is a nonzero probability that all five segments will be invariant for small θ for this model, we condition on the regions being variable and estimate Pr(asc) = Pr(S > 0) by rejection sampling as described above. The distribution of P-values for the CLRT in this case had a substantial excess of small P-values, with 49.8% of data sets having P ≤ 0.05. In other words, correcting for demography, but ignoring how loci are selected for testing, improves the false-positive rate by ∼23% (from 65% for the standard CLRT to 49.8%) when the CLRT is applied to regions ascertained from a bottlenecked population due to the observation of an invariant fragment. In other words, for the demographic model considered here, correcting for demographic effects alone results in an overly liberal test, whereas accounting for ascertainment, but not for demography, results in an overly conservative test. The intuitive explanation for this is that the procedure of identifying regions on the basis of small, invariant fragments specifically scans for regions with spatial patterns of variability that look like a recent sweep (i.e., qualitatively similar to Figure 4). While such spatial patterns are quite rare under the standard neutral model (i.e., there is a low probability of discovering such regions), they are enriched for under diversity-reducing neutral models compared to the standard neutral model. Thus while correcting for demography alone may result in a nonsignificant CLRT for individual examples (Table 1), this is not true in general (and holds only ∼50% of the time for the model studied here).
The utility of empirical distributions:
In the previous sections, we demonstrated that choosing loci from the tails of the empirical distribution of a summary statistic imposes an ascertainment bias that must be accounted for in subsequent analyses (e.g., Figures 3 and 5A). Of particular importance is how such loci are chosen from the tails in the first place. In practice, the choice is not made using the results of the CLRT/GOF (due to computational impracticability), but rather on the basis of a summary of the data (e.g., Harr et al. 2002; Glinka et al. 2003; Bauer-DuMont and Aquadro 2005; Beisswanger et al. 2006; Pool et al. 2006).
Teshima et al. (2006) have recently found that choosing outlier loci on the basis of low levels of diversity is more powerful than choosing on the basis of summaries of the site-frequency spectrum. They simulated genome-scan experiments consisting of 10-kb regions, with mutation rates appropriate for humans or maize. Diversity levels in maize are similar to those in D. melanogaster and are ∼10 times lower in humans. Thus, the economic efficiency of genome scans, in terms of SNPs discovered per dollar, depends on the organism. Further, the largest genome scans in flies have relied on ∼ 500-bp fragments (Glinka et al. 2003; Orengo and Aguade 2004; Ometto et al. 2005). Here, we explore the effect of region length on the power of genome scans, finding that longer regions have considerably more power. We have developed a novel simulation program that allows us to simulate a sweep in a derived, bottlenecked population in addition to the genealogy of the ancestral population. Our simulation allows us to explore the effect of sweeps on population differentiation, which was not considered in Teshima et al. (2006), but has been considered as a statistic in genome scans (e.g., Akey et al. 2004; Storz et al. 2004) because selective sweeps in structured populations are expected to increase population differentiation (Santiago and Caballero 2005).
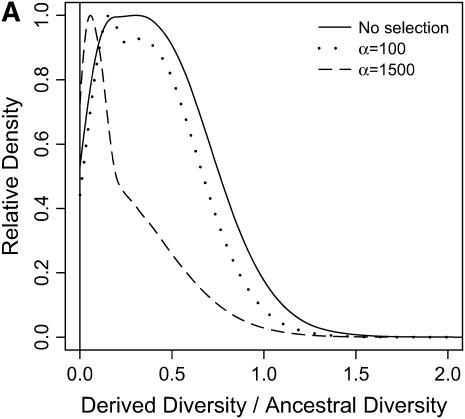
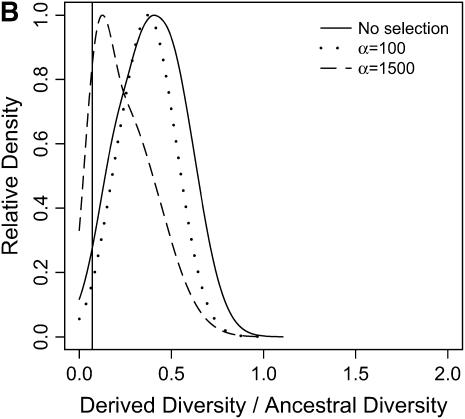
From our simulations, we estimated the distributions of three summary statistics—RH, FST, and H (Figures 6–8; see methods for descriptions of the statistics). In Figures 6–8, a vertical line is placed at the 5th quantile (95th for FST) of the distribution of the statistic under the bottleneck. Therefore, the density to the left (right for FST) of this line represents the amount by which selection in a bottlenecked population enriches the tail of an empirical distribution for selected loci.
