Abstract
BTN2 gene expression in the yeast Saccharomyces cerevisiae is up-regulated in response to the deletion of BTN1, which encodes the ortholog of a human Batten disease protein. We isolated Btn2 as a Snc1 v-SNARE binding protein using the two-hybrid assay and examined its role in intracellular protein trafficking. We show that Btn2 is an ortholog of theDrosophila and mammalian Hook1 proteins that interact with SNAREs, cargo proteins, and coat components involved in endosome-Golgi protein sorting. By immunoprecipitation, it was found that Btn2 bound the yeast endocytic SNARE complex (e.g., Snc1 and Snc2 [Snc1/2], Tlg1, Tlg2, and Vti1), the Snx4 sorting nexin, and retromer (e.g., Vps26 and Vps35). In in vitro binding assays, recombinant His6-tagged Btn2 bound glutathione S-transferase (GST)-Snc1 and GST-Vps26. Btn2-green fluorescent protein and Btn2-red fluorescent protein colocalize with Tlg2, Snx4, and Vps27 to a compartment adjacent to the vacuole that corresponds to a late endosome. The deletion of BTN2 blocks Yif1 retrieval back to the Golgi apparatus, while the localization of Ste2, Fur4, Snc1, Vps10, carboxypeptidases Y (CPY) and S (CPS), Sed5, and Sec7 is unaltered in btn2Δ cells. Yif1 delivery to the vacuole was observed in other late endosome-Golgi trafficking mutants, including ypt6Δ, snx4Δ, and vps26Δ cells. Thus, Btn2 facilitates specific protein retrieval from a late endosome to the Golgi apparatus, a process which may be adversely affected in patients with Batten disease.
Batten disease belongs to a family of autosomal recessive disorders called neuronal ceroid lipofuscinoses (NCLs) that lead to progressive neurodegeneration and early death in humans (30, 31, 54). At the cellular level, these diseases are typified by the abnormal accumulation of autofluorescent storage material in the lysosome, followed by subsequent neuron loss. The human CLN3 gene, which undergoes mutations that lead to juvenile-onset NCL (JNCL) (30, 31, 54), has a yeast ortholog called BTN1 (18, 59). Importantly, deletion of BTN1 leads to defects in vacuolar pH homeostasis and the up-regulation of BTN2 (12, 57), a gene of unclear function involved in cellular adaptation to the loss of BTN1 in yeast and, possibly, the pathogenesis of JNCL in humans.
The yeast Saccharomyces cerevisiae Btn2 protein has similarity to Hook1, a coiled-coil protein that associates with the cytoskeleton in mammalian cells (86) and with endocytic vesicles in Drosophila melanogaster (45). Hook1 in Drosophila is involved in the endocytosis of membrane receptors and their delivery to multivesicular bodies (MVBs) (45, 79). As MVBs represent a step in the maturation of late endosomes which fuse with lysosomes in higher organisms and vacuoles in yeast (32, 42, 63), it suggests a role for Hook proteins in lysosome/vacuole biogenesis. Interestingly, both the Drosophila and the mammalian Hook proteins interact with orthologs of the HOPS/class C vacuolar protein sorting complex (encoded by the vps gene) (composed of Vps11, Vps16, Vps18, and Vps33) (56, 69). This complex tethers membranes to both endosomes and vacuoles in yeast (61) and to early and late endosomes in mammals (64, 69). Thus, the HOPS complex mediates transport from either the Golgi apparatus or the endosomes to the lysosome/vacuole. The yeast HOPS complex was shown to interact with SNAREs involved in vacuolar fusion (i.e., Vam3, Nyv1, and Vti1) (61, 65, 72) and to fractionate with Pep12, a t-SNARE from late endosomes (4). Components of the mammalian HOPS complex interact with SNAREs involved in endosome fusion events (i.e., Syn6, Syn7, Syn13, and VAMP8) (69). Correspondingly, mammalian Hook1 was found in complexes containing these SNAREs, suggesting a possible role for the protein in membrane tethering and/or fusion at the endosomal level (56, 69).
Here we identify Btn2 as a SNARE- and retromer-binding protein that facilitates late endosome-Golgi protein sorting in yeast. By employing the two-hybrid assay, we found that Btn2 interacts with theSnc1 and Snc2 (Snc1/2) v-SNARE proteins, which mediate both exocytosis and endocytosis (34, 66). This suggested that Btn2 might have either SNARE regulatory or trafficking functions, like Vsm1 or Gcs1, which we identified earlier as Snc-binding proteins (49, 70). As the function of Btn2 is unclear, we examined the role of Btn2 in intracellular protein trafficking. First, we found that Btn2 binds to the assembled endocytic SNARE complex (e.g., Snc1/2, Tlg1, Tlg2, and Vti1), which facilitates protein recycling to endosomes and the trans Golgi apparatus (10). Second, we found that Btn2-red fluorescent protein (RFP) and Btn2-green fluorescent protein (GFP) colocalize with a variety of endosomal markers, including Snx4 (38), Tlg2 (2, 40), and Vps27. Vps27 is found on late endosomes and MVBs and is involved in the delivery of membrane proteins to the vacuole (62, 63). Third, the deletion of BTN2 results in the mislocalization of a Golgi marker, Yif1 (51), to the vacuole, as first shown earlier by others (14). We observed Yif1 mislocalization in other yeast mutant strains defective in late endosome-Golgi transport, such as ypt6Δ, snx4Δ, and vps26Δ cells, but not in mutants defective in endocytosis, early endosome-Golgi transport, late endosome-vacuole transport, and Golgi export. Thus, Btn2 may mediate the retrieval of cargo molecules from late endosomes to the Golgi apparatus. This effect is specific to certain Golgi proteins, as other markers, such as Sed5 (36) and Sec7 (26), are not mislocalized in the absence of BTN2. Similarly, Yif1 was unstable in btn2Δ cells, while other transient and resident Golgi markers were unaffected. Fourth, Btn2 immunoprecipitates the Snx4 sorting nexin (38) and components of retromer, including Vps26 and Vps35, which confer late endosome-Golgi transport (73, 74). In vitro binding assays confirm a direct interaction between Btn2 and Snc1 or Vps26. Since Btn2 interacts with components of the endosomal sorting machinery and a known cargo protein is mistrafficked in btn2Δ cells, it suggests that Btn2 plays an important role in endosomal protein sorting. We hypothesize that NCL onset and pathogenesis in humans may occur as a consequence of the loss of the Btn2/Hook1 function in endosomal protein sorting.
MATERIALS AND METHODS
Media, DNA, and genetic manipulations.
Yeast was grown on standard growth media containing either 2% glucose or 3.5% galactose. The preparation of synthetic complete and drop-out media was similar to the method described in reference 71. Standard methods were used for the introduction of DNA into yeast and the preparation of genomic DNA (71).
Growth tests.
Yeast was grown on synthetic and rich-growth media (71). For growth tests on plates, yeast was grown to log phase, normalized for optical density at 600 nm, diluted serially, and plated by drops onto solid medium preincubated at different temperatures. Calcofluor resistance was measured by adding 50 to 150 μg/ml fluorescent brightener 28 (Sigma) per plate and plating serial dilutions of yeast by drops.
Two-hybrid assay.
