Abstract
A comparative global proteomic screen identified factors required for COMPASS (complex of proteins associated with Set1)-mediated mono-, di-, and trimethylation of the fourth lysine of histone H3 (H3K4), which included components of a cyclin-dependent protein kinase (Ctk complex) that phosphorylates the C-terminal domain of the largest subunit of RNA polymerase II (Pol II). Our results indicate that histone H3K4 methylation levels are regulated by the Ctk1, Ctk2, and Ctk3 components of the Ctk complex. We show that loss of Ctk1 kinase activity results in reduced histone H3K4 monomethylation levels, followed by a global increase in histone H3K4 trimethylation levels on chromatin. Ctk1 loss does not appear to have a substantial effect on histone H2B monoubiquitination levels or COMPASS and Paf1 complex phosphorylation. Our chromatin immunoprecipitation studies demonstrate that histone H3 eviction during active transcription is decelerated in a CTK1 deletion strain in response to reduced levels of Pol II recruitment. Our in vitro studies show that the onset of monomethylation on an unmethylated histone H3 by COMPASS is virtually immediate, while the onset of trimethylation occurs upon extended time of association between the histone tail and COMPASS. Our study suggests a role for the Ctk complex in the regulation of the pattern of H3K4 mono-, di-, and trimethylation via COMPASS.
Transcriptional regulation by RNA polymerase II (Pol II) is a complex multistage process requiring the concerted action of many factors for proper synthesis of mRNA (44-46). Modification of histone tails within chromatin can affect transcriptional activation and repression, as well as the kinetic properties of transcriptional initiation and elongation (2, 8, 18, 20, 24, 44). Following proper transcriptional initiation, a host of histone-modifying enzymes associate with elongating Pol II as it travels from the promoter to the 3′ end of a gene (16, 17).
During active transcription, the histone H3K4 methyltransferase COMPASS (complex of proteins associated with Set1) associates with the elongation factor Paf1 complex (Paf1C) to interact with Pol II and chromatin (26, 31, 32, 50). The Paf1C appears to function as a platform for the recruitment of several methyltransferases such as COMPASS and Set2 (16, 17, 26, 44). Histone methylation by COMPASS is predominantly associated with the early 5′ coding regions of active genes, whereas Set2-mediated methylation is mostly found in the mid-to-late bodies of transcribed genes (26, 44). As Pol II initiates transcription, COMPASS can mono-, di-, and trimethylate H3K4 in the early 5′ coding regions of active genes (26, 32, 40, 50). Shortly after promoter clearance, the histone methyltransferase Set2 engages with the elongating Pol II to methylate H3K36 on the bodies of actively transcribed genes (27, 28, 29, 38, 44, 52). During the transition from the early elongating form to the processively elongating form, the phosphorylated residue of the Pol II carboxyl-terminal domain (CTD) shifts from serine 5 to serine 2. The CTD kinase I complex (Ctk complex), the kinase responsible for serine 2 phosphorylation in yeast, mediates the association of Set2 with Pol II through its ability to phosphorylate the CTD (9, 27, 34, 52).
While attempting to define the molecular mechanism of histone H3 methylation by COMPASS, we discovered that Ctk complex components regulate histone H3K4 methylation patterns. Deletion of any of the subunits of the Ctk complex not only abolishes Set2-meditaed methylation of H3 K36 but also affects the methylation state of H3K4. Systematic analysis of bulk histones in CTK1, CTK2, or CTK3 deletions demonstrated that the loss of these factors leads to global reduction of histone H3K4 monomethylation and elevated K4 di- and trimethylation levels. The Ctk complex does not appear to substantially regulate Rad6/Bre1-mediated histone H2B monoubiquitination (which is required for COMPASS function) (12, 48, 49). It was also recently demonstrated that monoubiquitination of histone H2B is not required for full histone H3 monomethylation by COMPASS (11, 40, 43). Our studies demonstrate that loss of Ctk1 activity can result in altered patterns of histone H3 eviction from transcribed regions.
Recent studies demonstrate that histone methylation (mono-, di-, or trimethylation) patterns may play a role in the regulation of gene expression or proper response to developmental or environmental signals (25, 37, 40, 41). Therefore, transition from mono- to dimethyl or from di- to trimethyl moieties on chromatin is likely to be a highly regulated event. Evidence presented here indicates a possible role for the Ctk complex in proper regulation of the pattern of H3K4 mono-, di-, and trimethylation.
MATERIALS AND METHODS
Functional genomic analyses of histone modification by methylation.
Global proteomic screen (GPS) analysis was performed as previously described (39). The GPS was carried out with antibodies specific for H3 Lys4 mono- and trimethylation.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed with wild-type (WT) and Δctk1 strains. ChIP experiments were then performed as previously described (4). Briefly, after formaldehyde fixation, cells were collected by centrifugation and lysed by agitation with glass beads. The cells were then sonicated to yield chromatin fragments between 300 and 500 bp in length. After clarification of the chromatin solution via high-speed centrifugation, cross-linked proteins were immunoprecipitated (IP) with the indicated antibodies. Following elution of the bound proteins, the protein was digested away from the DNA with proteinase K. Decrosslinking was then carried out at 65°C overnight, and the next day the DNA was isolated by phenol-chloroform extraction, followed by ethanol precipitation. The DNA fragments were then used for PCR with primers directed against the indicated gene regions. The primers used for PCR of the PHO84 and ADH1 loci as shown in Fig. 3 and 5 have been previously described (22, 23). The primers used for PCR were GAL1 (Core) (5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAG TATTAGTTAAAGTGGTTATGCA-3′), GAL1 (ORF1) (5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ and 5′-GGCAGCCTGATCCATACCGCCATT-3′, GAL1 (ORF2) (5′-CAGAGGGCTAAGCATGTGTATTCT-3′ and 5′-GTCAATCTCTGGACAAGAACATTC-3′), GAL1 [poly(A)] (5′GCATCACAAAATACGCAATAATAACGAG and 5′TTTTGTCCCTGTGTTTTAAAGTTTGTGG), ADH1 (ORF1) (5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-GCAGACTTCAAAGCCTTGTAGACG-3′), ADH1 (ORF2) (5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′), PHO84 (ORF1) (5′-TAGCTGATATTGTTGGTCGTAAGAG-3′ and 5′-TACCAATACCCATGACAAAACGGTA-3′), and PHO84 (ORF2) (5′-TCTGCAGACATTTTGGTCAATGGAA-3′ and 5′-AAACGTTTTTGGAACCGGCATAAC-3′).
