Abstract
The ATP-dependent molecular chaperone Hsp90 and partner cochaperone proteins are required for the folding and activity of diverse cellular client proteins, including steroid hormone receptors and multiple oncogenic kinases. Hsp90 undergoes nucleotide-dependent conformational changes, but little is known about how these changes are coupled to client protein activation. In order to clarify how nucleotides affect Hsp90 interactions with cochaperone proteins, we monitored assembly of wild-type and mutant Hsp90 with Sti1, Sba1, and Cpr6 in Saccharomyces cerevisiae cell extracts. Wild-type Hsp90 bound Sti1 in a nucleotide-independent manner, while Sba1 and Cpr6 specifically and independently interacted with Hsp90 in the presence of the nonhydrolyzable analog of ATP, AMP-PNP. Alterations in Hsp90 residues that contribute to ATP binding or hydrolysis prevented or altered Sba1 and Cpr6 interaction; additional alterations affected the specificity of Cpr6 interaction. Some mutant forms of Hsp90 also displayed reduced Sti1 interaction in the presence of a nucleotide. These studies indicate that cycling of Hsp90 between the nucleotide-free, open conformation and the ATP-bound, closed conformation is influenced by residues both within and outside the N-terminal ATPase domain and that these conformational changes have dramatic effects on interaction with cochaperone proteins.
The essential abundant molecular chaperone Hsp90 is critical for the folding and regulation of a wide array of cellular proteins, termed client proteins. Hsp90 and partner cochaperone proteins interact with client proteins in an ordered pathway that involves sequential ATP-dependent interactions of the client protein with Hsp70 and Hsp90. Because a number of oncogenic signaling proteins, including Akt, Raf-1, Bcr-Abl, mutant p53, and HER-2/Erb2, require Hsp90 for function, Hsp90 is a promising anticancer target and Hsp90 inhibitors are currently in clinical trials (27, 28, 38, 39).
Hsp90 may be divided into three domains: an N-terminal ATP-binding domain, a middle domain, and a carboxy-terminal domain, which contains the primary dimerization site. A model for Hsp90's ATPase cycle, termed a molecular clamp, is based on homology with ATPases such as the DNA gyrase GryB and the DNA mismatch repair protein MutL. In the absence of a nucleotide, Hsp90 dimerized only at the carboxy terminus is in an open conformation. Nucleotide binding induces closing of a lid over bound nucleotide and association of N-terminal domains. ATP hydrolysis is dependent on residues in a flexible loop from the middle segment that extend to the mouth of the nucleotide-binding pocket, and after hydrolysis Hsp90 returns to the open conformation (20, 29). Recent structural evidence confirmed the presence of a dimerization interface between the N termini in the presence of the nonhydrolyzable ATP analog, AMP-PNP, and demonstrated that a residue in the flexible loop contacts the bound nucleotide, supporting a role for this loop in catalysis (2). Structural analysis of the Hsp90 protein of the endoplasmic reticulum, GRP94, also provides evidence for nucleotide-dependent conformational changes (7, 13). The client binding site(s) and the relationship between Hsp90 client binding/release and ATP binding, hydrolysis, and release remain unclear (38, 39), but recent evidence suggests that bound Cdk4 kinase contacts both N-terminal and middle domains on the outer edge of Hsp90 rather than in between the two monomers (37).
Multiple cochaperone proteins, many of which interact in a mutually exclusive manner, have distinct effects on Hsp90 activity (27, 38, 39). In a current model, transfer of a client protein from Hsp70 to Hsp90 is facilitated by the tetratricopeptide repeat (TPR)-containing protein Sti1 (Hop in mammalian cells) (5), which binds the TPR acceptor site at the carboxy terminus of Hsp90 (34). Nucleotide binding results in Sba1 interaction, which stabilizes the dimerized amino terminus (2, 8, 32). Sti1 inhibits Hsp90 ATPase activity and N-terminal dimerization, and displacement by Cpr6, which also contains TPR domains, was proposed to activate Hsp90 (30, 31). A goal of our study was to determine how nucleotides affect the cycle of Sti1, Sba1, and Cpr6 interaction.
Analysis of Hsp90 in Saccharomyces cerevisiae has identified a number of mutations that disrupt growth, cochaperone interactions, client protein activity, and/or ATPase activity (15, 16, 20, 23-25, 29). We determined that the cochaperones Sba1 and Cpr6 stably interact with Hsp90 only in the presence of the nonhydrolyzable ATP analog, AMP-PNP. We then examined the effect of Hsp90 mutation on Sti1, Cpr6, and Sba1 interaction. Our results suggest that changes in the lid and the catalytic loop of the middle segment of Hsp90 play an important role in regulating Sba1 and Cpr6 interaction and identify Hsp90 mutations that exhibit reduced Sti1 interaction.
MATERIALS AND METHODS
Media, chemicals, antibodies, and plasmids.
Standard yeast genetic methods were employed (35). Five-floroorotic acid (5-FOA) was obtained from Toronto Research Chemicals. ATP, ADP, and 5′-adenylylimidodiphosphate (AMP-PNP), phosphocreatine, and creatine phosphokinase were obtained from Sigma. Polyclonal antisera were raised against keyhole limpet hemocyanin-conjugated peptides corresponding to amino acids 91 to 108 of Sti1 and 81 to 96 of Cpr6. Polyclonal antibodies against Hsc82/Hsp82 have been described (10). Polyclonal antibodies against Ssa1/2 and Sba1 were generous gifts of Elizabeth Craig and Brian Freeman, respectively.
S. cerevisiae strain JJ816 (hsc82::LEU2 hsp82::LEU2/YEp24-HSP82) is isogenic to W303 and has been described (9). Strain JJ40 (sba1::URA3 hsc82::LEU2 hsp82::LEU2/pRS313-His-Hsc82) was constructed by subsequent crosses and sporulations of an isogenic sba1::URA3 strain (8) to strain JJ816.
Hsc82 plasmids and constructs.
A plasmid expressing wild-type (WT) HSC82 was a gift from Susan Lindquist. Point mutations were constructed using site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA) or other PCR-based methods. Sequences of mutagenic primers are available on request. To construct His-Hsc82, the coding sequence of HSC82 was cloned into pRSETC (Invitrogen) to insert an amino-terminal His6 tag. The His6-Hsc82 coding sequence was subsequently cloned into pRS313GPD (22) to generate pRS313-GPDHISHSC82. The complete sequence inserted at the amino terminus of Hsc82 is NH2-MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDRWIRPRDLGTLVPRGS-Hsc82. The constitutively expressed His-Hsc82 fully rescues the lethality of an hsc82 hsp82 strain, and as judged by immunoblot analysis, the level of His-Hsc82 protein expressed is similar to that of Hsc82 expressed from the endogenous promoter (not shown).