The yeast two-hybrid assay was performed as described by Durfee et al. (22), using Snc13-94 as the bait and a yeast cDNA library as the prey, with Y153 cells. Out of 900,000 transformants, 24 positives were identified, of which 1 was positive and conferred resistance to both 3-aminotriazole (3-AT) and β-galactosidase activity. Cell growth in the presence of 25 mM 3-aminotriazole on synthetic medium lacking histidine, along with β-galactosidase activity on nitrocellulose filters, was measured using standard procedures (22).
Yeast strains and plasmids.
Yeast strains used are listed in Table 1. Standard yeast vectors included the following: pAD11 (CEN HIS3), pRS313 (CEN HIS3), pRS315 (CEN LEU2), pRS316 (CEN URA3), and YEp13M4 (2μm LEU2); pRS426 (2μm URA3); pAD4Δ (2μm LEU2 ADH1 promoter); and pAD54 and pAD6 (both are the same as pAD4Δ but contain sequences encoding thehemagglutinin [HA] or myc epitope, respectively, downstream of the ADH1 promoter). Plasmids used in this study are listed in Table 2. A BTN2 deletion construct was created by first cloning a PCR-amplified 4.3-kb genomic fragment of BTN2 (from −1,658 bp upstream of the start codon to +1,409 bp downstream of the stop codon) into pGEM-T Easy (Promega) to yield pGEM-BTN2. Next, pGEM-BTN2 was digested with BglII to remove a fragment of the BTN2 gene corresponding to the base pair sequence −164 to 1,194 of the coding region. Following that, either a LEU2 or a URA3 selectable marker was inserted into the BglII site to yield pGEM-BTN2-LEU2 or pGEM-BTN2-URA3. To disrupt BTN2, either pGEM-BTN2-LEU2 or pGEM-BTN2-URA3 was digested with NotI and transformed into yeast. Integration at the BTN2 locus was verified by PCR. To create a two-copy GFP [GFP(x2)] protein integration cassette for PCR amplification, plasmid pFA6a-GFP(S65T)-His3MX6 (48) was cut with BamHI and PacI, and a BamHI-PacI fragment of BTN2-GFP (derived from pAD6-BTN2-GFP by PCR amplification and lacking the stop codon) was inserted to yield plasmid pBTN2-GFP(x2)-His3MX6. Genomic tagging of BTN2 with GFP(x2) to yield strain RKY7 was accomplished by the transformation of BY4741 cells with the PCR product obtained by amplifying plasmid pBTN2-GFP(x2)-His3MX6 with a forward oligonucleotide specific to the 3′ end of BTN2 and a reverse oligonucleotide specific to the insertion cassette and the 3′ untranscribed region of BTN2. Integration was verified by PCR analysis.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Y153 | MATagal4 gal80 his3 trp-902 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ LYS2::Gal-HIS3 | S. Elledge |
| PP1a+RB control (in Y153) | MATagal4 gal80 his3 trp-902 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ LYS2::Gal-HIS3 pAS/N-RB and pPP1α | S. Elledge |
| W303-1a | MATacan1 his3 leu2 lys2 trp1 ura3 ade2 | J. Hirsch |
| W303-1b | MATαcan1 his3 leu2 lys2 trp1 ura3 ade2 | J. Hirsch |
| BY4741 | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 | Euroscarf |
| RSY979 | MATαura3-52 sec7-5 | R. Schekman |
| RSY1309 | MATahis3-Δleu2-3,112 lys2-801 suc2-9 sec21-2 | A. Spang |
| RSY1312 | MATaleu2-3,112 trp1 ura3-52 sec27-1 | A. Spang |
| RDY241 | MATαleu2 ura3 trp1 ade2 his3 lys2 sec28Δ::HIS3 | R. Duden |
| RDY260 | MATαleu2 ura3 sec33-1 | R. Duden |
| W3031a-SEC7RFP | MATa can1 his3 leu2 lys2 ura3 ade2 trp1::TRP1::TPI1-DsRed-SEC7 | This study |
| ATCC 201389 (Sik1-RFP) | MATαhis3Δ1 leu2Δmet15Δ0 ura3Δ0 sik1::mRFP-kanMX6 | W.-K. Huh |
| JG8 T15:85 (sncΔ) | MATacan1 his3 leu2 snc1::URA3 snc2::ADE8 pTGAL-SNC1 | J. Gerst |
| RKY4 | MATαcan1 his3 lys2 trp1 ura3 ade2 btn2Δ::LEU2 | This study |
| RKY5 | MATacan1 his3 lys2 trp1 ura3 ade2 btn2Δ::URA3 | This study |
| RKY6 | MATacan1 his3 lys2 ura3 ade2 trp1::TRP1::TPI1-DsRed-SEC7 btn2Δ::LEU2 | This study |
| RKY7 | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 BTN2::GFP(x2)::his5+ | This study |
| RH268-1C | MATacan1 his4 leu2 trp1 ura3 bar1-1 end4-1 | H. Riezman |
| MRY3 | MATacan1 his3 leu2 lys2 trp1 ura3 ade2 gcs1Δ::LEU2 | J. Gerst |
| JU28-1 | MATaade2 trp1 ura3 leu2 his3 can1-100 rhb1Δ::kanMX | F. Tamanoi |
| FvMY7 | MATaleu2-3,112 ura3-52 his3-200 ade2-101 trp1-901 suc2-9 mel− vti1-1 | G. Fischer von Mollard |
| FvMY21 | MATaleu2-3,112 ura3-52 his3-200 ade2-101 trp1-901 suc2-9 mel− vti1-11 | G. Fischer von Mollard |
| FvMY24 | MATaleu2-3,112 ura3-52 his3-200 ade2-101 trp1-901 suc2-9 mel− vti1-2 | G. Fischer von Mollard |
| btn2Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 btn2Δ::kanMX | Euroscarf |
| chs4Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 chs4Δ::kanMX | Euroscarf |
| snc1Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 snc1Δ::kanMX | Euroscarf |
| snx4Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 snx4::kanMX | Euroscarf |
| rcy1Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 rcy1Δ::kanMX | Euroscarf |
| rhb1Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 rhb1Δ::kanMX | Euroscarf |
| ric1Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 ric1Δ::kanMX | Euroscarf |
| tlg2Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 tlg2Δ::kanMX | Euroscarf |
| vam6Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vam6Δ::kanMX | Euroscarf |
| vps17Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vps17Δ::kanMX | Euroscarf |
| vps23Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vps23Δ::kanMX | Euroscarf |
| vps26Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vps26Δ::kanMX | Euroscarf |
| vps27Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vpa27Δ::kanMX | Euroscarf |
| vps28Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vps28Δ::kanMX | Euroscarf |
| vps51Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 vps51Δ::kanMX | Euroscarf |
| ypt6Δ | MATahis3Δ1 leu2Δmet15Δ0 ura3Δ0 ypt6Δ::kanMX | Euroscarf |
TABLE 2.