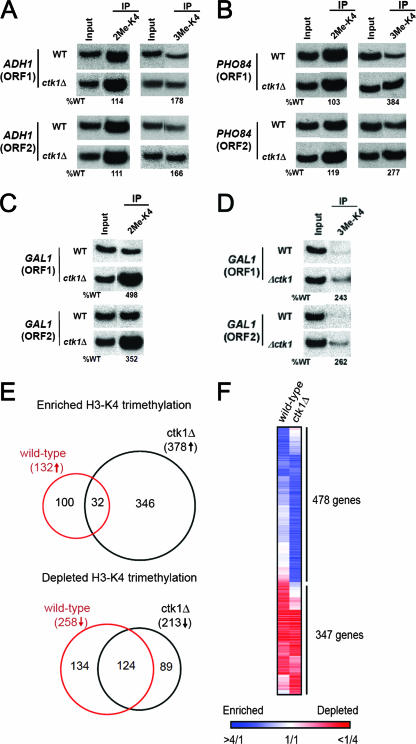
FIG. 3.
Analysis of H3K4 dimethylation and trimethylation on chromatin of active genes. (A and B) ChIP with antibodies directed against di- and trimethylated H3K4 was used to determine the enrichment of methylated histones at the constitutive ADH1 (A) and PHO84 (B) loci. (C and D) Analysis of levels of methylated H3K4 following induction of the GAL1 gene. During activation of the GAL1 locus, H3K4 dimethylation (C) and trimethylation (D) levels are increased in a ctk1 null strain compared to the WT. (E and F) Global levels of histone H3K4 trimethylation are elevated in the absence of Ctk1. A global ChIP-chip analysis was performed to compare the localization of H3K4 trimethylation between WT and CTK1 deletion strains. A genome-wide location analysis was performed to identify ORFs that were enriched (>2-fold IP/WCE ratio) or depleted (<0.5-fold IP/WCE ratio) for histone H3K4 trimethylation. Venn diagrams were used to compare the sets of genes that showed (E) enrichment or depletion of histone H3K4 trimethylation in the WT strain and a ctk1Δ strain. (F) The IP/WCE ratios from the WT and ctk1Δ strain genome-wide location analysis data are displayed for genes with enrichment or depletion of histone H3K4 trimethylation. Blue indicates enrichment of H3K4 trimethylation; red indicates depletion of H3K4 trimethylation.
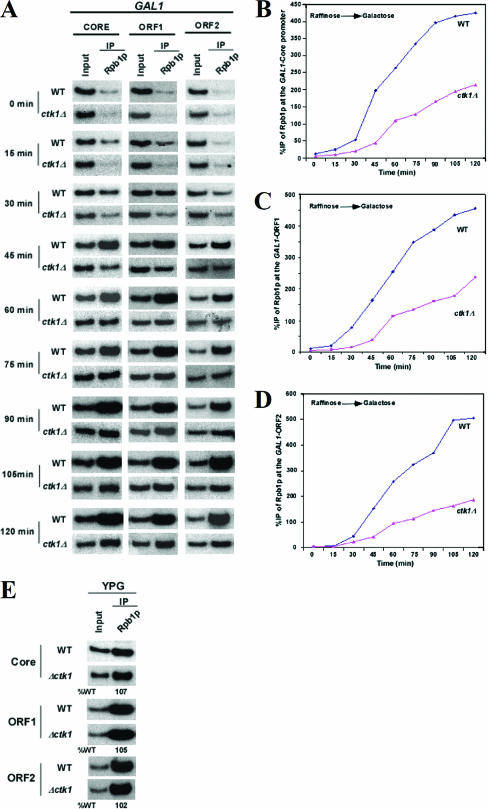
FIG. 5.
Kinetics of Pol II recruitment to the GAL1 gene during transcriptional activation. (A) After cells were grown in raffinose-containing medium, the cells were switched to galactose medium to induce expression of the GAL1 gene and equal amounts of cells were harvested at the indicated time points to determine Pol II localization in both WT and ctk1 null cells. ChIP was performed with 8WG16 antiserum against Rpb1. The PCRs used for analysis of Pol II localization at the GAL1 locus at the given time points are shown. (B, C, and D) The levels of Pol II found at each location of the GAL1 locus during activation are displayed schematically. DNA IP with respect to input is represented as %IP. (E) Cell were grown in YPG for an extended time to induce GAL1 gene expression. Following this extended induction of the GAL1 gene, ChIP analyses were performed to determine if the level of Pol II occupancy had risen to the WT level.
For ChIP of the GAL1 locus during activation, yeast strains were grown in raffinose-containing growth medium (yeast extract, peptone, raffinose [YPR]) to an optical density at 600 nm (OD600) of 0.9 and then shifted to galactose-containing growth medium (yeast extract, peptone, galactose [YPG]) for 90 min prior to formaldehyde cross-linking. Immunoprecipitations were performed with rabbit polyclonal antibodies against H3 dimethyl K4 (Upstate Biotechnology, Inc.) and trimethyl K4 (Abcam, Inc.). Primer pairs located at the core promoter and two different locations of the open reading frame (ORF) (ORF1 and ORF2) of the GAL1 gene were used for PCR analysis of the IP DNA samples. The percentage of DNA IP relative to WT DNA (%WT) is indicated below each band of the mutant strains. To measure RNA polymerase recruitment to the GAL1 locus during activation, both the CTK1 deletion mutant strain and its isogenic WT equivalent were grown in YPR to an OD600 of 0.9 and then shifted to YPG for the times indicated (Fig. 5) prior to formaldehyde cross-linking. ChIP was carried out as described previously (4). Primer pairs located in the core promoter and two different locations (ORF1 and ORF2, listed earlier) in the coding sequence of the GAL1 gene were used for PCR analysis of the IP DNA samples. Immunoprecipitation was performed with mouse monoclonal antibody 8WG16 (Covance) against the CTD domain of the Pol II large subunit (Rpb1p).
