Abstract
The anaerobic, syntrophic bacterium Syntrophus aciditrophicus grown in pure culture produced 1.4 ± 0.24 mol of acetate and 0.16 ± 0.02 mol of cyclohexane carboxylate per mole of crotonate metabolized. [U-13C]crotonate was metabolized to [1,2-13C]acetate and [1,2,3,4,5,7-13C]cyclohexane carboxylate. Cultures grown with unlabeled crotonate and [13C]sodium bicarbonate formed [6-13C]cyclohexane carboxylate. Trimethylsilyl (TMS) derivatives of cyclohexane carboxylate, cyclohex-1-ene carboxylate, benzoate, pimelate, glutarate, 3-hydroxybutyrate, and acetoacetate were detected as intermediates by comparison of retention times and mass spectral profiles to authentic standards. With [U-13C]crotonate, the m/z-15 ion of TMS-derivatized glutarate, 3-hydroxybutyrate, and acetoacetate each increased by +4 mass units, and the m/z-15 ion of TMS-derivatized pimelate, cyclohex-1-ene carboxylate, benzoate, and cyclohexane carboxylate each increased by +6 mass units. With [13C]sodium bicarbonate and unlabeled crotonate, the m/z-15 ion of TMS derivatives of glutarate, pimelate, cyclohex-1-ene carboxylate, benzoate, and cyclohexane carboxylate each increased by +1 mass unit, suggesting that carboxylation occurred after the synthesis of a four-carbon intermediate. With [1,2-13C]acetate and unlabeled crotonate, the m/z-15 ion of TMS-derivatized 3-hydroxybutyrate, acetoacetate, and glutarate each increased by +0, +2, and +4 mass units, respectively, and the m/z-15 ion of TMS-derivatized pimelate, cyclohex-1-ene carboxylate, benzoate, cyclohexane carboxylate, and 2-hydroxycyclohexane carboxylate each increased by +0, +2, +4, and +6 mass units. The data are consistent with a pathway for cyclohexane carboxylate formation involving the condensation of two-carbon units derived from crotonate degradation with CO2 addition, rather than the use of the intact four-carbon skeleton of crotonate.
Syntrophus aciditrophicus strain SB is a gram-negative bacterium that belongs to the δ subdivision of the Proteobacteria (21). S. aciditrophicus metabolizes various saturated and unsaturated fatty acids, methyl esters of butyrate and hexanoate, and benzoate in coculture with hydrogen/formate-using microorganisms. The anaerobic degradation of saturated fatty acids and aromatic acids in the absence of terminal electron processes necessitates the presence of a hydrogen-consuming organism to maintain a hydrogen partial pressure low enough for these reactions to be thermodynamically favorable (9, 28, 29, 34). This metabolic interdependence has made the study of the syntrophic microorganisms difficult. However, a number of fatty acid- and aromatic acid-degrading syntrophic bacteria grow in pure culture by fermenting crotonate (6, 24, 46, 48). More recently, S. aciditrophicus was shown to grow in pure culture by benzoate fermentation (15).
Crotonate was shown to be dismutated to acetate and butyrate in some clostridial species (2, 42), Eubacterium oxidoreducens (23), and Ilyobacter polytropus (38) and in other syntrophic microorganisms, such as Syntrophomonas wolfei (6) and Syntrophus buswellii (46). Jackson et al. (21) reported that crotonate-grown pure cultures of S. aciditrophicus produced acetate, butyrate, caproate, and hydrogen. However, recently, no butyrate or caproate were detected by gas chromatography or high-pressure liquid chromatography (HPLC) as end products of crotonate fermentation.
In this paper, we report the results of gas chromatography-mass spectrometry (GC-MS) and 13C nuclear magnetic resonance spectroscopy (13C-NMR) to elucidate the pathway for crotonate oxidation and cyclohexane carboxylate formation in S. aciditrophicus.
MATERIALS AND METHODS
Media and cultivation conditions.
Pure cultures of Syntrophus aciditrophicus strain SB (ATCC 700169) were maintained in basal medium as described by McInerney et al. (27), without rumen fluid and with 20 mM of crotonate (6, 14). The medium and stock solution were prepared according to the anaerobic techniques described by Balch and Wolfe (3). The headspace was pressurized to 172 kPa with a mixture of N2-CO2 (80:20, vol/vol), and the cultures were incubated at 37°C without shaking. Cells were harvested at the mid-log phase of growth by centrifugation (14,300 × g, 20 min, 4°C). The cell pellet was washed twice with 50 mM phosphate buffer (pH 7.2) with resazurin (10 mg/liter), cysteine-HCl (0.5 g/liter), and Na2S (0.5 g/liter). The final pellet was resuspended in basal medium without rumen fluid lacking substrate and was used as the inoculum for the different experiments. The culture purity was checked daily by microscopic examination and inoculation of a thioglycolate medium known not to support S. aciditrophicus growth.
The molar growth yield was determined in triplicate 50-ml serum bottles containing basal medium without rumen fluid supplemented with 20 mM crotonate. One-milliliter samples were taken immediately after inoculation and at the mid-log phase of growth to determine the protein concentration. One-milliliter samples were taken daily from the same cultures to measure growth by monitoring changes in optical density at 600 nm (27) and to measure substrate depletion and product formation to determine the mass balance.
To determine the end products of crotonate fermentation, duplicate 500-ml cultures of S. aciditrophicus were grown with 20 mM crotonate. Samples were taken immediately after inoculation and at the mid-log phase for GC-MS analysis. A heat-killed control and a substrate-unamended control were also analyzed. To confirm that the metabolites were made from crotonate, S. aciditrophicus was grown in 150-ml cultures containing 5.2 mM [U-13C]crotonate or unlabeled crotonate. Samples (75 ml) were taken initially and when 94% of the original substrate was depleted. A substrate-unamended culture and a 100-ml heat-killed culture containing 3.5 mM [U-13C]crotonate were included as negative controls.
To study the formation of cyclohexane carboxylate, a 300-ml culture of S. aciditrophicus was grown with 3.8 mM [U-13C]crotonate and 3.8 mM crotonate. Samples (75 ml) for GC-MS and NMR analyses were taken at three time points: immediately after inoculation, when 80% of the initial substrate was used, and when 99% of the initial substrate was used. In another experiment, S. aciditrophicus was grown with 20 mM crotonate and 40 mM unlabeled sodium acetate or 40 mM [1,2-13C]acetate. Triplicate 50-ml cultures in serum bottles with unlabeled sodium acetate and crotonate were used to test for the effect of acetate addition on growth. To detect intermediates and to study the labeling pattern of cyclohexane carboxylate, 300-ml cultures were used. One set of cultures contained 20 mM crotonate with 40 mM sodium acetate, and another set contained 20 mM crotonate and 40 mM [1,2-13C]acetate. A 100-ml heat-killed culture containing 20 mM crotonate and 40 mM [1,2-13C]acetate was included as a negative control. Samples (50 ml) for GC-MS analysis were taken immediately after inoculation and after 1, 2, and 4 days of incubation time.