Plasmids used for this study
| Plasmid name | Gene expressed or deleted | Promoter/terminator | Vector | Cloning site(s) | Type | Selectable marker | Source |
|---|---|---|---|---|---|---|---|
| pAD54-BTN2 | HA-BTN2 | ADH1/ADH1 | pAD54 | SalI | 2μm | LEU2 | This study |
| pGEM-BTN2 | BTN2 (w/o SacI)c | pGEM-T Easy | SalI-SacI | This study | |||
| pAD6-BTN2 (−Sac) | myc-BTN2 (w/o SacI) | ADH1/ADH1 | pAD6 | SalI-SacI | 2μm | LEU2 | This study |
| pAD6-BTN2-mGFP | GFP | ADH1/ADH1 | pAD6-BTN2 | SacI | 2μm | LEU2 | This study |
| pRS313-BTN2-mGFP | myc-BTN2-GFP (w/o SacI) | ADH1/ADH1 | pRS313 | BamHI | CEN | HIS3 | This study |
| pRS316-BTN2-mGFP | myc-BTN2-GFP (w/o SacI) | ADH1/ADH1 | pRS316 | BamHI | CEN | URA3 | This study |
| pRS313-BTN2-mRFP | myc-BTN2-mRFP (w/o SacI) | ADH1/ADH1 | pRS313 | BamHI | CEN | HIS3 | This study |
| pRS315-BTN2-mRFP | myc-BTN2-mRFP (w/o SacI) | ADH1/ADH1 | pRS315 | BamHI | CEN | LEU2 | This study |
| pRS316-BTN2-mRFP | myc-BTN2-mRFP (w/o SacI) | ADH1/ADH1 | pRS316 | BamHI | CEN | URA3 | This study |
| pRS313-BTN2 | myc-BTN2 | ADH1/ADH1 | pRS313 | BamHI | CEN | HIS3 | This study |
| pACT-BTN2a | BTN2 | ADH1/ADH1 | pACT | 2μm | LEU2 | This study | |
| pGEM-BTN2-LEU2b | btn2::LEU2 | pGEM-T Easy | LEU2 | This study | |||
| pGEM-BTN2-URA3b | btn2::URA3 | pGEM-T Easy | URA3 | This study | |||
| pHIS6-BTN2 | BTN2 | TRC (trpB-lacUV5) | pTrcHis B | XhoI/SalI | This study | ||
| pADHU-GFP-cSNC1 | GFP-cSNC1 | ADH1/ADH1 | YCp50 | BamHI | CEN | URA3 | This study |
| pAD54-GFP-cSNC1 | GFP-cSNC1 | ADH1/ADH1 | pAD54 | SalI-SacI | 2μm | LEU2 | J. Gerst |
| pGEX-SNC1 | SNC12-94 | tac | pETGEXCT | A. Spang | |||
| pAS1-cSNC13-94 | cSNC13-94 | ADH1/ADH1 | pAS1 | NcoI-BamHI | 2μm | TRP1 | This study |
| pHADH-mycSNC1 | mycSNC1 | ADH1/ADH1 | pAD11 | SalI-SacI | 2μm | HIS3 | J. Gerst |
| pAS1-SNC4-93 | SNC24-93 | ADH1/ADH1 | pAS1 | NcoI-BamHI | 2μm | TRP1 | J. Gerst |
| pHADH-mycSNC2 | mycSNC2 | ADH1/ADH1 | pAD11 | SalI-SacI | 2μm | LEU2 | J. Gerst |
| pRS315-GFP-SED5 | GFP-SED5 | ADH1/ADH1 | pRS315 | BamHI | CEN | LEU2 | J. Gerst |
| pRS315-GFP-cSNC1 | GFP-cSNC1 | ADH1/ADH1 | pRS315 | BamHI | CEN | LEU2 | This study |
| pRS315-GFP-TLG1 | GFP-TLG1 | ADH1/ADH1 | pRS315 | BamHI | CEN | LEU2 | This study |
| pRS315-GFP-TLG2 | GFP-TLG2 | ADH1/ADH1 | pRS315 | BamHI | CEN | LEU2 | This study |
| pGEX-TLG2 | TLG22-318 | tac | pGEX-4T-3 | BamHI-SalI | J. Gerst | ||
| pGO426-GFP-VPS27 | GFP-VPS27 | CPY1 | pRS426 | BamHI | 2μm | URA3 | S. Emr |
| pGO426-CPS1-GFP | CPS1-GFP | CPY1 | pRS426 | BamHI | 2μm | URA3 | S. Emr |
| pAD54-GFP-SNX4 | HA-GFP-SNX4 | ADH1/ADH1 | pAD54 | SalI | 2μm | LEU2 | This study |
| pGEX-SNX4 | SNX4 | tac | pGEX-4T-3 | SalI-NotI | This study | ||
| pAD54-VPS10-GFP | HA-VPS10-GFP | ADH1/ADH1 | pAD54 | SalI-SacI | 2μm | LEU2 | This study |
| pRS316-FUR4-GFP | HA-FUR4-GFP | ADH1/ADH1 | pRS316 | BamHI | CEN | URA3 | This study |
| pRS316-GFP-YIF1 | HA-GFP-YIF1 | ADH1/ADH1 | pRS316 | BamHI | CEN | URA3 | This study |
| pAD54-RFP-YIF1 | HA-RFP-YIF1 | ADH1/ADH1 | pAD54 | SalI-SacI | 2μm | LEU2 | This study |
| pGALΔBglII-CPY(1-50)GFP | CPY1-50-GFP | GAL1 | pGALΔBglII | CEN | URA3 | O. Deloche | |
| pRS314-STE2-GFP | STE2-GFP | STE2 | pRS314 | CEN | TRP1 | K. Blumer | |
| pAD54-VTI1 | VTI1 | ADH1/ADH1 | pAD54 | SalI-SacI | 2μm | LEU2 | J. Gerst |
| pGEX-VPS17 | VPS17 | tac | pGEX-4T-3 | SalI-NotI | This study | ||
| pGEX-VPS26 | VPS26 | tac | pGEX-4T-3 | SalI-NotI | This study | ||
| pGEX-SSO1 | SSO12-265 | tac | pGEX-4T-3 | BamHI-SalI | J. Gerst |
Isolated with a yeast two-hybrid screen.
See Materials and Methods for details on construction.
w/o, without.
Microscopy.
GFP and RFP fluorescence in strains expressing the appropriate GFP- and RFP-tagged fusion proteins was visualized by confocal microscopy. Nuclear staining with Hoechst dye (50 μg/ml Hoechst 33342; Molecular Probes) was carried out for 10 min at room temperature prior to visualization.
Immunoprecipitation and Western analysis.