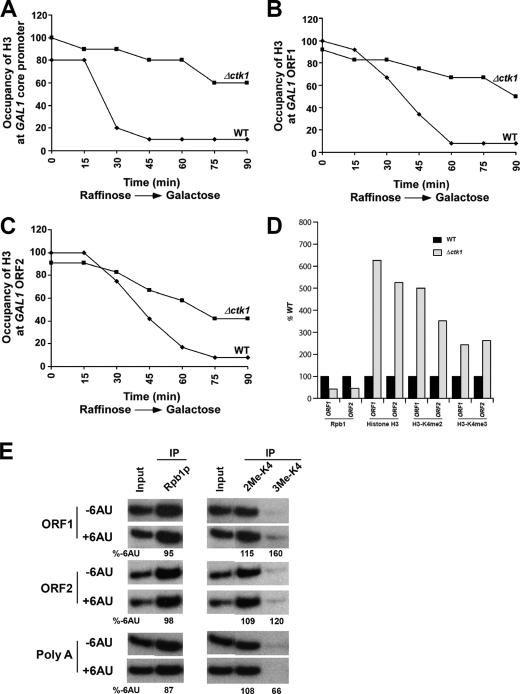
For ChIP done during 6-azauracil (6AU) treatment, the WT strain was grown in YPR to an OD600 of 0.9 and then transferred to YPG containing 6AU (112 μg/ml) for 90 min of induction prior to formaldehyde cross-linking. Immunoprecipitation was performed with 8WG16 antibody against Rpb1 and H3K4 dimethyl and trimethyl antibodies as indicated previously. IP DNAs were analyzed by PCR with specific primer pairs targeted to different regions of the GAL1 locus as indicated.
For analysis of histone H3 occupancy, both the WT and CTK1 deletion mutant strains were grown in YPR to an OD600 of 0.9 and then transferred to YPG for various induction times prior to formaldehyde cross-linking. Immunoprecipitation was performed with an anti-H3 antibody (Abcam 1791). IP DNAs were analyzed by PCR at the GAL1 core promoter, ORF1, and ORF2. Percent immunoprecipitation (%IP) of H3 at all induction time points from WT and CTK1 deletion mutant strains was calculated. The maximum %IP was set to 100%, and other %IP values were normalized with respect to the maximum %IP.
ChIP-on-chip methods.
ChIP-on-chip analysis was performed as described previously (36), with slight modifications. Briefly, triplicate cultures of WT and ctk1Δ strains were grown to mid-log phase (OD600, 0.4 to 0.5) and cross-linked with 1% formaldehyde. Cells were lysed with glass beads and sonicated to generate DNA fragment sizes of 150 to 400 bp. Cross-linked DNA fragments were enriched by immunoprecipitation with anti-histone H3 trimethyl K4 antibody (Abcam) bound to magnetic beads (Dynal Biotech). The beads were washed and then eluted, and the cross-links were reversed. IP DNA was amplified and labeled with Cy5 dye (Amersham Biosciences) by ligation-mediated PCR. As a control, a sample of whole-cell extract (WCE) DNA that was not IP was amplified by ligation-mediated PCR and labeled with Cy3 dye (Amersham Biosciences). Labeled PCR products were hybridized to yeast 6.4K ORF arrays (University Health Networks, Toronto, Ontario, Canada) and scanned with a ScanArray 4000XL scanner (Packard Bioscience).
The scanned images were quantitated with the QuantArray software (Packard Biosciences). The final spot intensity was calculated by subtracting the local background intensity from the spot intensity and then normalized by a global intensity normalization method. The reported gene intensities are averages of duplicate spots present in the yeast 6.4K ORF array. The average IP/WCE (Cy5/Cy3) ratio for each gene was calculated for the triplicate experimental data sets.
TAP tag purification of protein complexes.
Purification of tandem affinity purification (TAP)-tagged proteins was performed essentially as previously described (35). Briefly, 1.5 liters of yeast was grown to an OD600 of 1.2. After collection of the cells via centrifugation, the cells were lysed with glass beads in 25 ml of lysis buffer (10 mM Tris [pH 7.9], 150 mM NaCl, 0.1% NP-40, protease inhibitors [Roche]). After clarification of lysates by centrifugation, TAP-tagged proteins were bound to immunoglobulin G-Sepharose beads overnight at 4°C. Following cleavage of the TAP tag with tobacco etch virus protease, the calcium concentration of the eluent was adjusted to 3 mM CaCl2 and it was incubated with calmodulin-Sepharose 4B (Amersham). Following several washes with calmodulin binding buffer (10 mM Tris [pH 7.9], 150 mM NaCl, 0.1% NP-40, 5 mM β-mercaptoethanol, protease inhibitors [Roche complete mini, EDTA free], 3 mM CaCl2), bound proteins were eluted by the addition of EGTA elution buffer (10 mM Tris [pH 7.9], 150 mM NaCl, 4 mM EGTA, 5 mM β-mercaptoethanol). The purity of each fraction was then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining of the protein complexes.
Kinase assays using Ctk1.
Kinase assays were performed essentially as described before (51). Briefly, IP protein complexes were added to 28 μl of kinase assay buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, 0.5 mM dithiothreitol). After the addition of 5 μCi of α-32P, the reaction mixtures were incubated for 40 min at 30°C. Each reaction was quenched by the addition of SDS-PAGE loading buffer (preheated to 90°C) and brief heating of the samples at 90°C for 5 min. Reaction products were then immediately resolved by SDS-PAGE on 4 to 20% gradient gels and subjected to autoradiography.
RESULTS
Loss of yeast Ctk complex components alters global histone H3 lysine 4 monomethylation levels.