Figure 6.—
Reduction in diversity in nonequilibrium populations. The distribution of diversity (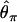 ) in the derived population, relative to
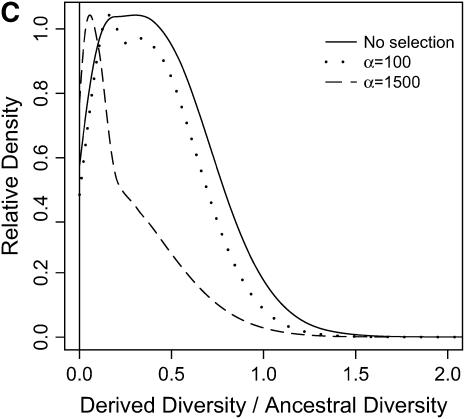
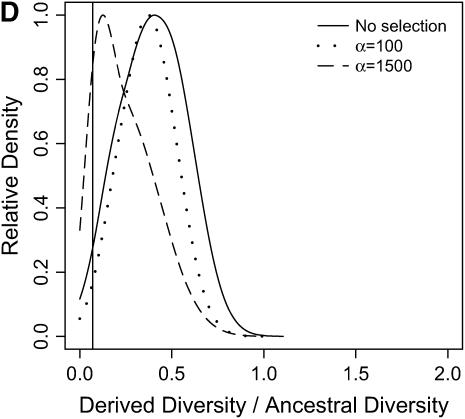
) in the derived population, relative to 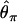 in the ancestral population is plotted for models of a bottleneck, as well as those of a bottleneck-plus-selection. For the cases including selection, a 10.5-kb region was simulated, and the position of the selected site was randomly placed (from a uniform distribution) in the region. The bottleneck parameters are given in methods. In each plot, the case of no selection is plotted as a reference and compared to two selection coefficients, α = 2fN0s = 100 and 1500. In addition, a vertical line is placed at the 5th percentile of the empirical distribution without selection. In A–D, different values of τ, the time of fixation of the beneficial allele, as well as different lengths of regions (500 or 2500 bp sampled from the middle of the 10.5 kb), are considered: (A) τ = 0.0041, 500-bp region; (B) τ = 0.0041, 2500-bp region; (C) τ = 0.015, 500-bp region; (D) τ = 0.015, 2500-bp region.
in the ancestral population is plotted for models of a bottleneck, as well as those of a bottleneck-plus-selection. For the cases including selection, a 10.5-kb region was simulated, and the position of the selected site was randomly placed (from a uniform distribution) in the region. The bottleneck parameters are given in methods. In each plot, the case of no selection is plotted as a reference and compared to two selection coefficients, α = 2fN0s = 100 and 1500. In addition, a vertical line is placed at the 5th percentile of the empirical distribution without selection. In A–D, different values of τ, the time of fixation of the beneficial allele, as well as different lengths of regions (500 or 2500 bp sampled from the middle of the 10.5 kb), are considered: (A) τ = 0.0041, 500-bp region; (B) τ = 0.0041, 2500-bp region; (C) τ = 0.015, 500-bp region; (D) τ = 0.015, 2500-bp region.