Interactions between the HA- or myc-tagged Btn2 and other proteins present in cell lysates were monitored by immunoprecipitation (IP) from cell extracts, as described previously (17). However, the following changes were made to the lysis and IP buffers. TE (10 mM Tris-HCl, pH 7.5, EDTA 1 mM, NaCl 150 mM) was used instead of phosphate-buffered saline, and 0.5% NP-40 was used instead of Triton X-100. The following protease inhibitors were added: 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml soybean trypsin inhibitor, 10 μg/ml pepstatin, and 1 mM PMSF (phenylmethylsulfonyl fluoride). IP antibodies included anti-myc antibodies (3 μl per reaction; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-HA antibodies (2.5 μl per reaction; Roche, Indianapolis, IN). Antibodies for protein detection in Western blots included polyclonal antibodies against carboxypeptidase Y (CPY), Mnn1, and Sec22 (gifts of S. Emr, UCSD, San Diego, CA); Kar2 (gift of C. Barlowe, Dartmouth University, Dartmouth, NH); Sec9 (gift of P. Brennwald, UNC, Chapel Hill, NC); Sed5 (gift of H. Pelham, MRC, Cambridge, United Kingdom); Snc1 (66); Sso1 (gift of S. Keranen, VTT, Espoo, Finland); Tlg1 (gift of H. Pelham, MRC, Cambridge, United Kingdom); Tlg2 (gift of H. Abeliovich, Faculty of Agriculture, HUJ, Rehovot, Israel); Vam3 (gift of A. Mayer, Université de Lausanne, Lausanne, Switzerland); Vam7 (gift of C. Ungermann, U. of Osnabrück, Osnabrück, Germany); Vti1 (gift of G. Fischer von Mollard, University of Göttingen, Göttingen, Germany); retromer (Vps26 and Vps35; gifts of M. Seaman, Cambridge Institute for Medical Research, Cambridge, United Kingdom); and monoclonal antibodies against the HA epitope (gift of M. Wigler, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), β-actin (MP Biomedicals, Aurora, OH), Dpm1 (Molecular Probes, Eugene, OR), GFP (Roche, Indianapolis, IL), glutathione S-transferase (GST) (Calbiochem, Darmstadt, Germany), and His6 (Sigma-Aldrich, St. Louis, MO). Samples of total cell lysates (TCLs) (20 μg protein per lane) and immunoprecipitates obtained from 500 μg protein of lysate (per IP reaction) were resolved by electrophoresis and detected by Western blotting. Detection was performed by chemiluminescence.
Recombinant protein purification and in vitro binding assay.
To perform in vitro binding assays, recombinant His6-Btn2, GST, GST-Snc1 (Snc12-94), GST-Snx4, GST-Vps17, GST-Vps26, GST-Tlg2 (Tlg22-318), and GST-Sso1 (Sso12-265) were generated with Escherichia coli strain BL21(DE3)-R3 from bacterial expression plasmids and purified using standard procedures. His6-Btn2 was purified using ProBond resin (Invitrogen, Carlsbad, CA) and a final elution with 200 mM imidazole, while GST-tagged proteins were purified using immobilized glutathione beads (Pierce, Rockford, IL) and a final elution with 50 mM glutathione. For binding, equal amounts of His6-Btn2 and GST-tagged proteins (10 μg each) were mixed together in binding buffer containing 0.1% NP-40 in phosphate-buffered saline (300 mM NaCl), pH 7.5. Following incubation at 4oC for 12 h, 50 μl of a 50% ProBond slurry in binding buffer was added, and the samples were incubated with rotation for an additional 2 h at 4oC. Then, samples were washed five times with binding buffer and eluted with the addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer prior to electrophoresis on 10% SDS-polyacrylamide gels.
RESULTS
Btn2 interacts with Snc1/2 v-SNAREs in the two-hybrid screen.
The yeast Snc1 v-SNARE (29) lacking its transmembrane domain (Snc13-94) was used as bait in the two-hybrid screen with a yeast cDNA library. Out of ∼900,000 transformants, one colony tested positive for both lacZ expression and resistance to 3-AT in repetitive assays (data not shown). DNA sequencing revealed that this clone carried the BTN2 gene, which encodes a protein that is overproduced in cells lacking BTN1, the yeast ortholog of the human CLN3 Batten disease gene (18, 59). Btn2 has been suggested to be similar to Hook1 (12), a coiled-coil protein of unclear function involved in endocytosis and protein sorting of MVBs (45, 79, 86). Sequence alignments performed using multiple sequence alignment with high accuracy and high throughput (MUSCLE) (23) indeed revealed that Btn2 from S. cerevisiae is orthologous to Drosophila and human Hook1 (Fig. 1). Btn2 was found to be 18% and 22% identical and 32% and 33% homologous to Drosophila and human Hook1, respectively, over its entire length (410 amino acids).
FIG. 1.
BTN2 encodes a Hook1 ortholog as shown by MUSCLE alignments of Btn2 from Saccharomyces cerevisiae (sc Btn2; GenBank accession no. NP_011658.1), Drosophila Hook1 (dm Hook1; GenBank accession no. NP_476573.1), and human Hook1 (hs Hook1; GenBank accession no. NP_056972.1). Amino acid identities shared between two or more alignments are shown in black boxes, while similarities are shown in light (less similar) and dark (more similar) gray boxes. Numbers correspond to the amino acid, while periods indicate gaps introduced into the sequence by the program.
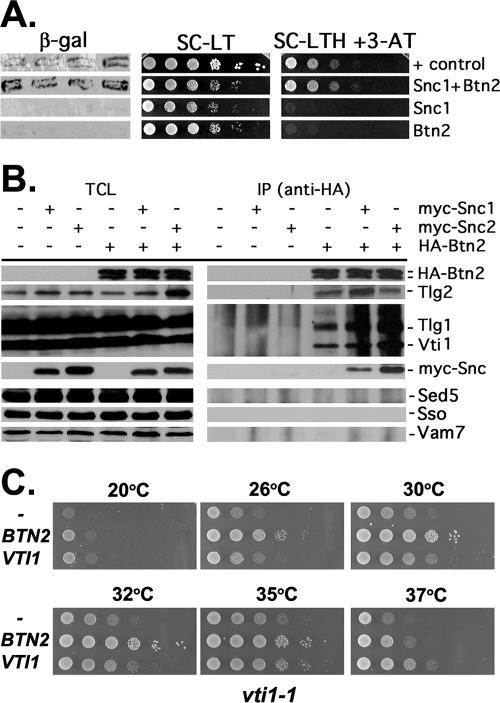
As the cellular functions of either Btn1 or Btn2 have not been fully resolved, we decided to characterize the Snc1-Btn2 interaction further. Similar analyses identified Vsm1 as a Snc2-interacting partner (49) and led to its characterization as a potential SNARE regulator (50). Likewise, we identified the Gcs1 Arf-GAP as a Snc2-interacting factor and demonstrated its involvement in Snc v-SNARE recycling to the Golgi apparatus (70). We verified the interaction seen between the Snc1 v-SNARE and Btn2 by using the yeast two-hybrid assay (Fig. 2A). As expected, the fusion of Btn2 to the transactivating domain of Gal4 induced both β-galactosidase activity and resistance to 3-aminotriazole, in combination with the fusion of Snc13-94 to the DNA binding domain of Gal4. Similar results were obtained with Snc24-93 (data not shown).
FIG. 2.