The involvement of a carboxylation step in cyclohexane carboxylate formation was tested by growing S. aciditrophicus with 10 mM crotonate and 41 mM [13C]sodium bicarbonate. Samples (50 ml) were taken for GC-MS and NMR analyses at three time points: initially, when 70% of the initial substrate was used, and when 95% of the initial substrate was used.
13C-NMR spectroscopy and GC-MS.
Samples were withdrawn at the time points mentioned above from S. aciditrophicus cultures that were grown with crotonate, [U-13C]crotonate, [13C]sodium bicarbonate with crotonate, and crotonate with [1,2-13C]acetate or nonlabeled sodium acetate and were analyzed by GC-MS. The samples were adjusted to a pH of 12 with 1 N NaOH to hydrolyze possible thioester bonds (14). Then, samples were acidified to a pH of <2 with 12 N HCl, extracted with ethyl acetate three times, and filtered through anhydrous sodium sulfate to remove water. The samples were concentrated under vacuum conditions at 47°C to a volume of 1 to 2 ml and split into two subsamples before they were evaporated to dryness under a stream of nitrogen gas. One subsample was used for GC-MS analysis, and the other was used for NMR analysis (except for the samples from the cultures grown with [1,2-13C]acetate and crotonate and the samples from the cultures grown on nonlabeled substrates). One subsample was resuspended in 200 μl ethyl acetate and derivatized with 100 μl of N,O-bis-(trimethylsilyl)trifluoroacetamide (BSTFA) for GC-MS analysis, and the other subsample was resuspended in 1 ml of deuterated chloroform for NMR analysis.
The derivatized samples were analyzed with an Agilent Technologies 6890N Network GC systems series gas chromatograph equipped with an Agilent Technologies 5973 network mass selective detector mass spectrometer and an HP-5MS capillary column (catalog no. 19091S-433; Agilent), 0.25 mm by 30 m by 0.25 μm (Wilmington, DE). The mass spectrometer was operated at 400 Hz. Helium was used as the carrier gas, with a flow of 1.2 ml/min. The oven temperature was held at 40°C for 5 min and then increased at a rate of 4°C/min until it reached a temperature of 250°C where it was held for 5 min. The metabolites were identified by comparison to the retention times and the mass spectra of BSTFA-derivatized chemical standards (14, 31).
Liquid 13C WALTZ-16 proton-decoupled NMR experiments were performed using 5-mm Wilmad glass NMR tubes on a Varian Mercury 300 MHz spectrometer. The one-dimensional spectra were obtained by using a 13C frequency of 75.45 MHz, a tip angle of 45.0o, a spectral width of 18,868 Hz, and an acquisition time of 1.815 s with a relaxation delay of 1.000 s. The 13C-13C correlation spectroscopy (COSY) spectra included 64 repetitions of 384 increments, using a spectral width of 18,868 Hz, an acquisition time of 0.217 s, and a relaxation delay of 1.000 s.
Analytical procedures.
The concentrations of crotonate, cyclohexane carboxylate, benzoate, cyclohex-1-ene carboxylate, and acetate were determined by HPLC with a Prevail organic acid column (250 by 4.6 mm; particle size 5 μm; Alltech Inc., Deerfield, IL). The isocratic mobile phase consisted of 25 mM KH2PO4 (pH 2.5) at a flow rate of 1 ml/min to measure acetate concentrations. To quantify crotonate, cyclohexane carboxylate, benzoate, and cyclohex-1-ene carboxylate, the mobile phase consisted of 60% (vol/vol) KH2PO4 (25 mM, pH 2.5) and 40% (vol/vol) acetonitrile. The UV absorbance detector was set at 210 nm to detect acetate and cyclohexane carboxylate and at 254 nm for crotonate, cyclohexane carboxylate, benzoate, and cyclohex-1-ene carboxylate.
Protein concentrations were determined by the method of Bradford (8) using a protocol and reagent from Pierce (Rockford, IL) with bovine serum albumin as the standard.
Thermodynamic calculations.
The ΔG°′ was calculated by using the ΔG° formation (ΔG°f) available from a study by Thauer et al. (43). The values of the ΔG°f of cyclohexane carboxylate and heptanoate were calculated by the group contribution method described by Mavrovouniotis (26).
Chemicals.
Stable isotopes of crotonic acid (U-13C, 99%), acetic acid (1,2-13C, 99%), sodium bicarbonate (13C, 99%), and chloroform-d (d, 99.8%) were obtained from Cambridge Isotope Laboratories, Inc. (Andover, MA). BSTFA was purchased from Pierce (Rockford, IL).
RESULTS
End products of crotonate fermentation.
Syntrophus aciditrophicus was grown in pure culture with crotonate with a specific growth rate of 0.025 ± 0.003 h−1. Approximately 6.6 ± 0.05 g of protein was obtained per mole of crotonate consumed. Assuming that 47% of the cell dry mass was protein (18), this corresponds to a cell yield of 14.0 ± 0.1 g dry weight per mole of crotonate. If the ATP yield is 10.5 g of biomass per mole of substrate (40), about 1.33 mol of ATP would be formed per mole of crotonate degraded.
Initial experiments recovered approximately 70% of the carbon and reducing equivalents as acetate from crotonate. Longer-chain fatty acids such as hexanoate or caproate were not detected by HPLC. A small amount of butyrate (0.112 mM) was detected by HPLC. However, its concentration did not change over the course of the experiment. To determine the fate of the missing crotonate carbon and reducing equivalents, cultures of S. aciditrophicus grown with crotonate were extracted, derivatized, and analyzed by gas chromatography-mass spectrometry. A large peak was detected in samples from crotonate-amended cultures and was not present in samples from the unamended or heat-killed controls. This peak had the same retention time (21.58 min) and the same mass fragmentation pattern as that of the TMS derivative of authentic cyclohexane carboxylate.