The following amino acid alterations were constructed within pRS313-GPDHISHSC82: E33A, D79N, T101I, A107N, W296A, G309S, F325A, F345A, N373A, R376A, E377K, Q380A, S481Y, T521I, A583T, F660A, I588A M589A, L647S L648S, and ΔMEEVD, which deletes the last five amino acids of Hsc82. The additional mutant, Hsc82-T22I, was expressed from the multicopy plasmid pRS423GPDHIS-HSC82. All mutant constructs were sequenced completely using automated DNA sequencing. The additional L487S mutation encoded in HSC82 was obtained in a STI1 synthetic lethal screen (9) and moved into the pRS313-GPDHISHSC82 construct using standard techniques.
Isolation of His-Hsc82 complexes.
A plasmid expressing His-tagged WT or mutant Hsc82 was transformed into strain JJ816, and resultant colonies were grown in the presence of 5-FOA to counterselect for the plasmid expressing WT untagged Hsp82 (YEp24-HSP82). His-Hsc82 was the only Hsp90 present in the cell unless the mutation was unable to support viability, in which case mutant His-Hsc82 was coexpressed with untagged WT Hsp82 as indicated. Strain JJ816 expressing mutant His-Hsc82 was grown overnight in selective media to an optical density at 600 nm of 1.2 to 2.0. Cells were harvested, washed with water, and resuspended in lysis buffer (20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2 containing a protease inhibitor mixture [Roche Applied Science]). Cells were disrupted in the presence of glass beads with eight 30-s pulses. After centrifugation, lysate was adjusted to contain (5 mM each) AMP-PNP, ATP plus an ATP regenerating system (ATP+RS) consisting of 4.5 mg phosphocreatine and 8 units of creatine phosphokinase per ml of yeast lysate (6), or ADP and incubated at 30°C for 5 min. Hsc82 complexes were isolated by incubation with nickel resin (1.5 h with rocking, 4°C), followed by washes with lysis buffer plus 0.1% Tween 20 and 35 mM imidazole. Nickel resin was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and protein complexes were separated by gel electrophoresis, followed by Coomassie blue staining or immunoblot analysis using antibodies against Sti1, Hsc82/Hsp82, Ssa1/2, Sba1, and/or Cpr6.
RESULTS
Recent structural evidence confirmed that the N termini of Hsp90 dimerize in the presence of AMP-PNP and identified the Sba1 binding site in the N-terminal domain of Hsp90 (2, 29). We examined which cochaperones in addition to Sba1 specifically interact with yeast Hsp90 in a nucleotide-dependent manner and then determined the effect of Hsp90 mutation on these interactions.
Analysis of Hsc82 mutations.
Saccharomyces cerevisiae contains two isoforms of Hsp90 that are largely assumed to be functionally identical, constitutively expressed Hsc82 and heat-inducible Hsp82 (97% identical at the amino acid level). Deletion of genes encoding both isoforms results in a lethal phenotype (3). hsp82 mutations that cause temperature-sensitive growth exhibit defects in Hsp90 client protein activity, but the effect of hsc82 mutation has not been described. We constructed mutant forms of Hsc82 containing an N-terminal His6 tag. Most of the mutants we tested have previously been analyzed in the context of Hsp82 (12, 15, 20, 23-25) (Table 1). This set of mutations includes residues in the ATPase domain (T22I, E33A, D79N, T101I, A107N), a flexible loop in the middle domain (N373A, R376A, E377K, and Q380A), a hydrophobic patch located near the flexible loop (F345A), a potential client-binding site (W296A and F325A), and another region of the middle domain (G309S). Three additional mutations cluster in the general vicinity of the carboxy-terminal dimerization interfaces (2). S481Y and T521I were previously studied in the context of Hsp82; we isolated the third mutation, L487S, in a STI1 synthetic lethal screen (9). We also targeted residues in the dimerization interface: A583T, I588A M589A, F660A, and L647S L648S (2, 11, 40). Finally, we deleted the binding site for TPR-containing cochaperones, such as Sti1 and Cpr6 (ΔMEEVD) (16, 34).
TABLE 1.
Hsc82 mutants used in this work
| Mutation | Location of mutation | Growth phenotype of hsc82 hsp82 straina
|
Reference(s) (protein) | |
|---|---|---|---|---|
| 30°C | 37°C | |||
| None | ++++ | ++++ | 2 | |
| T22I | ATPase | − | − | 23 (Hsp82-T22I) |
| E33A | ATPase | − | − | 24, 25 (Hsp82-E33A) |
| D79N | ATPase | − | − | 24, 25 (Hsp82-D79N) |
| T101I | ATPase | − | − | 23 (Hsp82-T101I) |
| A107N | ATPase | ++++ | +++ | 29 (Hsp82-A107N) |
| W296A | Middle | ++ | − | 20 (Hsp82-W300A) |
| G309S | Middle | ++++ | − | 23 (Hsp82-G313S) |
| F325A | Middle | ++++ | +++ | This study |
| F345A | Middle | ++++ | − | 20 (Hsp82-F349A) |
| N373A | Middle | ++++ | ++ | 20 (Hsp82-N377A) |
| R376A | Middle | − | − | 20 (Hsp82-R380A) |
| E377K | Middle | − | − | 23 (Hsp82-E381K) |
| Q380A | Middle | ++++ | − | 20 (Hsp82-Q384A) |
| S481Y | Middle | ++++ | − | 15 (Hsp82-S485Y) |
| L487S | Middle | ++++ | − | 10; this study |
| T521I | Middle | ++++ | − | 15 (Hsp82-T525I) |
| A583T | C terminus | ++++ | − | 23 (Hsp82-A587T) |
| I588A M589A | C terminus | ++++ | − | This study |
| L647S L648S | C terminus | ++ | − | 40; this study |
| F660A | C terminus | ++++ | ++++ | This study |
| ΔMEEVD | C terminus | ++++ | ++++ | 16 (Hsp82-ΔMEEVD) |
WT growth. Growth defects correspond to an approximate 10-fold (+++) and 100-fold (++) reduction in colony numbers observed upon serial dilation growth assays.
To assess the ability of mutant Hsc82 to support the viability of an hsc82 hsp82 strain when present as the only Hsp90 protein in the cell, plasmids expressing a wild-type (WT) or mutant form of Hsc82 were transformed into strain JJ816 (hsc82 hsp82/Yep24-HSP82). Resultant colonies were grown in the presence of 5-FOA, which counterselects for the plasmid expressing WT HSP82. Six alterations, T22I, E33A, D79N, T101I, R376A, and E377K, resulted in a lethal phenotype, and the growth of strains expressing remaining mutants is listed in Table 1. All mutant proteins were expressed at levels similar to that of WT His-Hsc82 (not shown). The growth phenotypes with hsc82 mutant alleles mirrored those with the respective hsp82 alleles except that hsc82-T22I, -T101I, and -E377K conferred a lethal phenotype, while the corresponding mutations in HSP82 conferred temperature-sensitive growth (23). Because the T22I, E33A, D79N, T101I, R376A, and E377K alterations disrupted essential functions, these His6-tagged mutants were coexpressed along with untagged WT HSP82 in the following studies. In a prior study, strong dominant-negative growth effects were observed upon coexpression of WT and mutant HSP82 alleles (25). In contrast, we observed only slight growth defects at 30°C or 37°C upon coexpression of WT HSP82 and mutant forms of HSC82 (not shown).