Btn2 interacts with Snc1 and the yeast endocytic SNARE complex. (A) Snc1 interacts with Btn2, as assayed using the two-hybrid β-galactosidase and 3-AT growth assays. Yeast (Y153 cells) were transformed with plasmids expressing the Gal4 DNA binding domain fused to Snc1 (pAS1-Snc1Δ) and full-length Btn2 fused to the Gal4 transactivating domain (pACT-Btn2), together or alone. Transformants were either grown as patches on plates and then replica plated onto nitrocellulose fibers (left panel) or grown to mid-log phase in liquid culture prior to serial dilution and plating by drops onto solid medium (middle and right panels). In addition, positive control (+control) cells expressing Rb and PP1 from plasmids (pAS/N-RB and pPP1α) were used in parallel. For the β-galactosidase (β-gal) assay, cells replica plated onto filters were lysed in liquid nitrogen and measured for β-galactosidase activity using standard techniques. For assaying resistance to 3-aminotriazole (3-AT) and growth in the absence of histidine, cells were plated onto either control selective medium (SC-LT) or medium lacking histidine and containing 25 mM 3-AT (SC-HLT +3-AT). Cells were grown for 60 h at 26°C. (B) The yeast endocytic SNARE complex coimmunoprecipitates with Btn2. Wild-type yeast (W303-1a) expressing HA-Btn2 from a multicopy plasmid (pAD54-Btn2) were transformed with single-copy plasmids expressing myc-Snc1 (pHADH-myc-cSnc1) or myc-Snc2 (pHADH-myc-Snc2), or empty vector alone (pAD11) as control. In addition, wild-type cells bearing a control multicopy plasmid (pAD54) were transformed with the same single-copy plasmids expressing myc-Snc1 or myc-Snc2 or an empty vector as controls. Cells were grown to mid-log phase and processed for immunoprecipitation with anti-HA antibodies. Precipitates (IP) formed from each reaction mixture (500 μg total protein per reaction) and TCLs (20 μg protein per lane) were resolved with SDS-PAGE and detected by Western blotting with a variety of antisera. Shown are blots detected with anti-HA antibody (1:7,000), which reveals that HA-Btn2 is expressed as a doublet. Also shown are blots detected with antisera to Tlg2 (1:1,000), Vti1 and Tlg1 (added together, 1:2,000 and 1:3,000, respectively), myc epitope (to detect myc-Snc1 or myc-Snc2, 1:1,000), Sed5 (1:3,000), Sso1/2 (1:3,000), and Vam7 (1:5,000). Based upon densitometric analysis of the individual Tlg1, Tlg2, and Vti1 signals in the three HA-Btn2-containing IP lanes, we estimate that an average of ∼4% of the endocytic SNARE complex coimmunoprecipitated with HA-Btn2 in the reaction mixtures after normalization for Btn2 precipitation (immunoprecipitated 3.8% Vti1, 3.1% Tlg1, and 4.2% Tlg2; data not shown). (C) Overexpression of BTN2 enhances the growth of vti1-1 cells. vti1-1 cells bearing a control vector (pAD54) or vectors overexpressing HA-BTN2 or HA-VTI1 (pAD54-BTN2 or pAD54-VTI1, respectively) were grown to mid-log phase at 26°C before serial dilution (10×) and plating by drops onto prewarmed solid medium. Plates were grown for 2 to 3 days at the indicated temperatures before photodocumentation.
Btn2 binds to members of the endocytic SNARE complex in yeast.
As Btn2 binds to the Snc1/2 v-SNAREs, we examined whether it interacts with other yeast SNAREs, particularly those that bind Snc1/2. A functional HA-tagged form of Btn2 (data not shown and Fig. 2C) was expressed in wild-type cells and used for immunoprecipitation with anti-HA antibodies. Western blotting of the precipitates after SDS-PAGE revealed that HA-Btn2 could precipitate the entire yeast endocytic SNARE complex (e.g., Snc1 or Snc2-Tlg1-Tlg2-Vti1) (10) but not the t-SNAREs of the exocytic SNARE complex (e.g., Sso1/2 and Sec9) (1, 9), the endoplasmic reticulum (ER)-Golgi SNARE complex (e.g., Sec22 and Sed5) (80), or the vacuolar SNARE complex (e.g., Vam3, Vam7) (82) (Fig. 2B and data not shown). Thus, Btn2 is a SNARE-interacting protein that associates with components of the yeast endocytic SNARE complex. This contrasts with Vsm1, which interacts preferentially with members of the exocytic SNARE complex (e.g., Snc1/2, Sso1/2, and Sec9) (49, 50; P. Brennwald, personal communication).
As Btn2 interacts with components of the endocytic SNARE complex, we examined whether BTN2 interacts genetically with mutations in VTI1, a t-SNARE that facilitates multiple endosomal transport routes leading to the vacuole (i.e., from the Golgi apparatus to the late endosome/prevacuole, the late endosome/prevacuole to the vacuole, cytosol to the vacuole, and retrograde late endosome/prevacuole to the Golgi transport) (25). We found that the overexpression of HA-BTN2 significantly enhanced the growth of temperature-sensitive vti1-1 mutants (Fig. 2C), which are defective principally in Golgi-to-late endosome/prevacuole and cytosol-to-vacuole transport at restrictive temperatures. This improvement in growth was observed at all temperatures and was more robust than that conferred by VTI1 overexpression at 32oC and 35oC (Fig. 2C). In contrast, HA-BTN2 overexpression did not enhance the growth of either vti1-2 cells, which are defective in all Vti1-mediated steps except for late endosome/prevacuole-to-Golgi retrograde transport, or vti1-11 cells, which are defective in all steps (25). Thus, the restoration of growth mediated by BTN2 overexpression is allele-specific and suggests a possible role for Btn2 in Golgi-to-late endosome/prevacuole transport. A role in cytosol-to-vacuole transport appears less likely given that Btn2 does not interact with Vam7 (Fig. 2B).
Btn2 localizes to the late endosome/MVB compartment.
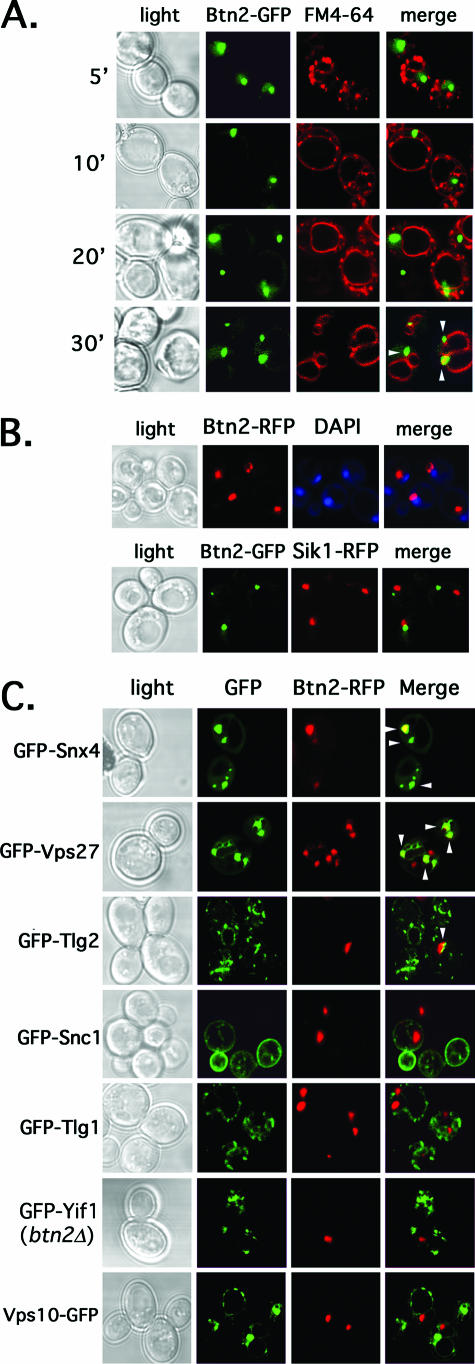
We next examined the localization of Btn2 in yeast. We determined whether Btn2 resides on the endocytic pathway by expressing Btn2-GFP from a single-copy plasmid and performing colabeling with the vacuolar marker FM4-64 (85). We first pulse-labeled cells with FM4-64 on ice, followed by a short chase (5 to 20 min) at 26oC, in order to label early endocytic compartments, and followed localization of the two markers. Btn2-GFP labeling produced individual, large punctate structures that did not colocalize with FM4-64 at any interval during the short chase (Fig. 3A). However, we did see colabeling in these large structures in cells that were incubated with FM4-64 at 26oC and then chased for 30 min at 26oC, to allow for vacuolar labeling (Fig. 3A, bottom panels). Thus, Btn2 does not appear to localize to an early compartment of the endocytic pathway but more likely localizes to a compartment located adjacent to the vacuole. This pattern is similar to the labeling seen with late endosomes/MVBs. We note that the Btn2-labeled compartment was often located near the nucleus, as revealed by DAPI (4′,6′-diamidino-2-phenylindole) staining (Fig. 3B). However, it did not overlap either with a nucleolar marker, Sik1, or with DAPI labeling itself (Fig. 3B). Thus, Btn2 appears to localize to the late endocytic pathway.