To confirm that cyclohexane carboxylate was made from crotonate, S. aciditrophicus was grown with 5.2 mM [U-13C]crotonate. The growth rate was similar to that observed with unlabeled crotonate (see above). Since a methyl group is easily lost from the TMS moiety of TMS-derivatized compounds, the total mass ion peak is often absent or very small and the m/z-15 ion is often used for identification purposes (31). The m/z-15 ion of the TMS-derivatized metabolite of the culture increased by +6 (185 to 191) mass units relative to the metabolite detected in the culture grown with unlabeled crotonate and relative to that of the TMS-derivatized authentic standard of cyclohexane carboxylate, showing that the metabolite was made from crotonate (Fig. 1 and Table 1). The major m/z ion fragments of the 13C-labeled metabolite were 58, 73, 87, 118, 132, 147, 161, 191, and 206, compared to 55, 73, 82, 117, 129, 145, 155, 185, and 200 for the TMS derivative of authentic cyclohexane carboxylate and to the peak detected for the culture fluid of cells grown with unlabeled crotonate (Fig. 1).
FIG. 1.
Mass spectrum profiles of the TMS-derivatized metabolite detected at the retention time of 21.58 min: (A) from a culture fluid of S. aciditrophicus grown with unlabeled crotonate; (B) from a culture fluid of S. aciditrophicus grown with [U-13C]crotonate; and (C) from a TMS-derivatized cyclohexane carboxylate authentic standard. The dashed arrows (panel C) represent the fission sites of the different fragments of cyclohexane carboxylate.
TABLE 1.
Comparison of the m/z-15 ions of TMS-derivatized metabolites to their corresponding authentic standardsa
| Metabolites | GC RTb (min) |
m/z-15 ion(s)
|
|||||
|---|---|---|---|---|---|---|---|
| Crotonate | [13C] crotonate | [13C]sodium bicarbonate | [13C]crotonate + crotonate | [13C]acetate + crotonate | Authentic standardc | ||
| Acetoacetate | 22.1 | 231 | 235 | 231 | 231, 233, 235 | 231, 233, 235 | 231 |
| 3-Hydroxybutyrate | 20.5 | 233 | 237 | 233 | 233, 235, 237 | 233, 235, 237 | 233 |
| Glutarate | 28.8 | 261 | 265 | 261, 262 | 261, 263, 265 | 261, 263, 265 | 261 |
| Pimelate | 34.8 | 289 | 295 | 289, 290 | 289, 291, 293, 295 | 289, 291, 293, 295 | 289 |
| Cyclohex-1-ene carboxylate | 24.4 | 183 | 189 | 183, 184 | 183, 185, 187, 189 | 183, 185, 187, 189 | 183 |
| Cyclohexane carboxylate | 21.6 | 185 | 191 | 185, 186 | 185, 187, 189, 191 | 185, 187, 189, 191 | 185 |
| Benzoate | 23.3 | 179 | 185 | 179, 180 | 179, 181, 183, 185 | 179, 181, 183, 185 | 179 |
| 2-Hydroxycyclohexane carboxylate | 30.0 | NDd | ND | ND | ND | 273, 275, 277, 279 | 273 |
The m/z-15 ions of TMS-derivatized metabolites were detected by GC-MS in culture fluids of Syntrophus aciditrophicus grown under different conditions. Metabolites were not detected in heat-killed or substrate-unamended controls.
Retention times (RT) of TMS-derivatized chemical authentic standards and the detected metabolites.
Chemical standards were commercially available.
ND, not detected.
13C-NMR analysis of the cell-free culture fluid of S. aciditrophicus grown with [U-13C]crotonate detected peaks that were consistent with the presence of cyclohexane carboxylate. Initially, the 13C-NMR spectrum had four peaks, at 18.0, 122.1, 147.7, and 171.5 ppm, which corresponded to the four carbons of [U-13C]crotonate (Table 2). When 94% of the substrate was depleted, the culture fluid was extracted and analyzed. The 13C-NMR spectrum had seven peak clusters, including the deuterated chloroform peak. The C-7 cluster corresponding to the carboxyl moiety of cyclohexane carboxylate was coupled to C-1 with a coupling constant of 55.2 Hz. The C-1 cluster corresponding to the peak at 43.0 ppm, a doublet of doublets, was further coupled to C-2 with a coupling constant of 33.2 Hz. The values of these coupling constants were in agreement with published values (11), and the correlations were further confirmed by off-diagonal peaks in the two-dimensional spectrum (data not shown). The integration of the one-dimensional spectrum showed that the partially resolved peaks near 25 ppm accounted for three carbons, C-3, C-4, and C-5, and for one carbon for each of the other peaks, C-1, C-2, and C-7. The coupling constants for C-3, C-4, and C-5 could not be resolved (39). No peak was detected for the C-6 position of the cyclohexane carboxylate. If the C-6 carbon was labeled and the cyclohexane carboxylate was fully labeled, the integration of the peak area at 29.0 ppm would indicate two carbons, corresponding to the C-2 position and the C-6 position, which was not observed. Moreover, the coupling pattern of C-1 would contain additional coupling resulting from the C-6 being 13C labeled. Since the C-6 was not 13C labeled, this indicates that a carboxylation step might be involved in the formation of cyclohexane carboxylate.
TABLE 2.
NMR data for compounds detected in cultures of Syntrophus aciditrophicus grown in pure culture with [U-13C]crotonate or with crotonate with [13C]sodium bicarbonate
| Growth condition and sampling time point | Compound detected | Position | δ 13C (ppm)
|
Jcc (Hz)c | |
|---|---|---|---|---|---|
| Expecteda | Observed | ||||
| [U-13C]crotonate at initial time point | Crotonate | C-1 | 172.4 | 171.4 | 77.7 (1-2) |
| C-2 | 122.4 | 122.1 | 77.7 (2-1) | ||
| 69.9 (2-3) | |||||
| 7.0 (2-4) | |||||
| C-3 | 147.6 | 147.7 | 69.9 (3-2) | ||
| 42.0 (3-4) | |||||
| C-4 | 18.1 | 18.0 | |||
| [U-13C]crotonate at final time pointb | Cyclohexane carboxylate | C-7 | 181.0 | 182.4 | 55.2 (7-1) |
| Ring C-1 | 41.5 | 43.0 | 33.2 (1-2) | ||
| 55.2 (1-7) | |||||
| Ring C-2 | 26.6 | 29.0 | 33.2 (2-1, 2-3) | ||
| Ring C-6 | 26.6 | Not observed | |||
| Ring C-4 | 27.1 | 25.0 | Unresolved peaks | ||
| Rings C-3 and C-5 | 24.3 | 25.0 | Unresolved peaks | ||
| Acetate | Carboxyl | 177.0 | 177.4 | 56.7 (1-2) | |
| Methyl | 20.7 | 21.0 | 56.7 (2-1) | ||
| [13C]sodium bicarbonate at final time point | Cyclohexane carboxylate | C-6 | 29.0 | 28.9 | |
Predicted chemical shifts (δ 13C) were obtained from ChemNMR (Upstream Solutions).