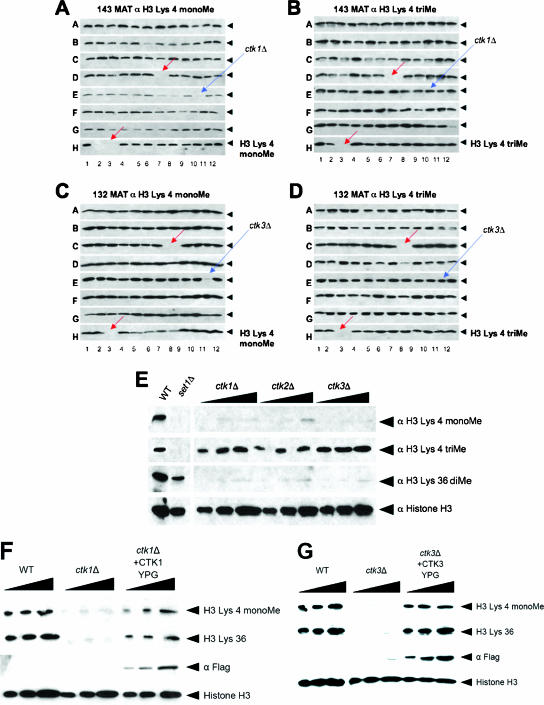
A comprehensive GPS in Saccharomyces cerevisiae surveying the entire nonessential yeast gene deletion consortium was used to seek out genes affecting global H3K4 monomethylation levels (39). With antibodies directed against monomethylated H3K4, the Ctk1 and Ctk3 components of the Ctk complex were found to be required for H3K4 monomethylation (Fig. 1A and C). The combination of Ctk1, Ctk2, and Ctk3 comprises the Ctk complex that catalyzes the phosphorylation of serine 2 of the Pol II CTD, a mark predominantly found on elongating Pol II (6, 45, 46). Ctk2 was not identified in the screen, as the CTK2 deletion does not exist in the collection. Although loss of Ctk1 and Ctk3 results in decreased histone H3K4 monomethylation levels, deletion of CTK1 or CTK3 does not reduce H3K4 trimethylation levels (Fig. 1B and D). Indeed, when bulk histone methylation is measured in a CTK1 deletion strain, it appears that global H3K4 trimethylation levels are increased in the absence of Ctk1 and Ctk3. This indicates that Ctk1 activity loss has a specific effect on the COMPASS-mediated transition from H3K4 monomethylation to trimethylation.
FIG. 1.
Loss of Ctk complex members affects histone H3K4 monomethylation. (A to D) GPS screen demonstrating that deletion of either CTK1 or CTK3 results in reduced levels of H3K4 monomethylation. (E) Deletion of any of the three members of the Ctk complex results in reduced H3K4 monomethylation and H3K36 methylation. (F and G) Loss of histone monomethylation can be genetically complemented by reintroduction of the missing Ctk complex member.
Since CTK2 deletion does not exist in the collection and were not tested in our screen, we analyzed the effect of CTK2 deletion on H3K4 mono- and trimethylation. Deletion of CTK2 has similar effects on H3K4 mono- and trimethylation as deletions of CTK1 and CTK3 (Fig. 1E). Furthermore, introduction of CTK1 or CTK3 genes into ctk1 and ctk3 null strains, respectively, can complement the loss of H3K4 monomethylation (Fig. 1F and G), indicating a role for the components of Ctk complex in the proper regulation of the pattern of H3K4 methylation via COMPASS.
Loss of Ctk complex impacts H3 lysine 4 monomethylation and appears to function in a Rad6/Bre1-independent manner.
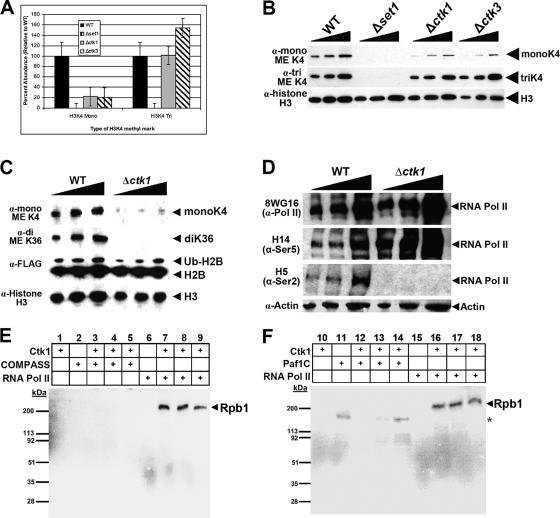
Although deletion of any of the three components of the Ctk complex leads to reduced histone H3K4 monomethylation levels, K4 dimethylation and trimethylation are not noticeably reduced. This observation indicates that the catalytic properties of COMPASS may shift toward di- or trimethylation in the absence of the functional Ctk complex. To test this, amounts of bulk H3K4 monomethylation and trimethylation in strains with CTK1 and CTK3 deleted were analyzed. Compared to the WT, ctk null strains present an about 60 to 70% reduction in global H3K4 monomethylation (Fig. 2A and B). Additionally, the H3K4 trimethylation found in bulk histone preparations of CTK1 and CTK3 deletion strains is slightly higher than that of WT cells (Fig. 2B). This suggests that COMPASS activity is indeed skewed toward dimethylation and trimethylation of H3K4 in a ctk1 null background.
FIG. 2.
Several histone modifications are altered by loss of Ctk1 kinase activity. (A) When components of the Ctk complex are deleted, the bulk levels of the histone H3K4 methylation pattern are altered. (B) WCE of either a WT strain or a strain with a single deletion of either SET1, CTK1, or CTK3 was resolved by SDS-PAGE and probed with antibodies against monomethylated and trimethylated histone H3K4. The extracts were also probed with antibody against histone H3 as a loading control. (C) The loss of CTK1 does not significantly affect histone H2B monoubiquitination by Rad6 but dramatically affects COMPASS- and Set2-mediated histone methyltransferase activity. (D) Loss of CTK1 does not result in reduced Rpb1 protein stability. Whole-cell lysates were prepared from either WT cells or those lacking CTK1. Extracts were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with antiserum against the CTD of Rpb1 (8WG16), serine 5-phosphorylated Rpb1 (H14), or serine 2-phosphorylated Rpb1 (H5). Loss of Ctk1 activity leads to a total absence of serine 2 phosphorylation. (E and F) COMPASS and the Paf1C are not targets of Ctk1 in vitro. (E) Both purified COMPASS and Pol II were tested for the ability to be phosphorylated by the Ctk complex. COMPASS or Pol II alone does not become phosphorylated (lanes 2 and 6). Lanes 3 to 5 and 7 to 9 had increasing amounts of COMPASS and Pol II, respectively, and were incubated in the presence of purified Ctk1. (F) The Paf1C does not appear to be phosphorylated by the Ctk complex in vitro. Increasing amounts of purified Paf1C were incubated with the purified Ctk complex (lanes 12 to 14) with no apparent change in the phosphorylation state of Paf1C subunits. The asterisk denotes the presence of a nonspecific band that is also found when the Paf1C is incubated by itself (lane 11). ME, methylated.