FIG. 3.
Btn2resides in a late endocytic compartment and colocalizes in parts with Snx4, Tlg2, and Vps27 (arrowheads). (A) Btn2-GFP resides in a late endocytic compartment. Wild-type cells (W303-1b) expressing Btn2-GFP from a single-copy plasmid (pRS316-myc-BTN2-GFP) were grown to mid-log phase on selective synthetic medium at 26°C. Cells were either pulse-labeled with FM4-64 (5.4 μM final concentration) on ice (45 min) and then chased for varying amounts of time (5 to 20 min) at 26°C or were pulse-labeled and chased (30 min) at 26°C. Elapsed chase times are shown in minutes (′). All labeling was performed in the dark. Merge indicates the merger of the GFP and FM4-64 fluorescence windows. (B) Btn2-GFP does not localize to the nucleus. Top panels: wild-type cells (W303-1b) expressing Btn2-RFP from a single-copy plasmid (pRS316-myc-BTN2-mRFP) were grown to mid-log phase on selective synthetic medium at 26°C, labeled briefly with Hoechst dye (50 μg/ml Hoechst 33342 for 10 min at room temperature; Molecular Probes), and visualized by confocal microscopy. Bottom panels: SIK1-RFP cells (ATCC 201389) were transformed with a single-copy plasmid (pRS316-myc-BTN2-GFP) expressing Btn2-GFP. Cells were grown to mid-log phase at 26°C and visualized by confocal microscopy. Merge indicates the merger of GFP and RFP or RFP and DAPI fluorescence windows. (C) Btn2-RFP colocalizes in part with the Snx4, Tlg2, and Vps27 endosomal markers (arrowheads). Wild-type cells (W303-1b) expressing Btn2-RFP from a single-copy plasmid (pRS313-myc-BTN2-mRFP) were transformed with plasmids expressing GFP-tagged forms of Snx4 (pAD54-GFP-SNX4), Vps27 (pGO426-GFP-VPS27), Tlg2 (pRS315-GFP-TLG2), Snc1 (pADHU-GFP-cSNC1), Tlg1 (pRS315-GFP-TLG1), Vps10 (pAD54-VPS10-GFP), and Yif1 (pRS316-GFP-YIF1) (see Table 2 for plasmid details). Cells were grown to mid-log phase at 26°C on selective synthetic medium prior to visualization by confocal microscopy. btn2Δ cells (RKY4) were used to coexpress GFP-Yif1 (pRS316-GFP-YIF1) and Btn2-RFP (pRS313-myc-BTN2-mRFP), instead of wild-type cells. Merge indicates the merger of the GFP and RFP fluorescence windows.
To verify this, we examined the colocalization of a functional monomeric Btn2-RFP with GFP-tagged markers of other organelles (Fig. 3C). We found that Btn2-RFP expressed from a single-copy plasmid colocalized partly with the endosomal marker, GFP-Snx4, as well as with the MVB marker GFP-Vps27 (38, 62). In addition, some colocalization with an endosomal t-SNARE, GFP-tagged Tlg2 (2, 40), was also observed. In contrast, Btn2-RFP did not colocalize (Fig. 3C) significantly with markers of early endosomes and the trans Golgiapparatus, such as GFP-Tlg1 and internalized GFP-Snc1 (15, 40, 47), respectively, nor with Yif1, a Golgi marker (51). Although Btn2 interacts physically with Tlg1 and Snc1 (Fig. 2), their apparent lack of colocalization may imply that the interaction is transient and does not occur at compartments where these SNAREs typically reside at steady state (i.e., in early endosomes) (15, 40, 47). We note that Btn2-RFP is functional in yeast, as it confers normal GFP-Yif1 localization to btn2Δ cells (Fig. 3C, compare to GFP-Yif1 localization in btn2Δ cells in Fig. 5A). Together, the results imply that Btn2 has an endosomal pattern of localization most similar to that of the late endosome/MVB. This pattern corresponds with that predicted for Hook1 in Drosophila and mammals (45, 69, 79). However, we note that Btn2 did not colocalize with Vps10-GFP (Fig. 3C), the CPY receptor that cycles through a late endosomal compartment to deliver CPY to the vacuole (19). This may indicate either that yeast cells contain multiple late endosomal compartments or that the late endosome is composed of separate domains. We also note that an earlier study demonstrated cytosolic labeling for Btn2-GFP (13); however, we were unable to recapitulate these results.
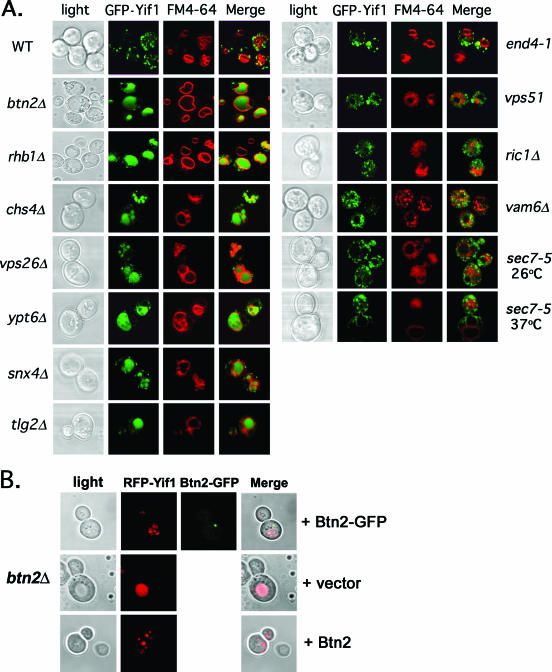
FIG. 5.