In vitro assembly of His-Hsc82 complexes.
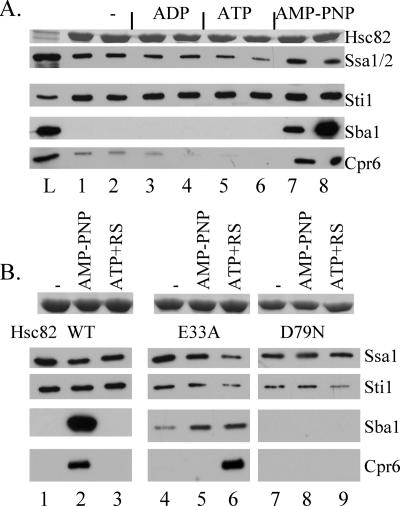
His-Hsp82 complexes were previously shown to contain Sti1, the Hsp70 Ssa1/2, and Cpr6 (4), and Hsp82 was shown to stably interact with Sba1 in the presence of AMP-PNP (8). We focused on the Hsc82 isoform because mutant forms of His-Hsp82 were inconsistently retained by the nickel resin (not shown). To examine the effect of nucleotides on His-Hsc82 complexes, yeast cells expressing WT His-Hsc82 as the only Hsp90 protein in the cell were lysed in a nondenaturing buffer. Cell extracts were supplemented with no exogenous nucleotide, ADP, ATP, or AMP-PNP (5 mM, final concentration) and then incubated on ice or at 30°C for 5 min (Fig. 1A, lanes 1 to 8). Cell lysates were incubated with nickel resin and washed with intermediate concentrations of imidazole to reduce levels of nonspecifically bound proteins. Proteins bound to the nickel resin were analyzed by SDS-PAGE followed by Coomassie blue staining (upper panel) or immunoblot analysis (lower panels). Hsc82 and associated proteins were specifically retained by nickel resin only in the presence of His-Hsc82 (not shown; also see Fig. 6). Similar levels of His-Hsc82 and Sti1 copurified under all conditions, while the recovery of Ssa1/2 was slightly reduced in the presence of ATP (lanes 5 and 6). As expected, stable Sba1 interaction was observed only in the presence of AMP-PNP (8). Interaction of Cpr6 with Hsc82 was dramatically increased in the presence of AMP-PNP, although some Cpr6 interaction was observed in the absence of a nucleotide (lanes 1 and 2), which is consistent with prior reports (17). Specific interaction of Sba1 and Cpr6 with His-Hsc82 in the presence of AMP-PNP was also observed when lysate was dialyzed to remove endogenous nucleotides prior to addition of a nucleotide (not shown). These studies indicate that a complex between Sba1, Cpr6, and Hsc82 forms in the presence of ATP, which is similar to a complex observed in mammalian cell lysates (14). We subsequently examined the composition of His-Hsc82 complexes after incubation of undialyzed cell extracts at 30°C for 5 min, since maximal Sba1 interaction was observed under these conditions (Fig. 1A, lane 8).
FIG. 1.
Interaction of cochaperone proteins with WT His-tagged Hsc82 and His-Hsc82 containing alterations of residues required for ATP binding and hydrolysis. A. Cell extracts were prepared from cells expressing His-Hsc82 as the only Hsp90 protein in the cell and supplemented with no exogenous nucleotide (lanes 1 and 2), 5 mM ADP (lanes 3 and 4), 5 mM ATP (lanes 5 and 6), or AMP-PNP (lanes 7 and 8). His-Hsc82 complexes were isolated after a 5-min incubation on ice (odd-number lanes) or at 30°C (even-number lanes). L, whole-cell extract. B. Cell extracts were prepared from cells expressing His-Hsc82 WT, -E33A, or -D79N along with WT untagged Hsp82. His-Hsc82 complexes were isolated from lysates incubated for 5 min at 30°C in the presence of no exogenous nucleotide (lanes 1, 4, and 7), 5 mM AMP-PNP (lanes 2, 5, and 8), or 5 mM ATP plus an ATP-regenerating system (ATP+RS, lanes 3, 6, and 9). Lanes 1 to 3, WT His-Hsc82; lanes 4 to 6, His-Hsc82-E33A; lanes 7 to 9, His-Hsc82-D79N. Nickel resin-bound protein complexes were separated by SDS-PAGE followed by Coomassie blue staining or immunoblot analysis. The Coomassie blue-stained band corresponding to His-Hsc82 is shown in the upper panel, and the lower panels represent immunoblot analysis using antibodies against the indicated proteins.
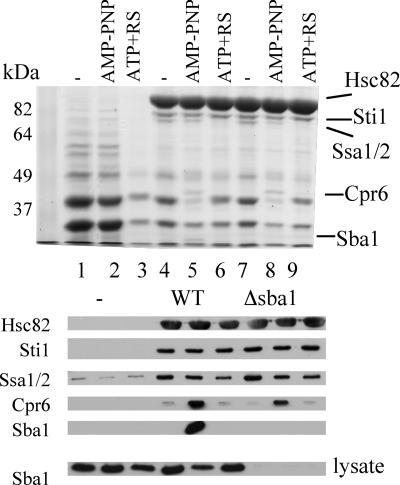
FIG. 6.
Cpr6 interacts with His-Hsc82 in a strain lacking Sba1. Untagged Hsc82 (lanes 1 to 3) or His-Hsc82 (lanes 4 to 6) was expressed in strain JJ816 (hsc82 hsp82). In lanes 7 to 9, His-Hsc82 was expressed in strain JJ40 (hsc82 hsp82 sba1). Cell lysates were isolated and supplemented with a nucleotide as described in the legend to Fig. 1: lanes 1, 4, and 7, no exogenous nucleotide; lanes 2, 5 and 8, 5 mM AMP-PNP; lanes 3, 6, and 9, 5 mM ATP plus an ATP regenerating system. For the upper panel, nickel resin-bound protein complexes were separated by SDS-PAGE (7.5% acrylamide) followed by staining with Coomassie blue. For the lower panels, protein complexes were subjected to SDS-PAGE followed by immunoblot analysis using antibodies against the indicated proteins. In the panel marked “lysate,” whole-cell extract was separated by SDS-PAGE and immunoblotted with an antibody specific for Sba1 to confirm the lack of expression of Sba1 in the hsc82 hsp82 sba1 strain.