Monoubiquitination of histone H2B by the Rad6/Bre1 complex serves as an upstream signal for histone H3K4 methylation (12, 32, 48, 49, 50, 51). These chromatin modifications are intimately tied to transcription (8, 10, 20, 26, 32, 50, 53). Since loss of any Ctk complex components can considerably impact methylated histone H3K4 levels, we wanted to determine whether the Ctk complex functions by regulating Rad6/Bre1 complex activity. Our studies demonstrate that the loss of Ctk1 function does not appear to significantly affect Rad6/Bre1's monoubiquitination of histone H2B (Fig. 2C), indicating that the Ctk complex-dependent effect observed on histone H3K4 methylation is perhaps Rad6/Bre1 independent. This is further substantiated by the finding that Ctk1 function is also required for Set2-mediated methylation of histone H3, which occurs independently of histone H2B monoubiquitination (52). We used anti H3 antibodies as the load control for this study. Although the level of H2B monoubiquitination appears to be slightly reduced in a ctk1 null background, histone H3 levels also appear to be slightly reduced (Fig. 2C). This observation is perhaps due to the fact that the strains with the components of the Ctk complex deleted demonstrate a slow-growth phenotype.
Since COMPASS activity is mediated through its association with Pol II via its interaction with the Paf1C (16, 17, 26), we wanted to ensure that the altered stability of the largest subunit of Pol II (Rpb1) in the absence of Ctk1 phosphorylation was not the cause of the observed H3K4 methylation changes. Therefore, we compared Rpb1 levels from WT and ctk1 null cells. This study indicated no apparent substantial changes in Rpb1 levels (Fig. 2D).
Changes in COMPASS activity do not appear to be the direct result of COMPASS phosphorylation by the Ctk complex.
Discrete subunits of COMPASS mediate individual H3K4 methyl marks (40). Phosphorylation of Rad6 and recruitment of the Paf1C to chromatin by the Bur1/Bur2 complex modulate H2B monoubiquitination levels (51, 53). Our observation that loss of Ctk1 activity results in an overall decrease in histone H3K4 monomethylation and an increase in H3K4 trimethylation from bulk histone may indicate that COMPASS processivity increases in the absence of Ctk1. Since COMPASS catalytic activity is altered in the absence of Ctk1, it is possible that Ctk1 activity may be directed toward a subunit of COMPASS. This activity could serve as a repressive mark to inhibit COMPASS activity or prevent the binding of a specific subunit required for proper regulation of histone methylation. To test this idea, COMPASS and the Ctk complex were affinity purified with the TAP tag on Cps60 and Ctk1, respectively, and the presence of subunit components was confirmed by MudPIT analyses (40). Using these reagents, we reconstituted an in vitro kinase assay. Pol II was purified with TAP-tagged Rpb3 as a positive control, since Ctk1 was previously shown to phosphorylate Rpb1 (9). When COMPASS is incubated in the presence of Ctk1, no observable kinase activity toward COMPASS is detected in vitro (Fig. 2E, lanes 3 to 5). However, addition of the Ctk complex to Pol II resulted in phosphorylation of the Rpb1 subunit (Fig. 2E, lanes 7 to 9), indicating that the purified Ctk complex is enzymatically active.
The Paf1C plays an integral role in COMPASS-mediated histone methylation (26, 32, 50). The affinity-purified Paf1C was also used in an in vitro kinase assay to determine if subunits of the Paf1C are targets for Ctk-mediated phosphorylation. While there is no specific Ctk-mediated activity toward any member of the Paf1C, a nonspecific activity was observed when the Paf1C was incubated alone or with the Ctk complex (Fig. 2F, compare lane 11 to lanes 12 to 14). Since the Ctk complex displays no observable specificity for subunits of COMPASS or the Paf1C, it seems likely that the aberrant histone H3 methylation observed in the absence of Ctk complex function may be directly linked to the phosphorylation state of other substrates such as the CTD of Pol II.
Deletion of CTK1 results in accumulation of histone H3K4 trimethylation on chromatin on constitutively active genes.
Histone H3K4 methylation and COMPASS are localized predominantly at the promoter and early 5′ coding regions of active genes in yeast (26, 33, 37). Since our studies have concentrated on analysis of bulk histones, we wanted to determine whether an increase in histone H3K4 di- and trimethylation occurs on chromatin of actively transcribed genes following the loss of Ctk1. Using antiserum directed against di- and trimethylated histone H3K4, ChIP was performed in a WT strain, as well as strains with CTK1 deleted. Interestingly, ChIP experiments revealed an increase in trimethylation levels in the coding region of the transcriptionally active genes tested (PHO84 and ADH1) (Fig. 3A and B). In the accompanying report, Strahl and colleagues have also demonstrated that an increase in trimethylation levels in a ctk1 null strain is manifested as a “spread” of these methyl marks from the 5′ end to the 3′ end of individual genes (54). Since Set2 does not associate with the elongating form of Pol II in the absence of Ctk1, it is possible that association of COMPASS with Pol II becomes more stable and COMPASS activity extends throughout active coding regions.
Both histone H3K4 dimethylation and trimethylation are increased during active transcription at an inducible gene in the absence of CTK1.
ChIP studies performed during galactose induction of the GAL1 gene demonstrated that both H3K4 di- and trimethylation dramatically increase in a CTK1 null strain compared to WT cells (Fig. 3C and D). The increase in histone methylation correlates with heightened times of transcriptional activity at the GAL1 locus, reaffirming the interplay between COMPASS activity and Pol II in H3K4 methylation regulation. Interestingly, although an increase in the levels of trimethylated H3K4 were observed at the constitutive ADH1 and PHO84 loci, the increase in both di- and trimethylation was greatly exacerbated during the switch from an inactive to an induced GAL1 gene (Fig. 3C and D).
Loss of CTK1 results in a global increase in H3K4 trimethylation levels.