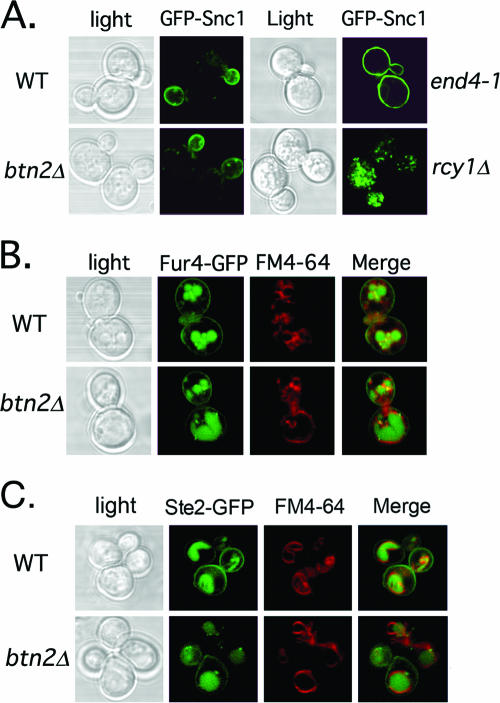
Yif1 is mislocalized to the vacuole in btn2Δ, rhb1Δ, vps26Δ, snx4Δ, chs4Δ, tlg2Δ, and ypt6Δ cells. (A) GFP-Yif1 is mislocalized to the vacuole in btn2Δ, chs4Δ, rhb1Δ, snx4Δ, tlg2Δ, vps26Δ, and ypt6Δ cells. A single-copy plasmid expressing GFP-Yif1 (pRS316-GFP-YIF1) was transformed into wild-type (WT) cells (both Euroscarf and W303-1b strains tested; Euroscarf cells shown) or cells bearing mutations of BTN2 (both Euroscarf and RKY4 btn2Δ strains tested; Euroscarf cells shown), RHB1 (both Euroscarf and JU28-1 rhb1Δ strains examined; Euroscarf strain shown), CHS4, VPS26, YPT6, SNX4, TLG2 VAM6, END4 (RH268-1c), RIC1, VPS51, and SEC7 (RSY979). Cells were grown to mid-log phase at 26°C on selective synthetic medium prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C) before visualization using confocal microscopy. sec7-5 (RSY979) cells were grown to mid-log phase at 26°C (permissive temperature) and either maintained and labeled with FM4-64 at 26°C or shifted to 37°C for 30 min prior to pulse-labeling with FM4-64 (30 min; 37°C) and chase (30 min; 37°C) before visualization. Note labeling of the vacuole by GFP in btn2Δ, chs4Δ, rhb1Δ, snx4Δ, tlg2Δ, vps26Δ, and ypt6Δ cells. See Table 3 for statistics regarding either Golgi or vacuolar GFP-Yif1 localization in the various strains examined. Merge indicates the merger of the GFP and FM4-64 fluorescence windows. (B) Btn2 and Btn2-GFP restore RFP-Yif1 localization to btn2Δ cells. btn2Δ cells expressing RFP-Yif1 from a multicopy plasmid (pAD54-RFP-YIF1) were transformed with either a control plasmid (pRS313) or single-copy plasmids expressing Btn2 or Btn2-GFP (pRS313-BTN2 or pRS313-BTN2-GFP). Cells were grown to mid-log phase at 26°C prior to visualization. Merge indicates the merger of light and fluorescence microscopy images.
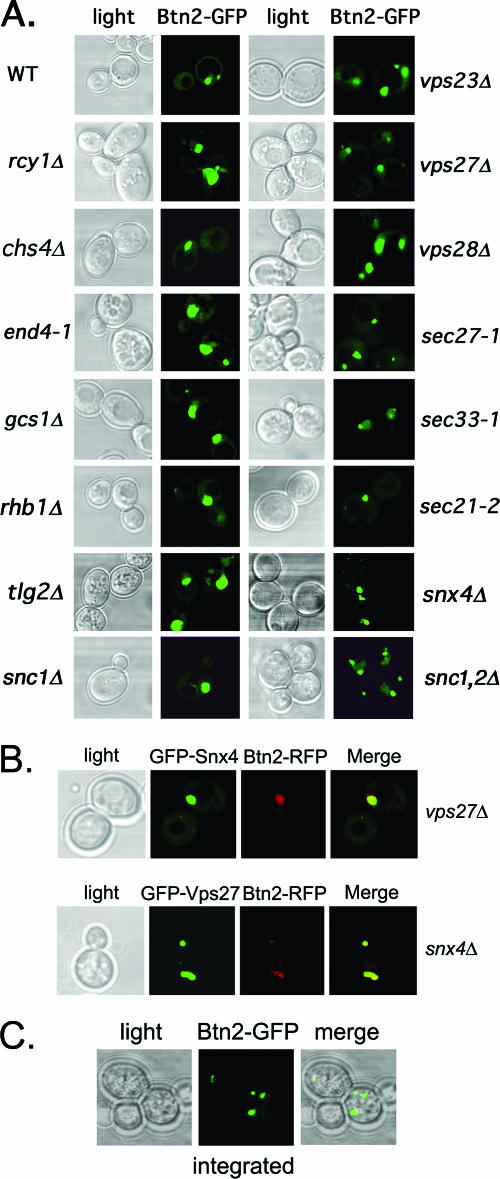
Since Btn2 may act at the level of the late endosome/MVB, as predicted for the function of Hook1 in Drosophila and mammals, we determined the localization of Btn2 in yeast mutants defective in Golgi-vacuole transport and MVB formation (Fig. 4A and data not shown). We found that the localization of a functional Btn2-GFP fusion (Fig. 5B) expressed from a single-copy plasmid was basically unchanged in all the mutants tested, including those involved in endocytosis (i.e., end4-1 and tlg2Δ) (2, 40, 47) protein recycling and export from endosomes (i.e., rcy1Δ, chs4Δ, gcs1Δ, and snx4Δ) (28, 38, 84), MVB formation (i.e., vps23Δ and vps27Δ) (41, 42), Golgi export (i.e., sec7-5 at 37oC) (26), COPI-mediated retrograde transport (i.e., sec21-2, sec27-1, and sec33-1) (20, 21, 46), and others (i.e., rsg1/rhb1; both Euroscarf and JU28-1 rhb1Δ strains tested) (83). We noted that there were more numerous and smaller Btn2-GFP-labeled structures in some cell types, particularly sncΔ cells which are defective in both endo- and exocytosis and have fragmented vacuoles (34, 66). Overall, though, Btn2-GFP localized to large perivacuolar structures in all transport mutants, suggesting that defects in intracellular protein sorting do not greatly affect Btn2 localization.
FIG. 4.