The ATPase activity of Hsp82 is essential, and two mutations known to disrupt this activity are Hsp82-E33A, which disrupted ATP hydrolysis, and Hsp82-D79N, which disrupted ATP binding. Purified WT Hsp82 stably interacted with Sba1 only in the presence of the nonhydrolyzable ATPγS, Hsp82-E33A interacted with Sba1 in the presence of either ATP or ATPγS, and Hsp82-D79N was unable to bind Sba1 (24, 25). We constructed the homologous mutations in His-Hsc82 (Hsc82-E33A and Hsc82-D79N) and examined their effect on cochaperone interactions. Because both mutations disrupt the essential in vivo functions of Hsc82 (Table 1), this experiment was conducted with cells coexpressing WT untagged Hsp82 (Fig. 1B). His-Hsc82 complexes were isolated as described above. The amount of Ssa1/2 recovered in complex with mutant Hsc82 was essentially unchanged relative to that with WT Hsc82, while Sti1 exhibited slightly reduced binding to E33A and D79N Hsc82 under all conditions. WT His-Hsc82 bound Sba1 and Cpr6 only in the presence of AMP-PNP. In contrast, Sba1 bound His-Hsc82-E33A under all conditions tested but did not bind Hsc82-D79N under any conditions (Fig. 1B, lanes 4 to 9). Surprisingly, stable interaction of Cpr6 with E33A Hsc82 was observed in the presence of ATP+RS but not AMP-PNP, suggesting that the nucleotides have differential effects on Hsc82 conformation.
Although the E33A and D79N alterations similarly disrupt the in vivo functions of either Hsc82 or Hsp82, additional studies will be required to determine whether these and additional amino acid alterations have similar effects on the enzymatic activities of the two isoforms of yeast Hsp90. However, nucleotide-dependent assembly of Hsc82 with Sba1 and Cpr6 was prevented by the D79N alteration, consistent with a defect in ATP binding. In addition, the pattern of interaction of Sba1 with Hsc82-E33A in our assay was similar to that observed with purified Sba1 and Hsp82-E33A (24), suggesting that the Hsc82-E33A mutation has the predicted effect of inhibiting ATP hydrolysis. Finally, although the presence of heterodimers between WT Hsp82 and mutant Hsc82 cannot be excluded, dramatic differences between WT and mutant forms of Hsc82 may be observed despite the presence of WT Hsp82 in cell extracts.
Effects of mutations predicted to affect lid closure and N-terminal dimerization.
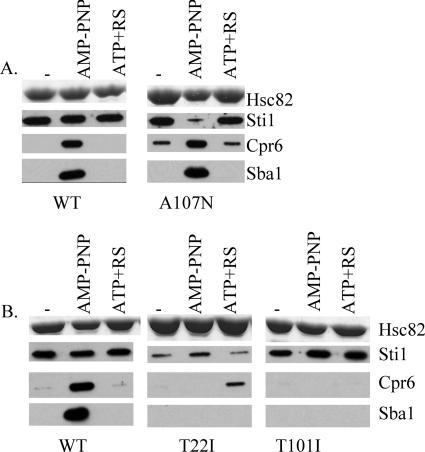
Structural studies indicate that nucleotide binding to Hsp82 is followed by closing of a lid over a bound nucleotide, association of N-terminal domains, and stabilized Sba1 interaction (2, 27). We constructed mutations in Hsc82 that were predicted, based on analysis of homologous Hsp82 mutants, to affect lid closing or association of N-terminal domains. Residue A107 of Hsp82 is located within the lid segment that closes over the mouth of the nucleotide-binding pocket upon ATP binding (2). The A107N alteration was predicted to stabilize lid closure, and purified Hsp82-A107N exhibited enhanced N-terminal dimerization and ATPase activity (29). We monitored the interaction of Hsc82-A107N with cochaperones (Fig. 2A). The interaction of Hsc82-A107N with Sba1 was unaltered. However, the interaction of Sti1 with Hsc82-A107N was specifically reduced in the presence of AMP-PNP, and significant levels of Cpr6 binding to Hsc82-A107N were observed in the absence of nucleotides and in the presence of ATP+RS. This suggests that while Hsc82 is in the closed conformation, Sti1 interaction is weakened and Cpr6 interaction is enhanced. This result is consistent with prior evidence that a mutant form of Hsp82 exhibited weaker binding to Sti1 in the ATP-bound form (31).
FIG. 2.
Effect of mutations predicted to affect lid closure and N-terminal dimerization. A. Cell extracts were prepared from yeast expressing His-Hsc82 WT or His-Hsc82-A107N. His-Hsc82 complexes were isolated from lysates incubated for 5 min at 30°C in the presence of no exogenous nucleotide, 5 mM AMP-PNP, or 5 mM ATP plus an ATP-regenerating system as indicated. Nickel-bound protein complexes were separated by SDS-PAGE and analyzed as described in the legend to Fig. 1. B. As above, except that WT His-Hsc82, His-Hsc82-T22I, or His-T101I was isolated from cells coexpressing WT untagged Hsp82, since hsc82-T22I and hsc82-T101I confer a lethal phenotype.
Next we examined the effect of mutations that affect N-terminal dimerization. In Hsp82, residue T22 is part of the N-terminal dimerization interface and also makes contact with residues in the catalytic loop. The T22I alteration was predicted to stabilize these contacts, and the Hsp82-T22I mutation displayed enhanced AMP-PNP-dependent N-terminal dimerization and ATPase activity (2, 29). In contrast, the Hsp82-T101I mutant, predicted to stabilize the open conformation, displayed reduced AMP-PNP-dependent N-terminal dimerization (29). We analyzed the interaction of Hsc82-T22I and Hsc82-T101I with cochaperones. Because each of these alterations disrupted the in vivo functions of Hsc82 (Table 1), they were coexpressed along with WT untagged Hsp82. Hsc82-T22I exhibited reduced Sti1 interaction, consistent with a weakened interaction of Sti1 with the closed conformation. As observed for Hsc82-E33A, Hsc82-T22I interacted with Cpr6 in the presence of ATP but not AMP-PNP, and surprisingly, Hsc82-T22I exhibited no Sba1 interaction (Fig. 2B). In contrast, consistent with a defect in obtaining the closed position, Hsc82-T101I exhibited WT levels of Sti1 interaction but failed to bind either Cpr6 or Sba1.
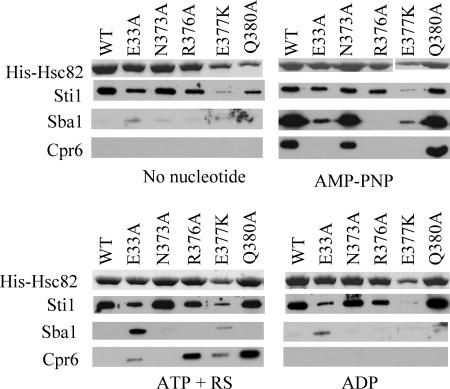
Another structural change upon ATP binding is docking of a flexible catalytic loop from the middle domain (residues 370 to 390) with the mouth of the nucleotide-binding pocket. Within the nucleotide-bound structure, Hsp82-R380 is located close to the catalytic residue E33, and the side chain of Hsp82-R380 makes a polar interaction with the γ-phosphate of bound AMP-PNP (2). Purified Hsp82-R380A and Hsp82-Q384A exhibited minimal ATPase activity, indicating a critical role for this loop in ATP hydrolysis (20).