When global ChIP-on-chip analyses were performed to observe regions where trimethylation increased in the absence of Ctk1 activity, the number of genes with enriched H3K4 trimethylation increased nearly threefold in the ctk1 null strain (378 genes) compared to the WT (132 genes; Fig. 3E and F). A similar increase in H3K4 trimethylation enrichment was observed independent of the cutoff used to identify enriched genes (e.g., 2-fold, 1.75-fold, and 1.5-fold). This result confirms that global levels of histone H3K4 trimethylation on chromatin are elevated in the ctk1 null mutant and that trimethylation is now occurring at loci not normally found to be trimethylated. In contrast, there is a slight decrease in the number of genes with depleted H3K4 trimethylation in the ctk1 null mutant compared to the WT (Fig. 3E and F). In the WT strain, regions depleted for H3K4 trimethylation contained a significant number of genes located in telomere-proximal chromosomal regions (86 genes, P = 2.4 × 10−57), as had been previously observed for H3K4 dimethylation (3). In the ctk1 null mutant, however, the number of telomere-proximal genes depleted for H3K4 trimethylation is decreased by 35% compared to the WT.
Histone occupancy and association of Pol II are altered on an actively transcribed gene in the absence of Ctk1 activity.
Since we observed an increase in the levels of H3K4 di- and trimethylation on actively transcribed gene GAL1 in the absence of Ctk1 (Fig. 3), we wanted to determine whether Ctk1 loss could alter the occupancy of histone H3. As shown in Fig. 4A to D, the occupancy of H3 on the core promoter and body of the GAL1 gene is inversely correlated with transcription. Histone H3 is evicted as the GAL1 gene is activated by switching the growth medium from raffinose (noninducing) to galactose (inducing). Such observations on the occupancy of H3 on promoters and transcribed regions of genes have recently been reported (1, 5). However, our ChIP studies demonstrate that histone H3 eviction during active transcription is reduced in a ctk1 null background (Fig. 4A to D). To determine whether this reduction in the rate of histone H3 eviction is through the reduced levels of Pol II recruitment to chromatin, we performed ChIP studies to determine the kinetics of recruitment of Pol II to the GAL1 gene upon activation (Fig. 5A to D). Our studies demonstrate that a reduced amount of Pol II was found to be associated with chromatin (compared to WT strains) when ChIP was used to determine if a buildup of Pol II occurred during the induction of the GAL1 gene in a CTK1 null strain (Fig. 5A to D). Approximately half as much Pol II is associated with the GAL1 locus in the CTK1 deletion cells after 90 min (Fig. 5A). However, in the same time period, H3K4 di- and trimethylation are increased drastically (Fig. 3C and D).
FIG. 4.
Nucleosome depletion from an activated gene is delayed in the absence of Ctk1. (A to C) Histone H3 occupancy in the core promoter (A), ORF1 (B), and ORF2 (C) regions of the GAL1 locus during transcriptional activation by ChIP as described in Materials and Methods. (D) During times of reduced recruitment of Pol II to the GAL1 locus, histone H3 levels and H3K4 dimethylation and trimethylation levels are elevated in the absence of Ctk1. (E) Transcriptional stress does not mirror the effects of a CTK1 deletion. To determine if the effects on histone occupancy and methylation could be a result of transcriptional “stress,” we used 6AU to chemically induce stress in the WT strain. Addition of 6AU does not result in a substantial increase in H3K4 di- and trimethylation compared to that in a ctk1 null strain. Similarly, Pol II recruitment is not altered in the presence of 6AU. The percentage of DNA IP in the presence of 6AU relative to the absence of 6AU (%-6AU) is shown below each lane.
If Ctk1 loss results in a slower association of Pol II with chromatin, then Pol II occupancy at the GAL1 locus could reach WT levels if given enough time. As shown in Fig. 5E, the amount of Pol II bound at the GAL1 locus is equivalent to that of WT cells induced for a longer period of time, indicating that the overall recruitment of Pol II to active genes is delayed in the absence of Ctk1 activity.
Ctk1 loss does not appear to specifically alter Pol II elongation properties on the GAL1 gene on the basis of our measurements; however, the overall rate of transcription, including transcription initiation, appears to be slowed down in the absence of Ctk1. Mason and Struhl recently demonstrated that the drug 6AU reduces both the elongation rate and the processivity of Pol II and that this processivity defect is exacerbated by mutations in Spt4, TFIIS, and the Ctk complex (the Ctk kinase complex) (30). Furthermore, induction of transcriptional stress, such as treatment with 6AU, can result in altered nucleosome occupancy on the TEF1 gene in yeast (55). We tested the effect of 6AU treatment on Pol II and nucleosome occupancy and H3K4 methylation on the GAL1 gene (Fig. 4E). Our study indicates that 6AU does not significantly alter the pattern of H3K4 trimethylation on the genes tested. In an accompanying report, Strahl and colleagues also demonstrated that 6AU treatment does not alter H3K4 trimethylation levels from bulk histone (54). Therefore, on the basis of these observations we conclude that Ctk1's regulation of K4 di- and trimethylation appears to be independent of 6AU-induced stress on the GAL1 gene.
Analysis of the role of factors required for histone eviction in the regulation of the pattern of H3K4 methylation.
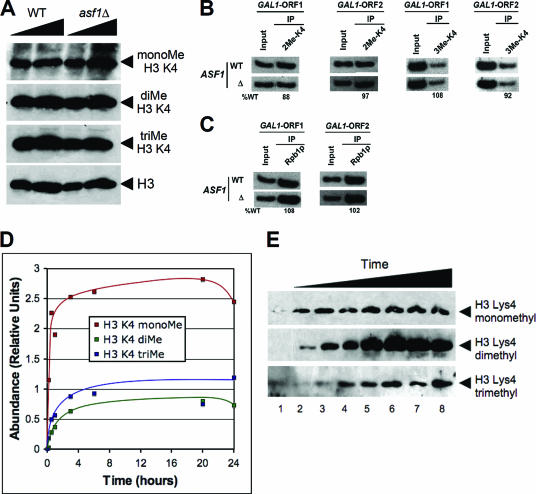
Our studies thus far indicate that loss of Ctk1 results in altered patterns of Pol II occupancy and H3 eviction from chromatin. These changes are accompanied by altered H3K4 methylation patterns. Pol II occupancy and histone eviction are functionally related effects resulting from Ctk1 loss. To determine whether defects in histone H3 eviction alone could cause the observed phenotypes, we tested the effect of an ASF1 deletion in this process. Recently, it was demonstrated that Asf1 specifically mediates histone H3 eviction (42). Therefore, we initially tested the effect of ASF1 deletion on bulk patterns of H3K4 methylation. As shown in Fig. 6A, ASF1 deletion does not significantly alter global H3K4 methylation patterns compared to CTK1, -2, and -3 deletions (compare Fig. 6A to Fig. 1E). To further demonstrate whether ASF1 deletion alters H3K4 methylation patterns on chromatin, we analyzed H3K4 methylation on the GAL1 gene in the presence and absence of Asf1 by ChIP (Fig. 6B). These studies also confirm our observation that Asf1 does not alter H3K4 methylation patterns (compare Fig. 6B to Fig. 3C and D). Furthermore, we did not observe defects in Pol II occupancy in an asf1 null strain (Fig. 6C). These studies together suggest that the defect in histone H3 eviction alone does not explicate the altered H3K4 methylation phenotype seen in the absence of Ctk complex components.