Figure 8.—
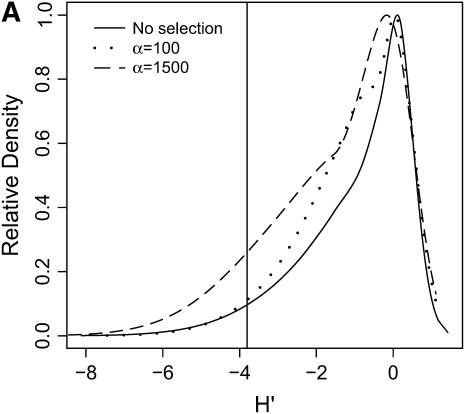
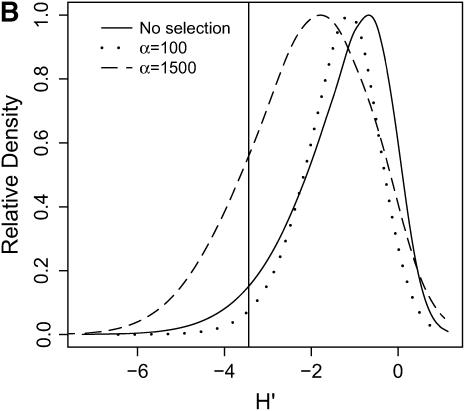
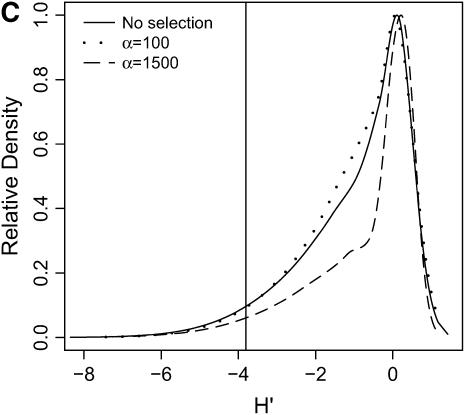
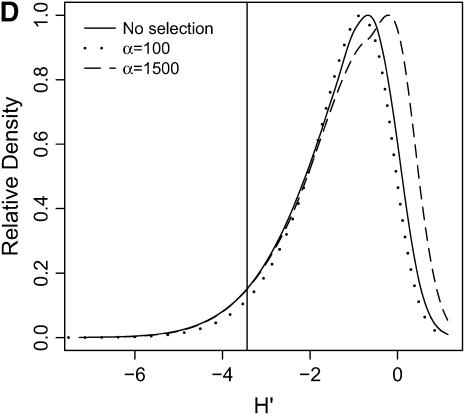
High-frequency, derived mutations in nonequilibrium populations. The same data analyzed in Figure 6 are analyzed here, calculating the distributions of H′ (Equation 2 of Thornton and Andolfatto 2006), a summary of high-frequency, derived alleles in the bottlenecked population. Again, a vertical line is placed at the 5th percentile of the distribution without selection. (A) τ = 0.0041, 500-bp region; (B) τ = 0.0041, 2500-bp region; (C) τ = 0.015, 500-bp region; (D) τ = 0.015, 2500-bp region.
Figure 7.—
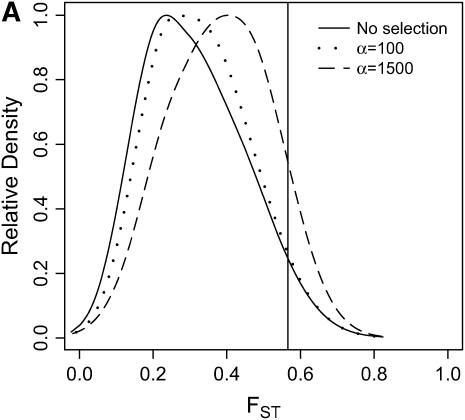
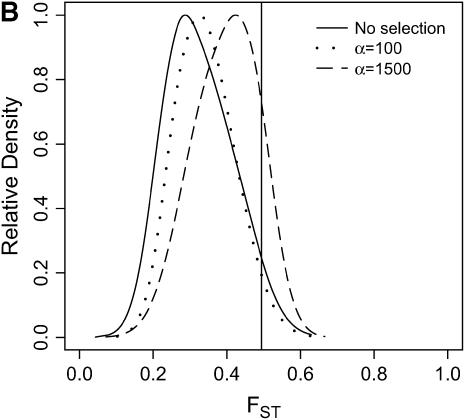
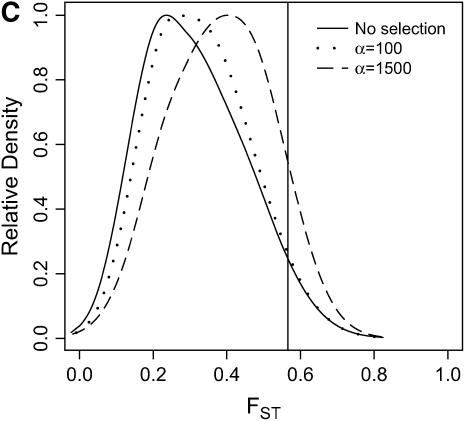
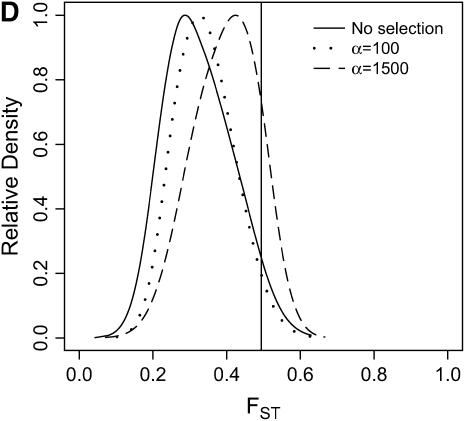
Differentiation between ancestral and derived populations in models of selection-and-demography. The same data analyzed in Figure 6 are analyzed here, calculating the distributions of FST (Hudson et al. 1992) between the ancestral and the derived population. As the position of the selected site is random a fixed difference between populations was added to the data at the position of the beneficial mutation before calculating FST. A vertical line is placed at the 95th percentile of the distribution without selection. (A) τ = 0.0041, 500-bp region; (B) τ = 0.0041, 2500-bp region; (C) τ = 0.015, 500-bp region; (D) τ = 0.015, 2500-bp region.
In general, when selection is both recent (τ = 0.0041) and strong (α = 1500), the tails of empirical distributions will be enriched for selected loci. However, the power to detect this pattern depends both on the statistic used and on the design of the genome-scan experiment. If one looks at RH, the reduction in diversity in the derived population, strong recent selection cannot be identified in the tails of a genome-scan experiment when 500-bp regions are surveyed, for the mutation and recombination rates considered here (Figure 6A). The reason for this is that, under this bottleneck model, a 500-bp region has an ∼10% chance of being invariant, and a selective sweep