We examined the effect of alterations of residues within the catalytic loop (Fig. 3). The biochemical defects of the Hsc82 mutants have not been determined, but the known biochemical properties of homologous mutations in Hsp82 are listed in Table 2. The hsc82-E33A, hsc82-R376A, and hsc82-E377K alterations caused a lethal phenotype and thus were coexpressed with WT Hsp82. Each of the other mutants analyzed was the only Hsp90 protein present in the cell. First we will focus on the interaction of Sti1 and Sba1 with these mutants. The E377K mutant exhibited reduced binding to Sti1 under all conditions, while the E33A mutant exhibited slightly reduced Sti1 interaction, as shown in Fig. 1. The E377K mutant also displayed a pattern of Sba1 interaction similar to that of the E33A mutant, with both mutants displaying stable, albeit reduced, Sba1 interaction in the presence of AMP-PNP or ATP+RS. The only other mutant with altered Sba1 interaction was the R376A mutant, which failed to interact with Sba1.
FIG. 3.
Effect of alteration of residues in the catalytic loop. Cell lysates from strains expressing indicated His-Hsc82 mutants were isolated, supplemented with a nucleotide, and incubated as described for Fig. 1, except that the effect of 5 mM ADP was also monitored. His-Hsc82 mutants unable to support viability of an hsc82 hsp82 strain (E33A, R376A, and E377K mutants) were coexpressed along with untagged WT Hsp82. In the remaining cases (WT and N373A and Q380A mutants), His-Hsc82 was the only Hsp90 protein expressed in the cell. Nickel-bound protein complexes were separated by SDS-PAGE. Upper panels, His-Hsc82 present in a Coomassie blue-strained gel. Lower panels, immunoblots using antibodies specific for Sti1, Cpr6, or Sba1.
TABLE 2.
Known properties of Hsp82 mutants and comparison with Hsc82 mutant interactionsa
| Hsp82 mutation | Relative ATPase activity (reference) | Sti1 interaction (reference) | Sba1 interaction (reference) | Hsc82 mutation | Interaction
|
||
|---|---|---|---|---|---|---|---|
| Sti1 | Sba1 | Cpr6 | |||||
| T22I | Enhanced (29) | WT (36) | WT (36) | T22I | ↓ | ↓ | +* |
| E33A | Decreased (24, 25) | ND | WT* (24) | E33A | + | +* | +* |
| D79N | No ATP binding (24, 25) | ND | ↓ (24) | D79N | + | ↓ | ↓ |
| T101I | Decreased (29) | WT (36) | ↓ (36) | T101I | + | ↓ | ↓ |
| A107N | Enhanced (29) | WT (36) | WT (36) | A107N | ↓ | + | +* |
| W300A | ∼WT (20) | ND | WT (12) | W296A | ↓ | + | +* |
| G313S | ND | ND | ND | G309S | + | + | + |
| F329A | ND | ND | ND | F325A | ↓ | + | +* |
| F349A | Decreased (20) | WT (36) | ↓ (36) | F345A | + | ↓ | ↓ |
| N377A | ∼WT (20) | ND | ND | N373A | + | + | + |
| R380A | Decreased (20) | ND | ND | R376A | + | ↓ | +* |
| E381K | ∼WT (20) | ND | ND | E377K | ↓ | +* | +* |
| Q384A | Decreased (20) | ND | ND | Q380A | + | + | +* |
| S485Y | Decreased (12) | ND | ↓ (8, 12) | S481Y | + | ↓ | ↓ |
| L491S | ND | ND | ND | L487S | + | ↓ | ↓ |
| T525I | Decreased (12) | ND | ↓ (8, 12) | T521I | + | ↓ | ↓ |
| A587T | WT (29) | ND | ND | A583T | + | ↓ | ↓ |
| I592A M593A | ND | ND | ND | I588A M589A | + | ↓ | ↓ |
| L651S L652S | ND | ND | ND | L647S L648S | + | ↓ | ↓ |
| F664A | ND | ND | ND | F660A | + | ↓ | ↓ |
| ΔMEEVD | ND | ↓(1) | ND | ΔMEEVD | ↓ | + | ↓ |
*, nucleotide dependence of the interaction was altered; +, WT level of interaction observed; ↓, reduced interaction relative to WT Hsc82.
Other than the N373A mutant, which exhibited a WT pattern of Cpr6 interaction, catalytic loop mutants had altered Cpr6 interaction. In the presence of AMP-PNP, only the N373A and Q380A mutants bound Cpr6. However, as observed for the E33A and T22I mutants, the R376A, E377A and Q380A mutants were able to interact with Cpr6 in the presence of ATP+RS. This result suggests that the conformation of the catalytic loop and nearby residues, such as T22 (2), is a key determinant of Cpr6 interaction.
Effect of additional hsc82 mutations on cochaperone interaction.
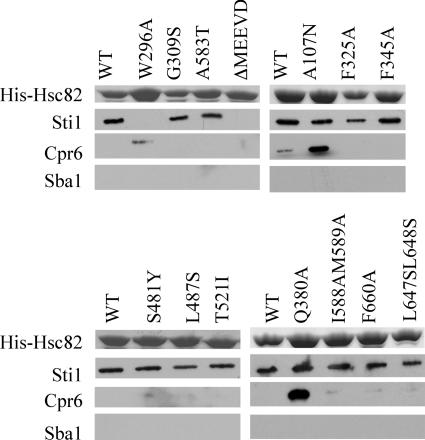
Next we monitored the effect of mutations outside the N-terminal domain or catalytic loop on Sti1, Cpr6, and Sba1 interaction. All of the mutants shown in Fig. 4 and 5 were able to support viability of the hsc82 hsp82 strain (Table 1) and were expressed as the only Hsp90 protein in the cell. First we examined the interactions of WT and mutant His-Hsc82 with cochaperones in the presence of AMP-PNP (Fig. 4). Three mutants besides the A107N mutant showed altered interaction with Sti1. Hsc82-ΔMEEVD was unable to bind Sti1 due to loss of the TPR binding site (1, 34), while Hsc82-W296A and -F325A exhibited reduced Sti1 interaction. With the exception of Hsc82-ΔMEEVD, which was able to bind Sba1 but not Cpr6, the remaining mutants displayed similar patterns of interaction with Sba1 and Cpr6. The W296A, G309S, F325A, and Q380A mutants were able to bind both Sba1 and Cpr6 and thus appear to be able to adopt the closed, ATP-bound conformation. The remaining mutants exhibited reduced or no interaction with Sba1 and Cpr6. This group includes mutations in both the middle domain, F345A, S481Y, L487S, and T521I, and the carboxy-terminal domain, A583T, I588A M589A, F660A, and L647S L648S.
FIG. 4.
Effect of additional Hsc82 mutations on cochaperone interaction in the presence of AMP-PNP. WT and mutant His-Hsc82 complexes were isolated and analyzed as described in the legend to Fig. 3 except that lysate was supplemented with 5 mM AMP-PNP. In all cases, WT or mutant His-Hsc82 was the only Hsp90 protein present in the cell.
FIG. 5.
Interaction of mutant Hsc82 with Sti1, Sba1, and Cpr6 in the presence of ATP plus an ATP-regenerating system. His-Hsc82 complexes were isolated and analyzed as for Fig. 4, except that samples were supplemented with 5 mM ATP+RS. In all cases, WT or mutant His-Hsc82 was the only Hsp90 protein present in the cell.