The culture was analyzed when 94% of the crotonate was degraded.
Jcc, coupling constants.
To test this, S. aciditrophicus was grown with unlabeled crotonate and [13C]sodium bicarbonate. The mass of the m/z-15 ion of the TMS derivative of cyclohexane carboxylate increased by +1 mass unit, from 185 to 186, relative to that of the TMS derivative of the authentic standard (Table 1). Moreover, the 13C-NMR spectrum of the culture fluid displayed a single, uncoupled peak at 28.9 ppm, which corresponded to the C-6 position of cyclohexane carboxylate (Table 2).
In addition to cyclohexane carboxylate, acetate was the only other major product detected when S. aciditrophicus was grown on [U-13C]crotonate (Table 2). The 13C-NMR spectrum contained two peaks, at 177.4 ppm and 21.0 ppm, which correspond to the carboxyl carbon and the methyl carbon of acetate, respectively. 13C-labeled metabolites were not detected in heat-killed or unamended controls. Except for acetate and cyclohexane carboxylate, no other compound was detected by 13C-NMR analysis.
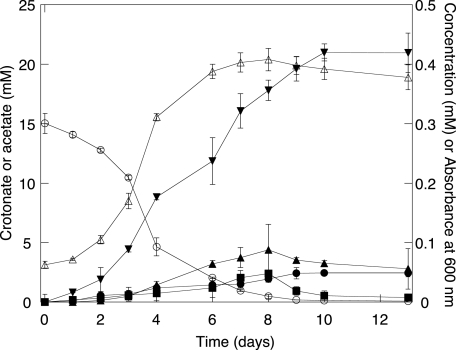
Now that the identities of the products of crotonate metabolism were known, the time course of crotonate metabolism and the carbon balance was determined by HPLC (Fig. 2). The pure culture reached a maximum absorbance of 0.41 within 8 days of incubation at 37°C. S. aciditrophicus metabolized crotonate to 1.41 ± 0.24 mol of acetate and 0.16 ± 0.02 mol of cyclohexane carboxylate per mole of crotonate degraded, giving a 99.4% carbon recovery and a 95.8% hydrogen recovery. The above values are consistent with the following stoichiometry for crotonate fermentation: 6C4H5O2− + HCO3− + 5H2O → 9C2H3O2− + C7H11O2− + 3H+.
FIG. 2.
Metabolism of crotonate by S. aciditrophicus pure cultures. ○, crotonate; ▾, acetate; □, cyclohexane carboxylate; •, cyclohex-1-ene carboxylate; ▪, benzoate; ▵, absorbance at 600 nm. The data are averages ± standard deviations of triplicate microcosms.
Metabolites detected during crotonate degradation.
During the growth of S. aciditrophicus with crotonate, GC-MS analyses of culture samples detected the presence of TMS derivatives of cyclohex-1-ene carboxylate, benzoate, pimelate, glutarate, 3-hydroxybutyrate, and acetoacetate, in addition to cyclohexane carboxylate. Each metabolite eluted at the same retention time and mass spectral profile as the TMS derivative of its respective authentic standard. None of these compounds was detected in substrate-unamended or heat-killed controls. To confirm that these metabolites were made from crotonate, samples from cultures grown with [U-13C]crotonate were analyzed. The m/z-15 ion of TMS derivatives of the metabolites corresponding to cyclohexane carboxylate, cyclohex-1-ene carboxylate, pimelate, and benzoate increased by +6 mass units, while those corresponding to glutarate, 3-hydroxybutyrate, and acetoacetate increased by +4 mass units relative to their respective authentic TMS-derivatized standards, confirming that these compounds are metabolites of crotonate degradation (Table 1).
3-Hydroxybutyrate and acetoacetate were detected only after alkaline hydrolysis of cells. Neither compound was detected when the samples were filtered to remove cells. This suggests that the above-mentioned metabolites were intracellular and present as their CoA derivatives. However, pimelate, glutarate, cyclohex-1-ene carboxylate, benzoate, and cyclohexane carboxylate were detected whether or not the cells were alkaline hydrolyzed and whether or not the sample was filtered to remove cells. These data imply that the latter compounds were excreted into the medium as their free acids.
The time course experiment showed that cyclohex-1-ene carboxylate and benzoate were transiently produced and degraded (Fig. 2). Each was detected at much lower concentrations (maximum concentration of 0.088 and 0.048 mM, respectively) than acetate and cyclohexane carboxylate were. Pimelate, glutarate, 3-hydroxybutyrate, and acetoacetate were not detected by HPLC (concentrations <0.05 mM). Nevertheless, GC-MS analysis showed that the peak areas corresponding to each compound increased and decreased during growth, supporting their roles as intermediates.
Pathway for cyclohexane carboxylate formation.
To resolve the major steps in the formation of cyclohexane carboxylate, we performed a series of experiments in which 13C-labeled compounds were used. In order to determine when the carboxylation step occurred, S. aciditrophicus was grown in basal medium with 20 mM of unlabeled crotonate and 41 mM of [13C]sodium bicarbonate. The m/z-15 ions of the TMS derivatives of cyclohexane carboxylate, benzoate, cyclohex-1-ene carboxylate, pimelate, and glutarate increased by +1 mass unit relative to the respective TMS-derivatized standards (Table 1). However, peaks corresponding to a potential three-carbon metabolite such as the TMS derivative of malonate were not detected by GC-MS. The m/z-15 ions of the TMS derivatives of 3-hydroxybutyrate and acetoacetate remained unchanged. These results are consistent with the carboxylation of a four-carbon intermediate.
To determine whether cyclohexane carboxylate is formed by the condensation of a four-carbon unit derived from crotonate without a carbon-carbon bond cleavage and a two-carbon unit or by the condensation of three two-carbon units, S. aciditrophicus was grown with equimolar concentrations of [U-13C]crotonate and nonlabeled crotonate as substrates. In the first model, part of the [U-13C]-labeled and nonlabeled crotonate is oxidized to form a mixture of [1,2-13C]acetate and nonlabeled acetate. If we assume that a four-carbon skeleton remains intact to form cyclohexane carboxylate and that no isotopic discrimination occurs, four possible labeling patterns to form cyclohexane carboxylate are possible. The U-13C four-carbon intermediate could condense with a U-13C-labeled or a nonlabeled two-carbon intermediate to give isotopomers of cyclohexane carboxylate with an increase of +6 or +4 mass units, respectively. The nonlabeled four-carbon intermediate could condense with a U-13C-labeled or a nonlabeled two-carbon intermediate to give isotopomers with an increase of +2 or +0 mass units, respectively. Each isotopomer occurs with the same probability. In the second model, a mixture of [1,2-13C]acetate and nonlabeled acetate is formed, and eight possible labeling patterns for cyclohexane carboxylate formation are possible, resulting in isotopomers with a m/z-15 ion increase of +0, +2, +4, and +6 mass units. The probabilities of having the m/z-15 ion of the TMS-derivatized cyclohexane carboxylate increase by +2 or +4 mass units would each be 0.375 and the probabilities of increasing by +0 or +6 mass units would each be 0.125, assuming that no isotopic discrimination occurred.