obviously cannot reduce diversity any further. However, if 2500-bp regions are surveyed in the genome scan, values of RH = 0 are unlikely under the demographic model, and strong selection can be detected, even relatively far back into the past (Figure 6, B and D). Interestingly, there is no apparent effect of sequence length on FST (Figure 7). There is a dramatic improvement in the efficiency of H to detect selection if longer regions are surveyed (compare Figure 8A to 8B), but the power vanishes rapidly with increasing τ (Figure 8, C and D).
DISCUSSION
We have studied the effect that the ascertainment of regions from genome-scan studies has on inferences of selection in subsequent analysis. When the true model is a nonequilibrium, neutral model, ascertainment of “unusual” regions for further analysis can lead to the false inference of selection (Figure 3) because the ascertainment procedure itself identifies regions with spatial patterns of variability mimicking what is expected from a selective sweep (Figure 4). For the parametric tests of selection considered here (Kim and Stephan 2002; Jensen et al. 2005), the false-positive rate can be controlled if both ascertainment and demography are accounted for when generating the null distribution for the tests (Figure 5A).
Uncertainty about demographic model:
While ascertainment is easily accounted for, the true demographic model for most populations of interest is unknown. In our analysis of the Beisswanger et al. (2006) data, we used bottleneck parameters inferred from the genome scan that identified the wapl region (Glinka et al. 2003; Thornton and Andolfatto 2006) and used those parameters as the demographic null model to analyze the new data. Although these parameter estimates are based on the simplifying assumptions that ρ/θ is constant across loci on the D. melanogaster X and that the African population is at demographic equilibrium (discussed in Thornton and Andolfatto 2006), our analysis suggests that correcting for the ascertainment of the wapl region in The Netherlands, and not attempting to account for demography, greatly weakens the evidence for a recent selective sweep (Table 1).