FIG. 6.
An increased time of association between COMPASS and histone H3 is necessary to achieve trimethylation. (A and B) Analysis of the role of Asf1 in the regulation of the H3K4 methylation pattern. Both WT and ASF1 deletion yeast strains were grown in either yeast extract-peptone-d-glucose (A) or YPR (B) to an OD600 of 0.9 and then switched to YPG prior to formaldehyde cross-linking. (A) Bulk H3K4 methylation patterns were analyzed by Western blot analysis with H3K4-specific antibodies. (B) H3K4 methylation patterns were also analyzed on the GAL1 gene in the presence and absence of Asf1 by ChIP. (C) Pol II occupancy on the GAL1 gene in the WT and asf1 null strains was also tested via ChIP. (D and E) H3K4 mono-, di-, and trimethylation by COMPASS as a function of time. In vitro methyltransferase assays were performed and allowed to progress for 0.16, 0.5, 1, 3, 6, 20, and 24 h. Methylated (Me) histones were resolved by SDS-PAGE, and the abundance of mono-, di-, and trimethylated histone H3 was determined by Western blot analysis with antiserum directed against mono-, di-, and trimethylated H3K4 as indicated. While monomethylation occurs rapidly, production of di- and trimethylated H3 requires extended incubation times.
Conversion of H3K4 monomethylation to trimethylation requires the extended time of association between COMPASS and histone H3.
To determine whether increased association of COMPASS with histone H3 is necessary for the transition of the H3K4 monomethyl moiety to the trimethylated form, we studied the enzymatic activity of purified COMPASS toward histone H3 as a function of time in vitro. As shown in Fig. 6D and E, the onset of monomethylation on an unmethylated template is almost immediate, with the methyltransferase assay reaching nearly saturated levels of monomethylation within the first hour. However, the onset of trimethylation does not occur until later time points (Fig. 6D and E).
Although trimethylation of H3K4 by COMPASS does not take place as rapidly as it is reported to do in vivo, this is to be expected as (i) the in vitro reaction is set up with limiting concentrations of enzyme and substrates for the purpose of studying the reaction itself in vitro and (ii) the in vitro methylation reaction with COMPASS and H3 is set up in the absence of monoubiquitinated H2B and chromatin, and therefore COMPASS will not be able to transition to trimethylation as rapidly. However, this study clearly demonstrates that a transition from mono- to dimethylation and then from di- to trimethylation is a sequential process and that COMPASS requires this extended time to process the conversion from unmethylated to monomethylated and then on to di- and trimethylated histone H3. Therefore, as more histone H3 becomes trimethylated, this means that less H3 in the reaction mixture is left in the monomethylated form. Indeed, toward the end of the assay one can observe that trimethylation is steadily increasing as monomethylation is starting to recede (Fig. 6D and E).
DISCUSSION
There is increasing evidence that histone modifications impact transcriptional elongation (14, 16, 45). We propose that transcriptional efficiency which is regulated by Pol II and the Ctk complex, and the increased time of COMPASS association with chromatin may impact the abundance and pattern of histone H3K4 methylation in the wake of transcribing Pol II. This observation provides a possible insight into the mechanism through which specific H3K4 methylation patterns are regulated on transcribed genes.
Since Ctk1 is the predominant CTD serine 2 kinase in yeast, any disruption of Ctk1 activity can have numerous outcomes for transcription efficiency (6, 9, 27, 34). Phosphorylation of the Pol II CTD by Ctk1 serves as a docking site for another histone methyltransferase, the Set2 protein. Deletion of Ctk1 not only abolishes interaction between Set2 and Pol II, it also ablates Set2's interaction with chromatin (27, 28, 29, 47, 52). Set2 histone methyltransferase activity remains associated with elongating polymerase until transcriptional termination (27). Unlike COMPASS-mediated methylation in the early 5′ coding regions of active genes, Set2-mediated methylation of histone H3 is linked to transcriptional repression (7, 19, 21, 47). Recently, several groups have shown that the bromodomain of Eaf3 binds methylated lysine 36 of histone H3, and this recruits the Rpd3 histone deacetylase complex (7, 19, 21). Thus, one of the main functions of CTD serine 2 phosphorylation by the Ctk complex is to regulate a subset of transcriptional repressors during the processive stages of transcription. In addition to regulation of Set2 histone methyltransferase activity by the Ctk complex, we suggest that the Ctk complex may also modulate the pattern of histone modifications mediated by Set1/COMPASS within active regions of chromatin. Although the Ctk complex is not capable of phosphorylating either COMPASS or the Paf1C in vitro (Fig. 2E and F), Ctk1 substrates other than the CTD of Pol II may regulate COMPASS-mediated H3K4 methylation patterns. We have recently initiated biochemical and genetic projects in the laboratory searching for a possible substrate.