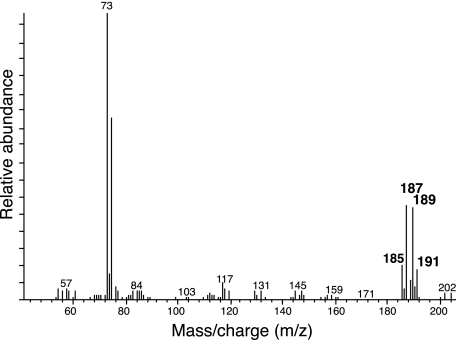
The mass spectrum profile of the TMS-derivatized cyclohexane carboxylate formed when S. aciditrophicus grown with equal amounts of labeled and unlabeled crotonate showed a mixture of differently labeled cyclohexane carboxylate, with m/z-15 ion mass increases of +0, +2, +4, and +6 mass units (Table 1). The relative abundance of those with the ion mass increases of +2 and +4 mass units (m/z-15 ions of 187 and 189) is around 2.9 times higher than those with mass increases of +0 or +6 units (m/z-15 ions of 185 and 191) (Fig. 3). These data are consistent with the model in which cyclohexane carboxylate formation occurs by the condensation of three two-carbon units.
FIG. 3.
Mass spectrum profile of the TMS-derivatized metabolite detected at a retention time of 21.58 min from cultures of S. aciditrophicus grown on equimolar mixtures of [U-13C]crotonate and unlabeled crotonate.
Further evidence to support the hypothesis that the formation of cyclohexane carboxylate occurs by the condensation of three two-carbon units was obtained by growing S. aciditrophicus in the presence of [1,2-13C]acetate and crotonate. Acetate does not support S. aciditrophicus growth in the absence of crotonate. The presence of 40 mM sodium acetate with 20 mM crotonate in the medium partially inhibited the growth of S. aciditrophicus compared to that with 20 mM crotonate alone. The growth rate was 0.006 ± 0.002 h−1 in the presence of 40 mM sodium acetate compared to 0.025 ± 0.003 h−1 without sodium acetate (data not shown). When S. aciditrophicus was grown with 40 mM [1,2-13C]acetate and 20 mM crotonate, samples of culture fluids were taken at different time points, derivatized, and submitted for GC-MS analysis. The metabolites detected by GC-MS when S. aciditrophicus was grown under these conditions were those detected previously (e.g., the TMS derivatives of cyclohexane carboxylate, cyclohex-1-ene carboxylate, benzoate, pimelate, glutarate, 3-hydroxybutyrate, and acetoacetate), along with one additional metabolite, the TMS derivative of 2-hydroxycyclohexane carboxylate. Multiple m/z-15 ions were detected for each metabolite. The m/z-15 ions of the TMS derivatives of 3-hydroxybutyrate, acetoacetate, and glutarate each showed increases of +0, +2, and +4 mass units relative to the TMS derivatives of their respective authentic standards. The m/z-15 ions of TMS derivatives of pimelate, benzoate, cyclohex-1-ene carboxylate, 2-hydroxycyclohexane carboxylate, and cyclohexane carboxylate each showed increases of +0, +2, +4, and +6 mass units relative to the TMS derivatives of their respective authentic standards (Table 1). The cyclohexane carboxylate formed in cultures after 24 h of incubation had a mass spectrum profile with a mixture of m/z-15 ions of 185, 187, 189, and 191 corresponding to a mass increase of +0, +2, +4, and +6 units, respectively. The relative abundance of the 191 ion was higher than the other ions. To form this ion, cyclohexane carboxylate must have been formed by the condensation of three molecules of [1,2-13C]acetate. Over the course of the experiment, the relative abundance of the different m/z-15 ions changed, and at the end of the experiment, the m/z-15 ion with a mass of 185 increased in abundance. This was probably due to the utilization of nascent, nonlabeled acetate derived from crotonate oxidation for cyclohexane carboxylate formation. These data confirm that cyclohexane carboxylate is formed from the condensation of two-carbon units.
DISCUSSION
The conversion of crotonate to acetate and cyclohexane carboxylate by a microorganism has never been reported before. A number of anaerobes including syntrophic bacteria are known to dismutate crotonate to 1 mol of acetate and 0.5 mol of butyrate (2, 6, 10, 24, 32, 42, 48, 49). Some organisms will also form caproate in addition to butyrate. Jackson et al. (21) reported that S. aciditrophicus degraded 1 mol of crotonate to 0.1 mol of butyrate, 0.13 mol of caproate, and 1.4 mol of acetate, which differs from the results presented here. This discrepancy may be due to variations in the growth conditions of S. aciditrophicus. However, since S. aciditrophicus has been repeatedly transferred in liquid culture for a number of years, it is possible that a genetic change occurred that resulted in this change in physiology.
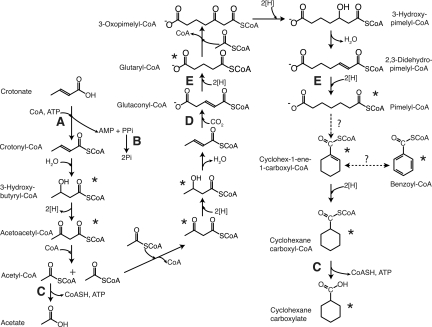
Our results obtained from GC-MS, 13C-NMR, and HPLC analyses demonstrated the synthesis of a cyclic molecule from a straight-chain unsaturated fatty acid (Fig. 4). Approximately two-thirds of the crotonate present is degraded to acetate. The rest of the crotonate (probably as acetyl-coenzyme A [CoA]) is used to form cyclohexane carboxylate and reoxidize the reducing equivalents generated during crotonate oxidation to acetate. The labeling patterns observed in cyclohexane carboxylate when S. aciditrophicus was grown with [1,2-13C]acetate or [U-13C]crotonate in the presence of nonlabeled crotonate are consistent with the synthesis of cyclohexane carboxylate by the condensation of two-carbon units derived from crotonate rather than the use of the intact four-carbon skeleton of crotonate (Table 1; Fig. 3). Thus, cyclohexane carboxylate formation probably serves a function analogous to the formation of butyrate and caproate by Clostridium kluyveri (4, 37, 42). In the case of C. kluyveri, the reducing equivalents generated during ethanol oxidation to acetate are used to reduce two or three equivalents of acetate to form butyrate or caproate.