We examined the same set of mutants under conditions that support ATP hydrolysis (Fig. 5). Under these conditions, the W296A mutant still exhibited a defect in Sti1 interaction, while the A107N and F325A mutants displayed WT Sti1 interaction. Sba1 interaction was not observed for any mutants, suggesting that Sba1 release was not affected. Finally, as observed with the A107N and Q380A mutants, discussed earlier, the W296A mutant exhibited enhanced interaction with Cpr6 in the presence of ATP+RS.
Interaction with Cpr6 is not dependent on Sba1 interaction.
Sba1 interaction with Hsc82 is not dependent on stable Sti1 or Cpr6 interaction, since His-Hsc82-ΔMEEVD exhibited a WT pattern of Sba1 interaction. In order to determine if Sba1 was required for Cpr6 interaction, we examined whether Cpr6 interacted with His-Hsc82 in a strain lacking Sba1 (Δsba1 hsc82 hsp82). As shown in Fig. 6, no Sba1 binding to Hsc82 was observed in the SBA1 deletion strain (lanes 7 to 9). However, the pattern of Cpr6 binding to WT His-Hsc82 was unaffected (lower panels, compare lanes 4 to 6 with lanes 7 to 9). Thus, Cpr6 and Sba1 independently bind the ATP-bound form of Hsc82.
As shown in Fig. 6, we also demonstrated that retention of Hsc82 and cochaperones by nickel resin was dependent on the presence of His-Hsc82. Yeast lysates were prepared from cells expressing untagged WT Hsc82 (lanes 1 to 3) or His-Hsc82 (lanes 4 to 9), supplemented with a nucleotide, bound to nickel resin, and analyzed by SDS-PAGE followed by Coomassie blue staining (upper panel) or immunoblot analysis (lower panels). As seen in the stained gel, the predominant copurifying protein is Sti1, and the Hsp70 Ssa1/2 migrates just below Sti1. In the absence of His-Hsc82 (lanes 1 to 3), background binding of two predominant unidentified proteins that migrate at approximately 30 and 40 kDa was observed. In the presence of His-Hsc82, the binding levels of these unidentified proteins were reduced, and significant amounts of Hsc82, Sti1, and Ssa1/2 were retained only in the presence of His-Hsc82. The levels of copurifying Sti1 were similar under the three conditions, while the level of Ssa1/2 was slightly reduced in the presence of ATP+RS. Upon comparison of His-Hsc82 complexes isolated in the presence of no nucleotide, AMP-PNP, or ATP+RS (lanes 4 to 6), two protein bands are observed only in the presence of AMP-PNP (lane 5), with one migrating at approximately 45 kDa and another vary faint band migrating just above the dye front. The size of these bands, coupled with the pattern of Sba1 and Cpr6 obtained by immunoblot analysis (lower panels), strongly suggests that these bands correspond to Cpr6 (42 kDa) and Sba1 (24 kDa), particularly since the smaller band did not copurify with His-Hsc82 isolated out of a sba1 strain (lane 8; also data not shown). We did not observe specific interaction of additional proteins in the presence of AMP-PNP, which suggests that there are not other predominant Hsp90 cochaperones that stably and specifically interact with His-Hsc82 in the ATP-bound, closed conformation.
DISCUSSION
Our studies provide an outline for how nucleotides modulate the interaction of Hsp90 with Sti1, Sba1, and Cpr6. We observed nucleotide-dependent interaction of Hsc82 with Sba1 (yeast p23) and Cpr6 (yeast Cyp40), which is similar to a complex observed in mammalian cell extracts (14). As evidence of the specificity of these interactions, a mutant form of Hsc82 predicted to be unable to bind a nucleotide, D79N (24, 25), was unable to bind Cpr6 or Sba1. We further demonstrated that Sba1 and Cpr6 independently interact with yeast Hsp90 and that mutations throughout Hsp90 have differential effects on the nucleotide-dependent interactions of Sti1, Cpr6, and Sba1.
Identification of Hsc82 mutations with altered Sti1 interaction.
The primary site of Sti1 interaction is the TPR acceptor site at the carboxy terminus of Hsp90 (34), but multiple lines of evidence suggest that Sti1 has additional contacts with Hsp90: Hop (mammalian Sti1) exhibited reduced binding to Hsp90 lacking part of the N-terminal domain (6); binding of Sti1 to Hsp82 inhibited ATPase activity and displaced bound geldanamycin from the ATP-binding pocket (30); and Sti1 was shown to prevent N-terminal dimerization of Hsp82 (31). Although WT Hsc82 bound Sti1 under all conditions, our results support a prior study that demonstrated that Sti1 has a weakened interaction with the ATP-bound form of a mutant Hsp90 protein (31). Two Hsc82 mutants predicted to favor the closed conformation displayed reduced Sti1 interaction: the Hsc82-A107N mutant exhibited a specific defect in the presence of AMP-PNP, while the T22I mutant exhibited reduced overall interaction. Two additional Hsc82 mutants also exhibited reduced Sti1 interaction in the presence of AMP-PNP: the W296A mutant, which caused a dramatic reduction, and Hsc82-F325A, which resulted in slightly reduced Sti1 interaction. Within the isolated middle domain of Hsp82, the homologous residues W300 and F329 are located within an exposed hydrophobic patch proposed to play a role in protein-protein interactions (20). A mutation in the catalytic loop, E377K, also resulted in reduced Sti1 interaction under all conditions. In a prior study (36), purified Sti1 did not exhibit reduced interaction with Hsp82-A107N, -T22I, -T101I, or -F349A, but those studies were conducted in the absence of a nucleotide, and with the exception of the T22I mutant, we did not observe reduced Sti1 interaction under those conditions. Further studies will be required to determine whether these differences are isoform specific or whether the presence of a nucleotide results in reduced interaction of purified Sti1 with Hsc82/Hsp82 containing these alterations. Further studies will be required to determine whether the exposed hydrophobic patch containing Hsc82 residues W296 and F325 is a second site of contact between Hsp90 and Sti1.
Identification of Hsc82 mutations with altered Sba1 interaction.
Binding of ATP to Hsp90 induces the closed conformation characterized by Sba1 interaction with the N-terminal domains (2). Our results suggest that Sba1 interaction is specifically linked to ATP binding and hydrolysis. With the exception of the E33A mutant, Sba1 interaction was not observed in the absence of a nucleotide, and only two Hsc82 mutants, the E33A and E377K mutants, demonstrated stable Sba1 interaction in the presence of ATP. Hsc82-E33A is predicted to have a defect in ATP hydrolysis (24, 25), and E377 is located within the catalytic loop (20), suggesting the E377K alteration may affect hydrolysis. Further studies will be required to determine if these mutations cause specific defects in ATP hydrolysis or Sba1 release after ATP hydrolysis. As expected, Hsc82-T101I, predicted to favor the open conformation (29), did not interact with Sba1. Two additional mutants that disrupted Sba1 interaction, the T22I and R376A mutants, are located near contact sites between Hsp90 and Sba1 (2), and thus, it is possible that these alterations cause local conformational changes that disrupt Sba1 interaction. The remaining mutations that disrupted Sba1 interaction (F345A, S481Y, L487S, T521I, A583T, I588A M589A, L647S L648S, and F660A) are located outside the vicinity of N-terminal domain and thus are not expected to directly affect nucleotide binding. It seems likely that these mutations disrupt Sba1 interaction either by restricting the conformational changes required to adopt the closed conformation or by stabilizing alternative conformations of Hsp90. These results are generally consistent with prior studies of Hsp82-Sba1 interaction, since the T101I, F349A, S485Y, and T525I mutations in Hsp82 were shown to disrupt Sba1 interaction and Hsp82-S485Y and Hsp82-T525I exhibit reduced ATPase activity (8, 12, 36). In contrast, purified Hsp82-T22I did not exhibit a defect in Sba1 interaction (36). Additional studies will be required to determine if this is due to isoform-specific differences or assay-specific differences.