When correcting for demographic effects, our approach made use only of point estimates of demographic parameters and failed to account for uncertainty in the estimates. However, Equations 10 and 11 are easily extended to simulating from the full, joint posterior distribution of parameters. Further, the approach can be extended to multiple demographic models. For example, Pritchard et al. (1999) implemented a summary-statistic Bayesian method with equal prior weight on different demographic scenarios, and the acceptance rate from each model was proportional to the posterior probability that the data were drawn from that model. In principle, a similar approach could be used to generate null distributions for the CLRT that take into account uncertainty about demography. The power of any of these approaches, and their computational feasibility, however, remains an open question.
Empirical distributions:
Although we have shown how ascertainment from the tails of empirical distributions can be accounted for, power of the outlier detection approach to identify selected loci depends on the design of the genome-scan experiment. In the simulations we conducted, we find that summaries of diversity (Figure 6) or population differentiation (Figure 7) are likely to be of more use in reliably identifying outlier loci that are under selection than summaries of the site-frequency spectrum (Figure 8). Teshima et al. (2006) have recently reached similar conclusions in a simulation study focusing on demographic models believed to be plausible for humans and maize.
Of particular interest is the effect that the size of the regions surveyed has in a genome scan. For the models explored here, we find that there is a substantial practical benefit to surveying longer regions (Figures 6–8). To date, the largest genome-scan data sets in D. melanogaster have consisted of fragments that are short enough to sequence across in a single pass (Glinka et al. 2003; Ometto et al. 2005). However, such regions may be too short, such that selected loci are not more extreme than neutral loci, as measured by levels of diversity (Figure 6), although such an effect is not observed when looking at FST (Figure 7). The advantage of sequencing larger fragments is also important in post-genome-scan analyses, since sequencing small, dispersed fragments leads to poor estimates of selection parameters (J. D. Jensen, K. R. Thornton and C. F. Aquadro, unpublished results).
Important caveats:
In our simulations of selection, we have assumed a specific model where adaptation occurs from new mutations sweeping to fixation (Maynard-Smith and Haigh 1974). Przeworski et al. (2005) have recently used simulations to show that selection on standing variation (i.e., a previously neutral mutation that becomes beneficial) results in selective sweeps with less pronounced effects on variability at linked, neutral sites. Further, selection on standing variability does not enrich the tails of empirical distributions to the same extent as positive selection acting on new mutations (Teshima et al. 2006). Thus, while genome scans will identify interesting candidate loci, the false-positive and false-negative rates depend on details both of the demographic history of the populations in question and of the nature of beneficial mutations (Teshima et al. 2006).
In this study, we have considered only the case of ascertainment bias imposed by studying a region because of prior knowledge of levels of polymorphism. Our simulations assumed that the polymorphic markers themselves are randomly sampled from the population. While this is appropriate for the large SNP data sets that currently exist for Drosophila (Glinka et al. 2003; Ometto et al. 2005), they do not mimic the sampling schemes that are currently being applied to the largest data sets for humans, where SNPs are first identified in a small discovery panel and then later genotyped in larger samples (Hinds et al. 2005; International HapMap Consortium 2005). Ascertainment of markers is straightforward to account for in simple cases (Nielsen et al. 2004), and accurate inferences of levels of diversity and population structure depend on applying such corrections (Clark et al. 2005). Attempts to identify recent directional selection in the human genome by outlier analysis therefore have two types of ascertainment to account for, that of markers and that of outlier regions (e.g., Carlson et al. 2005; Kelley et al. 2006).
Future prospects:
We have focused on keeping the false-positive rate under control in genome-scan studies. Further progress requires both evaluation of the power of existing methods and the development of procedures to increase the power to detect selection in nonequilibrium populations. Alternatively, methods that are robust to demographic effects will provide an important, complementary approach. The recent method of Nielsen et al. (2005) is an important advance in this regard, although it relies on the assumption that a class of neutral DNA exists in the genome, which may not be the case in Drosophila (Akashi 1995; Halligan et al. 2004; Andolfatto 2005; Haddrill et al. 2005a). In addition, the current study and that of Teshima et al. (2006) have conducted simulations under the simplifying assumption that mutation rates are constant across loci. Given that looking for outliers in terms of levels of diversity may be the most promising approach to identify selected loci, variation in mutation rates is a particular concern, as regions of low variability will contain both selected loci and loci with low mutation rates. In principle, examining statistics like RH and ln RH should control for variation in mutation rates (Schlotterer 2002; Kauer et al. 2003), but the issue of power remains.