FIG. 4.
Proposed pathway for the formation of acetate and cyclohexane carboxylate from crotonate by S. aciditrophicus. Asterisks indicate 13C-labeled intermediates that were detected. Letters A through E indicate potential sites of energy conservation or use as discussed in the text.
The carboxylation step probably occurs after the four-carbon intermediate is made, since the mass units of TMS derivatives of glutarate, pimelate, and cyclohex-1-ene carboxylate each increase by 1 unit relative to their respective TMS-derivatized standards (Table 1) when S. aciditrophicus was grown with [13C]sodium bicarbonate. In the biosynthesis of membrane lipids, the fatty acid elongation step involves the use of malonyl-acyl carrier protein (ACP), which is decarboxylated during the transfer of the acetyl group to the growing fatty acid molecule (25). One possible pathway for cyclohexane carboxylate formation would be the synthesis of a five-carbon intermediate by the condensation of malonyl-CoA and acetyl-CoA (or as their ACP derivatives). However, no labeled malonyl-CoA was detected when S. aciditrophicus was grown in the presence of unlabeled crotonate and [13C]sodium bicarbonate. Also, the m/z-15 ions of the TMS derivatives of 3-hydroxybutyrate and acetoacetate increased by +0, +2, and +4 mass units relative to the TMS derivatives of their respective standards when S. aciditrophicus was grown in the presence of [1,2-13C]acetate (Table 1), consistent with their formation from two-carbon intermediates (Fig. 4).
GC-MS analysis allowed the identification of acetoacetate, 3-hydroxybutyrate, glutarate, pimelate, 2-hydroxycyclohexane carboxylate, and cyclohex-1-ene carboxylate as intermediates of cyclohexane carboxylate formation from crotonate. With the exception of the TMS derivatives of 3-hydroxybutyrate and acetoacetate, all of the above-mentioned compounds were detected in low amounts in cell-free culture broth. The GC peak areas increased and then subsequently decreased during growth, consistent with their roles as intermediates in crotonate metabolism. Given that the enzymes involved in benzoate and cyclohexane carboxylate metabolism in S. aciditrophicus and Rhodopseudomonas palustris (14, 19) use CoA derivatives as their substrates, it is likely that cyclohexane carboxylate formation does as well.
The above intermediates are the same as those detected during syntrophic (14) and fermentative benzoate metabolism (15) by S. aciditrophicus, suggesting that cyclohexane carboxylate formation may involve the same pathway as that used for benzoate degradation. However, analysis of the genome sequence of S. aciditrophicus (GenBank accession no. CP000252) (McInerney et al., submitted for publication; 33) suggests that benzoate is degraded to 3-hydroxypimelyl-CoA by a pathway involving cyclohex-1,5-diene-1-carboxyl-CoA, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA, and 6-oxocyclohex-1-ene-1-carboxyl-CoA as intermediates rather than those shown in Fig. 4. The S. aciditrophicus genome contains three genes (SYN1653 to SYN1655) whose gene products have high sequence similarities (>47% identity) to cyclohex-1,5-diene-1-carboxyl-CoA hydratase, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase, and 6-oxocyclohex-1-ene-1-carboxyl-CoA hydrolase of Azoarcus strain CIB (5). S. aciditrophicus may use separate pathways to degrade cyclohexane carboxylate and benzoate to 3-hydroxypimelyl-CoA. Cyclohexane carboxylate formation (Fig. 4) may occur by the reversal of the pathway used to degrade cyclohexane carboxylate, which is similar to the pathway used by R. palustris to degrade benzoate and cyclohexane carboxylate (19). The pathway in each organism has several identical intermediates, and cell extracts of syntrophically grown S. aciditrophicus contain the enzyme activities for the conversion of cyclohex-1-ene carboxyl-CoA to pimelyl-CoA (14). Interestingly, genes homologous to those involved in benzoate and cyclohexane carboxylate degradation in R. palustris (13, 19) were not detected in the S. aciditrophicus genome (<30% amino acid sequence identity). The unfavorable carboxylation of crotonyl-CoA to glutaconyl-CoA could be driven by a sodium gradient, since membrane-bound decarboxylation enzymes have been shown to be reversible (12, 36).
There are several possible mechanisms for the formation of the alicyclic ring of cyclohexane carboxylate. The ring formation from short-chain fatty acids is already well established for polyketide biosynthesis (20). Here, the correct alignment of carbonyl and methylene groups of poly-β-ketone intermediates leads to cyclization. The formation of cyclohexane carboxylate may be analogous to the synthesis of 1,4-dihydroxy-2-naphthoyl-CoA from O-succinylbenzoyl-CoA (OSB) by 1,4-dihydroxy-2-naphthoyl-CoA synthase (MenB) in menaquinone biosynthesis (44). This is essentially the back reaction of 2-oxocyclohexane-1-carboxyl-CoA hydrolase in R. palustris (30), and both enzymes have conserved Asp and Tyr at the active site (44). As mentioned above, a gene homologous to the R. palustris gene was not detected in the S. aciditrophicus genome.
The transient accumulation of benzoate (probably as its CoA derivative) from crotonate is surprising. However, previous studies have shown the formation of aromatic compounds from alicyclic compounds such as cyclohexane carboxylate or 4-oxocyclohexane carboxylate in animals and microorganisms. Enzymes purified from the liver of guinea pigs can convert cyclohexane carboxyl-CoA to benzoyl-CoA with cyclohex-1-ene carboxylate as an intermediate (1). Cyclohexane carboxylate can be converted to p-hydroxybenzoate aerobically by Arthrobacter sp. (7) or Alcaligenes strain W1 (41). Corynebacterium cyclohexanicum is able to convert 4-oxocyclohexane carboxylate to 4-hydroxybenzoate (22). The S. aciditrophicus genome contains two gene clusters with high similarities (>50% identity at the amino acid level) to the benzoate-induced gene cluster in Geobacter metallireducens (47) but lacks two of the four genes needed to encode the ATP-dependent benzoyl-CoA reductase found in denitrifiers and anaerobic phototrophs (19). The presence of two putative benzoyl-CoA reductase systems is consistent with the benzoate dismutation hypothesis (15) where one reductase functions in a pathway that oxidizes benzoate to acetate and carbon dioxide and the other reductase functions in a pathway that forms cyclohexane carboxylate.