Identification of Hsc82 mutations with altered Cpr6 interaction.
A number of Hsc82 mutations similarly disrupted the interactions of both Sba1 and Cpr6 (Table 2). However, some mutations that did not disrupt Sba1 interaction altered Cpr6 interaction, indicating that the two cochaperones have distinct requirements for binding to Hsc82. Since the only known Cpr6 binding site is located in the carboxy-terminal domain, the signal of ATP binding and associated conformational changes in the N terminus must be transmitted to the carboxy-terminal domain. Mutations predicted to alter nucleotide-induced conformational changes apparently disrupted transmission of the signal of nucleotide interaction to the carboxy terminus. A few mutants (E33A, T22I, R376A, and E377K) stably bound Cpr6 in the presence of ATP but not AMP-PNP. A prior study suggested that AMP-PNP is not as efficient as ATP in inducing conformational changes in Hsp90 (32). Since the affected residues are located in or near the catalytic loop (2), it seems probable that conformational changes in the loop are not as efficient in the presence of AMP-PNP and that this effect is exacerbated by the mutations we tested. Further studies will be required to establish that these changes correspond to a proposed rate-limiting conformational change in Hsp90 that is required for ATP hydrolysis (18, 19).
Cpr6 binds Hsp90 in the closed conformation, and the A107N mutation, which favors the closed conformation and exhibits enhanced ATPase activity (29), bound low levels of Cpr6 both in the absence of an exogenous nucleotide and in the presence of ATP. Similar results were obtained with W296A (Fig. 4 and 5; also not shown), suggesting that W296A also favors the closed conformation. Neither of these mutants exhibited altered Sba1 binding. Thus, is appears that these mutants have a specific defect in Cpr6 release or the return of Hsp90 to the open conformation after ATP hydrolysis, although it is also possible that altered patterns of Cpr6 interaction reflect decreased interaction of these mutants with Sti1.
In summary, our results indicate that mutations in the catalytic loop, lid, or N-terminal dimerization domain result in altered specificity of the Cpr6-Hsc82 interaction. Since mutations in chick Hsp90 containing deletions of the amino terminus or catalytic loop resulted in altered patterns of interaction of multiple TPR-containing proteins (6), it is possible that these conformational changes regulate Hsp90 interactions with multiple TPR-containing cochaperones.
Model for nucleotide-dependent interactions of Hsc82 with cochaperone proteins.
Our results extend existing models of interaction of Hsp90 with Sti1, Sba1, and Cpr6 (27, 31, 32) to include novel information about how mutation of Hsc82 affects cochaperone interactions and conformational changes (Fig. 7). In the absence of a nucleotide, Sti1 interacts with both the TPR acceptor site at the carboxy terminus and a second site in or near the ATPase domain, possibly involving residue Hsc82-W296. Nucleotide binding results in a shift from the open conformation to the closed conformation, weakened Sti1 interaction, and enhanced Sba1 interaction. Formation of a stable Hsc82-Sti1-Sba1 complex has not been observed, and there is evidence for competitive interactions of Sba1 and Sti1 (32). Thus, it seems likely that Sti1 dissociates upon Sba1 interaction. A conformational change involving the catalytic loop results in stable Cpr6 interaction. Sba1 appears to be released upon ATP hydrolysis, but Cpr6 release and Sti1 binding appear to require additional steps as Hsp90 returns to the open conformation. This cycle between the open and closed forms of Hsp90 is disrupted by mutations in or near the carboxy terminus of Hsp90.
FIG. 7.
Model of Hsc82 interaction with Sba1 and Cpr6. Our results suggest the presence of two intermediate complexes during the ATPase cycle: Sba1 interaction prior to Cpr6 interaction (as observed with the E33A and E377K mutants) and Sba1 release prior to Cpr6 release (as observed with the A107N and W296A mutants). See the text for details.
These conformational switches likely have dramatic effects on Hsp90 activity. While bound to the open conformation of Hsp90, Sti1 inhibits N-terminal dimerization and ATPase activity (30, 31). The Hsc82-Sba1-Cpr6 complex, which interacts with client proteins late in the folding pathway (14, 28, 39), was proposed to be the complex capable of hydrolysis (32). However, purified Sba1 inhibited the ATPase activity of purified Hsp82, although the effects were variable (18, 26, 32, 41). Because we observed interaction of Hsc82-E33A and -E377K with Sba1 but not Cpr6 in the presence of AMP-PNP, we propose an intermediate step in which Sba1 interacts with Hsc82 prior to interacting with Cpr6 (Fig. 7). Sequential interaction of Sba1 and Cpr6 suggests that Sba1 may inhibit activity of Hsp90 until formation of the specific Hsc82-Sba1-Cpr6 ternary complex.
Conformational changes in the N-terminal domain, such as dimerization, lid closing, and catalytic loop repositioning, appear to regulate Sti1, Cpr6, and Sti1 interaction. N-terminal dimerization and conformational changes in the catalytic loop also regulate the interaction of Hsp90 with the cochaperones Cdc37 and Aha1, respectively (21, 33). Additional studies will be required to determine how these mutations affect the direct interaction of Hsp90 with these and other Hsp90 cochaperone proteins. In addition, it is important to note that the interaction of Hsp90 with cochaperone proteins is very dynamic, and there are likely distinct complexes at different stages of the ATPase cycle, which may explain why we do not always see reduced Sti1 interaction in conjunction with enhanced Cpr6 interaction.
Relationship of cochaperone interaction to overall Hsp90 function.
Our study provides novel information linking the conformational changes that occur during the ATPase cycle of Hsp90 to cochaperone interactions. For simplicity we included only three cochaperones here, and studies are under way to monitor the interaction of additional cochaperones with WT and mutant forms of Hsp90. It will be interesting to determine whether Hsc82-G309S, which caused a temperature-sensitive phenotype but did not exhibit defects in interaction with Sti1, Sba1, or Cpr6, has defects in interaction with other cochaperone proteins. In addition, we are curious about how the interaction of cochaperone proteins is affected by client protein interaction and vice versa. A recent report suggests that the stoichiometry of the Hsp90-Cdc37 interaction changes in the presence of the kinase Cdk4 (37), and thus, it will be important to incorporate assays of client interaction into our studies to determine the effect on cochaperone interaction.