Numerous studies in the last few years have investigated the regulation of COMPASS histone methyltransferase activity, and here we indicate a unique role for the Ctk complex in this process. Many of the previously described mechanisms involving Rad6/Bre1, the Paf1C, and the Bur1/Bur2 complex demonstrate a reduction in COMPASS activity. However, deletion of Ctk complex components results in a change in COMPASS-mediated histone methylation and a global shift toward dimethylation and trimethylation of H3K4 as monomethylation levels diminish in vivo. This effect is quite different from that observed when histone H2B monoubiquitination is altered. An increase in histone H2B monoubiquitination results in increased COMPASS activity, and monomethylation becomes enriched (10). It is possible that increased histone monoubiquitination allows transcription to proceed more rapidly; therefore, the time of association between COMPASS and the histone H3 tail is reduced and monomethylation becomes the predominant form of methylated H3K4. This is similar to in vitro studies performed with COMPASS and H3 (Fig. 6).
Early in our studies, it was uncertain whether Ctk1 loss results in an increase in the levels of histone H3K4 trimethylation in a manner by which COMPASS histone methyltransferase activity becomes hyperactivated. Loss of Ctk1 affects several events in the transcriptional cycle, i.e., (i) the histone methyl mark left by Set2, (ii) serine 2 phosphorylation of the CTD, (iii) the timely recruitment of polymerase to activated genes, and (iv) the loss of proper transcriptional efficiency. Additionally, we demonstrated with our ChIP studies that histone H3 eviction during transcription is reduced in a CTK1 deletion strain. However, this delay does not fully explain the observed increase in H3K4 trimethylation levels, as the loss Asf1 (which is also required for histone H3 eviction [42]) does not result in H3K4 monomethylation loss or an increase in trimethylation similar to that of ctk1 null strains. Our in vitro studies demonstrated the necessity of increased times of association between COMPASS and the histone H3 tail in order to properly catalyze the pattern of mono, di-, and trimethylation of H3K4.
On the basis of the above observations, one can envision a scenario in which the transcription initiation, promoter clearance, and transcription elongation stages of the transcription cycle are sluggish in the absence of a functional Ctk complex. Furthermore, Ctk1 loss results in a decrease in the rate of histone H3 eviction and creates an increase in the concentration of the substrate for COMPASS. As a result of the “sluggish polymerase,” the local histone H3 concentration can increase, as does the time of association between COMPASS and histone H3 on chromatin.
In addition to the importance of methylation on particular lysine residues within histones, patterns of methylation (such as mono-, di-, or trimethylation) are required for a proper response to extracellular and intracellular signals (25, 37, 40, 41). Therefore, cells must somehow regulate the patterns of methylation on chromatin. Our studies suggest a possible mechanism for the regulation of the pattern of H3K4 methylation via COMPASS by the Ctk complex. If the pattern of H3K4 methylation on chromatin could be regulated by the rate of the elongating Pol II, therefore, elongation factors such as ELL or elongin A, which alter the Vmax of transcription elongation (13, 15, 45), could play a role in the regulation of the pattern of histone methylation. Future genetic and biological studies aiming to define roles for histone mono-, di-, and trimethylation patterns on chromatin and factors required for the implementation of such marks should be very informative.
Acknowledgments
J.D.S. and J.J.W. were supported by American Cancer Society grant RSG-03-181-01-GMC. S.R.B. is supported by American Heart Association Scientist Development grant 0635008N, American Cancer Society Research Scholar grant 06-52, and a Central Research Committee grant from the Southern Illinois University School of Medicine. The work in A. Shilatifard's laboratory is supported by grants from the National Institutes of Health (2R01CA089455 and 1R01GM069905) and the American Cancer Society. A. Wood is supported by an American Heart Association predoctoral fellowship. A. Shilatifard is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., and M. R. Green. 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370:445-454. [DOI] [PubMed] [Google Scholar]
- 5.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10:679-680. [DOI] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W.-J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 8.Carvin, C. D., and M. P. Kladde. 2004. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J. Biol. Chem. 279:33057-33062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, E.-J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 11.Dehé, P.-M., M. Pamblanco, P. Luciano, R. Lebrun, D. Moinier, R. Sendra, A. Verreault, V. Tordera, and V. Geli. 2005. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 353:477-484. [DOI] [PubMed] [Google Scholar]
- 12.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 13.Eissenberg, J. C., J. Ma, M. A. Gerber, A. Christensen, J. A. Kennison, and A. Shilatifard. 2002. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc. Natl. Acad. Sci. USA 99:9894-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eissenberg, J. C., and Shilatifard. 2006. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 16:184-190. [DOI] [PubMed] [Google Scholar]
- 15.Gerber, M., J. Ma, K. Dean, J. C. Eissenberg, and A. Shilatifard. 2001. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 20:6104-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 17.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 18.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 20.Kao, C. F., C. Hillyer, T. Tsukuda, K. Henry, S. Berger, and M. A. Osley. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keogh, M.-C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, and N. J. Thompson. 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 25.Koyanagi, M., A. Baguet, J. Martens, R. Margueron, T. Jenuwein, and M. Bix. 2005. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in TH1 cells. J. Biol. Chem. 280:31470-31477. [DOI] [PubMed] [Google Scholar]
- 26.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 30.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 31.Miller, T., N. J. Krogan, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. B. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 34.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 274:27823-27828. [DOI] [PubMed] [Google Scholar]
- 35.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 36.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 38.Schaft, D., A. Roguev, K. M. Kotovic, A. Shevchenko, M. Sarov, A. Shevchenko, K. M. Neugebauer, and A. F. Stewart. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, J., J. Dover, M. Johnston, and A. Shilatifard. 2004. Global proteomic analysis of S. cerevisiae (GPS) to identify proteins required for histone modifications. Methods Enzymol. 377:227-234. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, J., A. Wood, J. S. Lee, R. Schuster, J. Dueker, C. Maguire, S. K. Swanson, L. Florens, M. P. Washburn, and A. Shilatifard. 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19:849-856. [DOI] [PubMed] [Google Scholar]
- 41.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 43.Shahbazian, M. D., K. Zhang, and M. Grunstein. 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19:271-277. [DOI] [PubMed] [Google Scholar]
- 44.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 45.Shilatifard, A., R. C. Conaway, and J. W. Conaway. 2003. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72:693-715. [DOI] [PubMed] [Google Scholar]
- 46.Sims, R. J. III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 47.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 49.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 50.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 51.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20:589-599. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao, T., Y. Shibata, B. Rao, R. N. Laribee, R. O'Rourke, M. J. Buck, J. F. Greenblatt, N. J. Krogan, J. D. Lieb, and B. D. Strahl. 2007. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol. Cell. Biol. 27:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]