Thermodynamic considerations show that the formation of cyclohexane carboxylate and acetate from crotonate is favorable, with a standard free energy of −48.1 kJ/mol (Table 3). The free-energy change for cyclohexane carboxylate formation is similar to the standard free-energy changes for butyrate, caproate, or heptanoate formation from crotonate fermentation (Table 3). The molar growth yield suggests that about 1.33 mol of ATP are made per mole of crotonate. The formation of 1.41 mol of acetate and 0.16 mol of cyclohexane carboxylate per mole of crotonate could result in the formation of 1.57 mol of ATP by substrate-level phosphorylation reactions involving the CoA derivatives of these compounds (Fig. 4, reaction C). However, energy must be used to activate crotonate to crotonyl-CoA. No acetyl-CoA-crotonyl-CoA transferase activity was detected when S. aciditrophicus was grown with crotonate (14), suggesting that a CoA ligase reaction is used (Fig. 4, reaction A). Crotonyl-CoA ligase activity has been detected in crotonate-grown cells (N. Q. Wofford and M. J. McInerney, unpublished data). This reaction forms AMP and pyrophosphate in addition to crotonyl-CoA (16, 17) and would consume 2 ATP equivalents if pyrophosphate is hydrolyzed. These calculations suggest that S. aciditrophicus must also use the energy in ion gradients for growth. Energy could be conserved by a membrane-bound, proton-translocating pyrophosphatase as shown in Syntrophus gentianae (35) (Fig. 4, reaction B). In addition, the redox reactions leading to the formation of cyclohexane carboxylate may be energy-yielding reactions. During benzoate oxidation to acetate, the production of hydrogen (E°′ of −414 mV) from high-redox electrons derived during pimelyl-CoA and glutaryl-CoA oxidation (E°′ of −10 mV) is coupled to a reverse electron transport and probably consumes the equivalent of two-thirds of an ATP (36, 45). Since glutarate and pimelate were detected as intermediates, the redox reactions involved in the formation of their respective CoA substrates may be energy yielding as well (Fig. 4, reaction E).
TABLE 3.
Standard free-energy changes for various crotonate fermentations
| Equation no. | End product | Equation | ΔG°′ (kJ/mol)a |
|---|---|---|---|
| 1 | Butyrate | 2C4H5O2− + 2H2O → 2C2H3O2− + C4H7O2− + H+ | −51.1 |
| 2 | Caproate | 4C4H5O2− + 4H2O → 5C2H3O2− + C6H11O2− + 2H+ | −51.1 |
| 3 | Heptanoate | 7C4H5O2− + HCO3− + 7H2O → 11C2H3O2− + C7H13O2− + 4H+ | −52.1 |
| 4 | Cyclohexane carboxylate | 6C4H5O2− + HCO3− + 5H2O → 9C2H3O2− + C7H11O2− + 3H+ | −48.1 |
Acknowledgments
We thank N. Q. Wofford for technical assistance.
This work was supported by contract DE-FG02-96ER20214 from the U.S. Department of Energy.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Babior, B. M., and K. Bloch. 1966. Aromatization of cyclohexanecarboxylic acid. J. Biol. Chem. 241:3643-3651. [PubMed] [Google Scholar]
- 2.Bader, J., H. Gunther, E. Schleicher, H. Simon, S. Pohl, and W. Mannheim. 1980. Utilization of (E)-2-butenoate (crotonate) by Clostridium kluyveri and some other Clostridium species. Arch. Microbiol. 125:159-165. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, H. A. 1956. Bacterial fermentation, p. 95. John Wiley Inc., New York, NY.
- 5.Barragán, M. J. L., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaty, P. S., and M. J. McInerney. 1987. Growth of Syntrophomonas wolfei in pure culture on crotonate. Arch. Microbiol. 147:389-393. [Google Scholar]
- 7.Blakley, E. R. 1974. The microbial degradation of cyclohexane carboxylate: a pathway involving aromatization to form p-hydroxybenzoate. Can. J. Microbiol. 20:1297-1306. [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Bryant, M. P. 1976. The microbiology of anaerobic digestion and methanogenesis with special reference to sewage, p. 107-118. In H. G. Schlegel and J. Barnes (ed.), Microbial energy conversion. Verlag Erich Goltze KG, Göttingen, Germany.
- 10.Cao, X., X. Liu, and X. Dong. 2003. Alkaliphilus crotonatoxidans sp. nov., strictly anaerobic, crotonate-dismutating bacterium, isolated from a methanogenic environment. Int. J. Syst. Evol. Microbiol. 53:971-975. [DOI] [PubMed] [Google Scholar]
- 11.Dean, J. A. 1999. Lange's handbook of chemistry, vol. 7, p. 7.108-7.109. McGraw-Hill, New York, NY. [Google Scholar]
- 12.Dimroth, P., and W. Hilpert. 1984. Carboxylation of pyruvate and acetyl coenzyme A by reversal of the Na+ pumps oxaloacetate decarboxylase and methylmalonyl-CoA decarboxylase. Biochemistry 23:5360-5366. [Google Scholar]
- 13.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elshahed, M. S., V. K. Bhupathiraju, N. Q. Wofford, M. A. Nanny, and M. J. McInerney. 2001. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” strain SB in syntrophic association with H2-using microorganisms. Appl. Environ. Microbiol. 67:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elshahed, M. S., and M. J. McInerney. 2001. Benzoate fermentation by the anaerobic bacterium Syntrophus aciditrophicus in the absence of hydrogen-using microorganisms. Appl. Environ. Microbiol. 67:5520-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, G., M. E. S. Mohamed, U. Altenschmidt, J. Koch, A. Lack, R. Brackmann, C. Lochmeyer, and B. Oswald. 1994. Biochemistry of anaerobic biodegradation of aromatic compounds, p. 513-553. In C. Ratledge (ed.), Biochemistry of microbial degradation. Kluwer, Dordrecht, The Netherlands.
- 17.Geissler, J. F., C. S. Harwood, and J. Gibson. 1988. Purification and properties of benzoate-coenzyme A ligase: a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J. Bacteriol. 170:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg. 1994. Methods for general and molecular bacteriology, p. 227-248. American Society for Microbiology Press, Washington, DC.