The mechanism by which Hsp90 and cochaperones recognize and interact with diverse client proteins in order to mediate their folding and regulation is becoming clearer as structural studies indicate the range of conformational changes in Hsp90 (2, 7, 13). The location of the client-binding site has been elusive, but a recent report characterized a complex between Hsp90, the cochaperone Cdc37, and the Cdk4 kinase. In this complex, both the N-terminal and middle domains of Hsp90 contact the Cdk4 kinase (37). Intriguingly, one site of Cdk4 interaction was centered on Hsp82-W300. Since mutation of homologous Hsc82-W296A caused reduced Sti1 interaction, analysis of this mutation may provide valuable information about the Sti1-mediated transfer of client proteins from Hsp70 to Hsp90 and the ability of Hsp90 to couple nucleotide-induced conformational changes in Hsp90 to client protein activation.
Acknowledgments
We thank Elizabeth Craig, Susan Lindquist, Avrom Caplan, and Brian Freeman for reagents. We also thank David Toft and Elizabeth Craig for helpful advice pertaining to the manuscript.
This publication was made possible by grant no. P20 RR15587 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). This project was also funded in part by an American Cancer Society seed grant through Washington State University, IRG-77-003-26.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Abbas-Terki, T., O. Donze, P. A. Briand, and D. Picard. 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21:7569-7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, M. M., S. M. Roe, C. K. Vaughan, P. Meyer, B. Panaretou, P. W. Piper, C. Prodromou, and L. H. Pearl. 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440:1013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, H. C., D. F. Nathan, and S. Lindquist. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, S., and D. F. Smith. 1998. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 273:35194-35200. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., W. P. Sullivan, D. O. Toft, and D. F. Smith. 1998. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones 3:118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dollins, D. E., R. M. Immormino, and D. T. Gewirth. 2005. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J. Biol. Chem. 280:30438-30447. [DOI] [PubMed] [Google Scholar]
- 8.Fang, Y., A. E. Fliss, J. Rao, and A. J. Caplan. 1998. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18:3727-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flom, G., J. Weekes, and J. L. Johnson. 2005. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 47:368-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flom, G., J. Weekes, J. J. Williams, and J. L. Johnson. 2006. Effect of mutation of the tetratricopeptide repeat and asparatate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics 172:41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, S. F., A. K. Shiau, and D. A. Agard. 2004. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure (Cambridge) 12:1087-1097. [DOI] [PubMed] [Google Scholar]
- 12.Hawle, P., M. Siepmann, A. Harst, M. Siderius, P. H. Reusch, and W. M. Obermann. 2006. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol. Cell Biol. 26:8385-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Immormino, R. M., D. E. Dollins, P. L. Shaffer, K. L. Soldano, M. A. Walker, and D. T. Gewirth. 2004. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J. Biol. Chem. 279:46162-46171. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J. L., and D. O. Toft. 1995. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol. Endocrinol. 9:670-678. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, Y., S. Matsumoto, and I. Yahara. 1994. Temperature-sensitive mutants of hsp82 of the budding yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 242:517-527. [DOI] [PubMed] [Google Scholar]
- 16.Louvion, J. F., R. Warth, and D. Picard. 1996. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. USA 93:13937-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayr, C., K. Richter, H. Lilie, and J. Buchner. 2000. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275:34140-34146. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin, S. H., F. Sobott, Z. P. Yao, W. Zhang, P. R. Nielsen, J. G. Grossmann, E. D. Laue, C. V. Robinson, and S. E. Jackson. 2006. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 356:746-758. [DOI] [PubMed] [Google Scholar]
- 19.McLaughlin, S. H., L. A. Ventouras, B. Lobbezoo, and S. E. Jackson. 2004. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J. Mol. Biol. 344:813-826. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, P., C. Prodromou, B. Hu, C. Vaughan, S. M. Roe, B. Panaretou, P. W. Piper, and L. H. Pearl. 2003. Structural and functional analysis of the middle segment of hsp90. Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11:647-658. [DOI] [PubMed] [Google Scholar]
- 21.Meyer, P., C. Prodromou, C. Liao, B. Hu, S. Mark Roe, C. K. Vaughan, I. Vlasic, B. Panaretou, P. W. Piper, and L. H. Pearl. 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23:511-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 23.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obermann, W. M., H. Sondermann, A. A. Russo, N. P. Pavletich, and F. U. Hartl. 1998. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 143:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panaretou, B., C. Prodromou, S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panaretou, B., G. Siligardi, P. Meyer, A. Maloney, J. K. Sullivan, S. Singh, S. H. Millson, P. A. Clarke, S. Naaby-Hansen, R. Stein, R. Cramer, M. Mollapour, P. Workman, P. W. Piper, L. H. Pearl, and C. Prodromou. 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10:1307-1318. [DOI] [PubMed] [Google Scholar]
- 27.Pearl, L. H., and C. Prodromou. 2006. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75:271-294. [DOI] [PubMed] [Google Scholar]
- 28.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228:111-133. [DOI] [PubMed] [Google Scholar]
- 29.Prodromou, C., B. Panaretou, S. Chohan, G. Siligardi, R. O'Brien, J. E. Ladbury, S. M. Roe, P. W. Piper, and L. H. Pearl. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19:4383-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan, B. Panaretou, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter, K., P. Muschler, O. Hainzl, J. Reinstein, and J. Buchner. 2003. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the ATPase cycle. J. Biol. Chem. 278:10328-10333. [DOI] [PubMed] [Google Scholar]
- 32.Richter, K., S. Walter, and J. Buchner. 2004. The co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 342:1403-1413. [DOI] [PubMed] [Google Scholar]
- 33.Roe, S. M., M. M. Ali, P. Meyer, C. K. Vaughan, B. Panaretou, P. W. Piper, C. Prodromou, and L. H. Pearl. 2004. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37). Cell 116:87-98. [DOI] [PubMed] [Google Scholar]
- 34.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 35.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Siligardi, G., B. Hu, B. Panaretou, P. W. Piper, L. H. Pearl, and C. Prodromou. 2004. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 279:51989-51998. [DOI] [PubMed] [Google Scholar]
- 37.Vaughan, C. K., U. Gohlke, F. Sobott, V. M. Good, M. M. Ali, C. Prodromou, C. V. Robinson, H. R. Saibil, and L. H. Pearl. 2006. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell 23:697-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegele, H., L. Muller, and J. Buchner. 2004. Hsp70 and Hsp90—a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151:1-44. [DOI] [PubMed] [Google Scholar]
- 39.Whitesell, L., and S. L. Lindquist. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5:761-772. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, S., T. Ono, A. Mizuno, and T. K. Nemoto. 2003. A hydrophobic segment within the C-terminal domain is essential for both client-binding and dimer formation of the HSP90-family molecular chaperone. Eur. J. Biochem. 270:146-154. [DOI] [PubMed] [Google Scholar]
- 41.Young, J. C., and F. U. Hartl. 2000. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the cochaperone p23. EMBO J. 19:5930-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]