Acknowledgments
The authors thank Peter Andolfatto, Celine Becquet, and members of the Clark lab for fruitful discussions and Steffen Beisswanger for sharing data. We are grateful to Andrew Clark, Chip Aquadro, and two anonymous reviewers for comments on the manuscript and to Molly Przeworski for sharing the Teshima et al. (2006) manuscript ahead of publication. K.R.T. is supported by National Institutes of Health (NIH) grant GM065509 to A. G. Clark, and J.D.J. is supported by NIH grant GM36431 to C. F. Aquadro. and National Science Foundation grant DMS-0201037 to R. Durrett, C. F. Aquadro, and R. Nielsen.
APPENDIX: SIMULATING THE NULL DISTRIBUTION UNDER ASCERTAINMENT
We describe here an algorithm to simulate a null distribution of samples from a model where loci are not randomly sampled. The algorithm is general and requires the following ingredients:
The parameters of a demographic model: For example, one may use point estimates obtained from fitting a demographic model to the initial genome-scan data (e.g., Ometto et al. 2005; Thornton and Andolfatto 2006).
A means of generating coalescent samples from the demographic model: If the initial genome scan was done by surveying single-nucleotide polymorphisms, a perl script to run ms (Hudson 2002) would be sufficient. In this article, we wrote the simulations directly, using libsequence (Thornton 2003). If the genome scan were performed using microsatellites, and unusual regions then followed up on by surveying SNPs (e.g., Bauer-DuMont and Aquadro 2005; Pool et al. 2006), a program like simcoal (Excoffier et al. 2000; Laval and Excoffier 2004) could be used, which is capable of simulating linked SNP and microsatellite data.
A function that checks if a simulated sample is compatible with the ascertainment criteria: We label this function ascertain(data) and assume it returns 1 (true) if the ascertainment criteria are met and 0 (false) otherwise. For example, the function may return 1 if the middle 500 bp of a region are invariant or if Tajima's D < −1.5 (if the 5th quantile of the empirical distribution of D in the genome scan were −1.5). If the empirical data are sampled sparsely over large regions (e.g., Beisswanger et al. 2006), then care must be taken to analyze only the portions of the simulated sample corresponding to the regions sampled in the data.
Given the above ingredients, the algorithm to generate n samples from the null distribution under ascertainment is:
set k = 0
set m = 0
while m < n do
set k = k + 1
Generate a single coalescent sample, data, from the demographic model
if (ascertain(data) = = 1) then
set m = m + 1
save data to a file
end if
end while
return
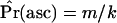 .
.
At the end of the algorithm, an estimate of the ascertainment probability under the model, 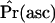 , is obtained. Further, the n instances of data that were stored are samples from the null distribution under ascertainment. The ascertainment-corrected distribution of λ, the CLRT test statistic, would then be obtained as previously described (Kim and Stephan 2002), and the P-value of the test would be calculated from Equation 10. Note that, while we discuss processing the stored instances of data for the CLRT, the method of generating samples from the corrected null distribution is general and in principle should be applied to any follow-up analysis of loci that are nonrandomly sampled from the genome.
, is obtained. Further, the n instances of data that were stored are samples from the null distribution under ascertainment. The ascertainment-corrected distribution of λ, the CLRT test statistic, would then be obtained as previously described (Kim and Stephan 2002), and the P-value of the test would be calculated from Equation 10. Note that, while we discuss processing the stored instances of data for the CLRT, the method of generating samples from the corrected null distribution is general and in principle should be applied to any follow-up analysis of loci that are nonrandomly sampled from the genome.