- 19.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 20.Hutchinson, C. R., and I. Fuji. 1995. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu. Rev. Microbiol. 49:201-238. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, B. E., V. K. Bhupathiraju, R. S. Tanner, C. R. Woese, and M. J. McInerney. 1999. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch. Microbiol. 171:107-114. [DOI] [PubMed] [Google Scholar]
- 22.Kaneda, T., H. Obata, and M. Tokumoto. 1993. Aromatization of 4-oxocyclohexanecarboxylic acid to 4-hydroxybenzoate by two distinctive desaturases from Corynebacterium cyclohexanum. Eur. J. Biochem. 218:997-1003. [DOI] [PubMed] [Google Scholar]
- 23.Krumholz, L. R., and M. P. Bryant. 1986. Eubacterium oxidoreducens sp. nov., requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol, and guercetin. Arch. Microbiol. 144:8-14. [Google Scholar]
- 24.Liu, Y., D. L. Balkwill, H. C. Aldrich, G. R. Drake, and D. R. Boone. 1999. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov., and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 49:545-556. [DOI] [PubMed] [Google Scholar]
- 25.Magnuson, K., S. Jackowski, C. O. Rock, and J. E. Cronan, Jr. 1993. Regulation of fatty acids biosynthesis in Escherichia coli. Microbiol. Rev. 57:522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrovouniotis, M. L. 1990. Group contributions for estimating standard Gibbs energies of formation of biochemical compounds in aqueous solution. Biotechnol. Bioeng. 36:1070-1082. [DOI] [PubMed] [Google Scholar]
- 27.McInerney, M. J., M. P. Bryant, and N. Pfennig. 1979. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch. Microbiol. 122:129-135. [DOI] [PubMed] [Google Scholar]
- 28.McInerney, M. J., and M. P. Bryant. 1980. Metabolic stages and energetics of microbial anaerobic digestion, p. 91-98. In D. Stafford, B. Wheatley, and D. Hughes (ed.), Anaerobic digestion. Applied Science Publishers, London, United Kingdom.
- 29.McInerney, M. J. 1988. Anaerobic degradation of proteins and lipids, p. 373-415. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, Inc., New York, NY.
- 30.Pelletier, D. A., and C. S. Harwood. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 180:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters, F., Y. Shinoda, M. J. McInerney, and M. Boll. 22 November 2006. Cyclohexa-1,5-diene-1-carbonyl-coenzyme A hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: evidence for a common benzoyl-CoA degradation pathway in facultative and strict anaerobes. J. Bacteriol. doi: 10.1128/JB.01467-06. [DOI] [PMC free article] [PubMed]
- 32.Pierce, A. E. 1968. Silylation of organic compounds, p. 33-39. Pierce Chemical Co., Rockford, IL.
- 33.Qiu, Y. L., Y. Sekiguchi, H. Imachi, Y. Kamagata, I. C. Tseng, S. S. Cheng, A. Ohashi, and H. Harada. 2003. Sporotomaculum syntrophicum sp. nov., a novel anaerobic, syntrophic benzoate-degrading bacterium isolated from methanogenic sludge treating wastewater from terephthalate manufacturing. Arch. Microbiol. 179:242-249. (Erratum, 179:457.) [DOI] [PubMed] [Google Scholar]
- 34.Schink, B. 1992. Syntrophism among prokaryotes, p. 276-299. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, a handbook of the biology of bacteria: ecophysiology, isolation, identification, and applications, 2nd ed. Springer, Berlin, Germany.
- 35.Schöcke, L., and B. Schink. 1998. Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium. Eur. J. Biochem. 256:589-594. [DOI] [PubMed] [Google Scholar]
- 36.Schöcke, L., and B. Schink. 1999. Energetics and biochemistry of fermentative benzoate degradation by Syntrophus gentianae. Arch. Microbiol. 171:331-337. [DOI] [PubMed] [Google Scholar]
- 37.Smith, G. M., B. W. Kim, A. A. Franke, and J. D. Roberts. 1985. 13C-NMR studies of butyric fermentation in Clostridium kluyveri. J. Biol. Chem. 260:13509-13512. [PubMed] [Google Scholar]
- 38.Stieb, M., and B. Schink. 1984. A new 3-hydroxybutyric acid-fermenting anaerobe, Ilyobacter polytropus, gen. nov. sp. nov., possessing various fermentation pathways. Arch. Microbiol. 140:387-390. [Google Scholar]
- 39.Stoecker, M. 1981. Carbon-13-carbon-13 coupling constants of cycloalkane carboxylic acids. J. Chem. Res. Synop. 1981:52-53. [Google Scholar]
- 40.Stouthamer, A. H., and C. Bettenhaussen. 1973. Utilization of energy for growth and maintenance in continuous and batch cultures of microorganisms. A reevaluation of the method for the determination of ATP production by measuring molar growth yields. Biochim. Biophys. Acta 301:53-70. [DOI] [PubMed] [Google Scholar]
- 41.Taylor, D. G., and P. W. Trudgill. 1978. Metabolism of cyclohexane carboxylic acid by Alcaligenes strain W1. J. Bacteriol. 134:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thauer, R. K., K. Jungermann, H. Henninger, J. Wenning, and K. Decker. 1968. The energy metabolism of Clostridium kluyveri. Eur. J. Biochem. 4:173-180. [DOI] [PubMed] [Google Scholar]
- 43.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truglio, J. T., K. Theis, Y. Feng, R. Gajda, C. Machutta, P. J. Tonge, and C. Kisker. 2003. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J. Biol. Chem. 278:42352-42360. [DOI] [PubMed] [Google Scholar]
- 45.Wallrabenstein, C. N., and B. Schink. 1994. Evidence of reverse electron transport in syntrophic butyrate or benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii. Arch. Microbiol. 162:136-142. [Google Scholar]
- 46.Wallrabenstein, C. N., N. Gorny, N. Springer, W. Ludwig, and B. Schink. 1995. Pure cultures of Syntrophus buswellii, definition of its phylogenic status, and description of Syntrophus gentianae sp. nov. Syst. Appl. Microbiol. 18:62-66. [Google Scholar]
- 47.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. Van Dorsselaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238-1252. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, C., X. Liu, and X. Dong. 2005. Syntrophomonas erecta sp. nov., a novel anaerobe that syntrophically degrades short-chain fatty acids. Int. J. Syst. Evol. Microbiol. 55:799-803. [DOI] [PubMed] [Google Scholar]
- 49.Zou, B. Z., K. Takeda, A. Tonouchi, S. Akada, and T. Fujita. 2003. Characteristics of an anaerobic syntrophic butyric acid-degrading bacterium in paddy field soil. Biosci. Biotechnol. Biochem. 67:2059-2067. [DOI] [PubMed] [Google Scholar]