References
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey, J. M., M. A. Eberle, M. J. Rieder, C. S. Carlson, M. D. Shriver et al., 2004. Population history and natural selection shape patterns of genetic variation in 132 genes. PloS Biol. 2(10): 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosohila. Nature 437: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Barton, N., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Bauer-DuMont, V., and C. F. Aquadro, 2005. Multiple signatures of positive selection downstream of Notch on the tip X chromosome in Drosophila melanogaster. Genetics 171: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswanger, S., W. Stephan and D. DeLorenzo, 2006. Evidence for a selective sweep in the wapl region of Drosophila melanogaster. Genetics 172: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, J. M., R. R. Hudson, N. L. Kaplan, C. H. Langley and W. Stephan, 1995. The hitchhiking effect on the site frequency-spectrum of DNA polymorphisms. Genetics 140: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C. S., D. J. Thomans, M. A. Eberle, J. E. Swanson, R. J. Livingston et al., 2005. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 15: 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., M. J. Hubisz, C. D. Bustamante, S. H. Williamson and R. Nielsen, 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15: 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L., J. Novembre and S. Schneider, 2000. A general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J. Hered. 91: 506–510. [DOI] [PubMed] [Google Scholar]
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. DeLorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multilocus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P., B. Charlesworth, D. Halligan and P. Andolfatto, 2005. a Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 6: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P., K. Thornton, P. Andolfatto and B. Charlesworth, 2005. b Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D. L., A. Eyre-Walker, P. Andolfatto and P. D. Keightley, 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., M. Kauer and C. Schlotterer, 2002. Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99(20): 12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds, D. A., L. L. Stuve, G. B. Nilson, E. Halperin, E. Eskin et al., 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 1983. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. Estimation of levels of gene flow from DNA-sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J., Y. Kim, V. B. DuMont, C. Aquadro and C. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170: 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N. L., T. Darden and R. R. Hudson, 1988. The coalescent process in models with selection. Genetics 120: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M. O., B. Zangerl, D. Dieringer and C. Schlotterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M. O., D. Dieringer and C. Schlotterer, 2003. A microsatellite variability screen for positive selection associated with the “Out of Africa” habitat expansion of Drosophila melanogaster. Genetics 165: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, J. L., J. Madeoy, J. C. Calhoun, W. Swanson and J. M. Akey, 2006. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 16: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and R. Nielsen, 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval, G., and L. Excoffier, 2004. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 20: 2485–2487. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Nielsen, R., M. J. Hubisz and A. G. Clark, 2004. Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics 168: 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., S. Williamson, Y. Kim, M. J. Hubisz, A. G. Clark et al., 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto, L., S. Glinka, D. DeLorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22: 2119–2130. [DOI] [PubMed] [Google Scholar]
- Orengo, D., and M. Aguade, 2004. Detecting the footprint of positive selection in a European population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics 167: 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool, J. E., V. Bauer-DuMont, J. L. Mueller and C. F. Aquadro, 2006. A scan of molecular variation leads to the narrow localization of a selective sweep affecting both Afrotropical and cosmopolitan populations of Drosophila melanogaster. Genetics 172: 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. T. Seielstad, A. Perez-Lezaun and M. W. Feldman, 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Przeworski, M., G. Coop and J. D. Wall, 2005. Signature of positive selection on standing variation. Evolution 59: 2312–2323. [PubMed] [Google Scholar]
- Santiago, E., and A. Caballero, 2005. Variation after a selective sweep in a subdivided population. Genetics 169: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke, T., and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer, C., 2002. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics 160: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., T. Wiehe and M. W. Lenz, 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. [Google Scholar]
- Storz, J. F., B. A. Payseur and M. W. Nachman, 2004. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol. Biol. Evol. 21: 1800–1811. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., J. U'Ren, O. Tenaillon and B. S. Gaut, 2004. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21(7): 1214–1225. [DOI] [PubMed] [Google Scholar]
- Teshima, K. M., G. Coop and M. Przeworski, 2006. How reliable are empirical genome scans for selective sweeps? Genome Res. 16: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., 2003. libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19: 2325–2327. [DOI] [PubMed] [Google Scholar]
- Thornton, K., and P. Andolfatto, 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PloS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Williamson, S. H., R. Hernandez, A. Fledel-Alon, L. Zhu, R. Nielsen et al., 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA 102: 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., I. V. Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]