Btn2 localization is not altered in cells deficient in endocytosis and endosomal protein sorting. (A) Btn2 localization is not altered in cells deficient in endocytosis and endosomal protein sorting. A single-copy plasmid (pRS316-myc-BTN2-GFP) expressing Btn2-GFP was transformed into wild-type (WT) cells (W303-1b) or a variety of yeast mutants. The latter includes strains bearing deletions of RCY1, CHS4, GCS1, RHB1 (both Euroscarf and JU28-1 rhb1Δ strains tested; Euroscarf strain shown), SNC1, SNC1/2 (sncΔ; JG8 T15:85), VPS23, VPS27, or VPS28, as well as strains bearing temperature-sensitive mutations in genes SEC27 (RSY1312), SEC33 (RDY260), and SEC21 (RSY1309) COPI subunits or in END4 (RH268-1c). Cells were grown to mid-log phase at 26°C (permissive temperatures) on selective synthetic medium prior to visualization by confocal microscopy. (B) The colocalization between Btn2 and Snx4 or Vps27 is not altered in vps27Δ and snx4Δ cells, respectively. vps27Δ and snx4Δ cells expressing Btn2-RFP from a single-copy plasmid (pRS313-myc-BTN2-mRFP) were transformed with plasmids expressing either GFP-Snx4 (pAD54-GFP-SNX4) or GFP-Vps27 (pGO426-GFP-VPS27), respectively. Merge indicates the merger of the GFP and RFP fluorescence windows. (C) Btn2-GFP(x2) expressed from the genomic BTN2 locus labels multiple large compartments. Yeast bearing BTN2 tagged with GFP(x2) at its genomic locus (RKY7 cells) were grown to mid-log phase at 26°C prior to visualization by confocal microscopy. Merge indicates the merger between the light microscopy and GFP fluorescence windows.
Since Btn2 colocalizes with Snx4 and Vps27, we examined the colocalization of Btn2-RFP and either GFP-Snx4 or GFP-Vps27 in vps27Δ and snx4Δ cells, respectively (Fig. 4B). As predicted from Fig. 4A, no change in the localization of Btn2-RFP was found, and likewise, no change in the abilities of Btn2 and either Vps27 or Snx4 to colocalize was observed. Finally, we examined the localization of Btn2-GFP(x2) expressed from its genomic locus under the native promoter (Fig. 4C). We found that few cells (∼3%) had significantly visible GFP labeling, but all cells that did had a pattern of localization very similar to that observed for either Btn2-GFP or Btn2-RFP expressed from single-copy plasmids. We note that Btn2-GFP expressed from its genomic locus by the native promoter and bearing only one copy of GFP did not yield visible labeling (data not shown).
Retrieval of Yif1 to the Golgi apparatus is blocked in btn2Δ cells.
Because Btn2 has homology to Hook1 (Fig. 1), localizeslate on the endocytic pathway (Fig. 3), and interacts with SNAREs involved in endocytic retrieval (Fig. 2), we examined whether the deletion of BTN2 regulates the intracellular trafficking of proteins. We first examined the localization of Yif1, a Golgi protein involved in the recruitment of Ypt1 and the consumption of COPII vesicles (51). This protein was demonstrated to mislocalize to the vacuole in btn2Δ cells (14), although the manner and mechanism were not resolved. We found that GFP-tagged Yif1 (GFP-Yif1) expressed from a single-copy plasmid gave punctate Golgi-like labeling in wild-type cells (Fig. 5A), as previously shown (14, 51). We examined Yif1 localization in a variety of cell types, including cells lacking BTN2 or RSG/RHB1, a small GTPase of unknown function (83) that was found to interact with Btn2, using the yeast two-hybrid screen (13). Interestingly, instead of the punctate labeling seen in wild-type cells, GFP-Yif1 was found to accumulate in the vacuole in both btn2Δ and rhb1Δ cells (Fig. 5A; Table 3). This effect was observed in the Euroscarf btn2Δ and rhb1Δ deletion mutants, as well as in the RKY4 btn2Δ and JU28-1 rhb1Δ strains (data not shown). GFP-Yif1 mislocalization was also observed in other cells defective in endosomal protein sorting. For example, cells lacking CHS4, which encodes a protein required for the efficient export of chitin synthase III (Chs3) from endosomes (84), or those lacking YPT6, which encodes a small GTPase involved in endosome-Golgi and intra-Golgi transport (81) gave identical patterns of localization (Fig. 5A). Likewise, cells bearing a deletion of VPS26, which encodes a component of the retromer complex that resides on late endosomes and mediates late endosome-Golgi transport (73, 74), gave the same pattern (Fig. 5A). Finally, cells lacking SNX4, which encodes a sorting nexin required for endosome-Golgi transport (38), or TLG2, which encodes a t-SNARE involved in both endocytosis and endosomal transport (2, 40) and which binds Btn2 (Fig. 2B), mislocalized GFP-Yif1 to the vacuole (Fig. 5A). Thus, many cellular components known to confer protein sorting and retrieval from endosomes to the Golgi apparatus affect GFP-Yif1 localization and recycling to the Golgi apparatus. We note that some mutations had more dramatic effects upon GFP-Yif1 localization. For example, the relative amount of mislocalization observed (based upon counting cells with vacuole-localized GFP-Yif1) was rhb1Δ ∼ btn2Δ ∼ ypt6Δ ∼ snx4Δ ≥ vps26Δ > snc1Δ > tlg2Δ > snc2Δ ∼ chs4Δ ≫ vps10Δ ∼ wild-type cells (Table 3). Interestingly, the deletion of VPS10, which confers CPY trafficking to the vacuole via late endosomes in a retromer-dependent manner (73, 74), had no effect on GFP-Yif1 localization (Table 3). This suggests that Vps10 is not involved in Yif1 retrieval.
TABLE 3.
Localization of GFP-Yif1 in yeast
| Strain (Euroscarf) | % GFP-Yif1 localized in thea:
|
|
|---|---|---|
| Vacuole | Golgi apparatus | |
| WTb | 10 | 90 |
| btn2Δ | 87 | 13 |
| chs4Δ | 41 | 59 |
| rhb1Δ | 93 | 7 |
| snc1Δ | 57 | 43 |
| snc2Δ | 40 | 60 |
| snx4Δ | 87 | 13 |
| tlg2Δ | 48 | 52 |
| vps10Δ | 13 | 87 |
| vps26Δ | 74 | 26 |
| ypt6Δ | 91 | 9 |
Three hundred cells were counted per experiment.
WT, wild type.
In contrast to this observation, GFP-Yif1 localization was basically unaltered in mutant cells defective in endocytosis (e.g., end4-1 cells) (67), early endosome-Golgi transport (e.g., ric1Δ and vps51Δ cells) (6, 16, 68, 75), late endosome-vacuole transport (e.g., vam6Δ cells) (55), and Golgi export (e.g., sec7-5 cells at 37oC) (26) (Fig. 5A). We note that the vacuoles observed in ric1Δ and vps51Δ cells are fragmented, while they are apparently absent from vam6Δ cells, as detected using FM4-64 labeling. Nonetheless, GFP-Yif1 does not access the FM4-64-labeled compartments in these cells. Together, the results suggest that Btn2, along with Rhb1, functions upon a late endosome-to-Golgi recycling pathway that is regulated by Ypt6, Snx4, and retromer.
To verify the functionality of Btn2-GFP, we examined its ability to confer RFP-Yif1 trafficking in btn2Δ cells (Fig. 5B). RFP-Yif1 is functional as its expression was shown to confer temperature-resistant growth to yif1-2 cells at restrictive temperatures (data not shown). In cells lacking BTN2, we found that RFP-Yif1 expressed from a single-copy plasmid mislocalized to large structures that corresponded to vacuoles, as seen for GFP-Yif1 in Fig. 5A. In contrast, RFP-Yif1 labeling appeared as numerous small punctate structures in cells expressing either Btn2-GFP or Btn2 from single-copy plasmids. Our results imply that Btn2-GFP is functional and can correct Yif1 mislocalization.
Golgi-, endosome-, and vacuole-targeted proteins are not generally mislocalized in btn2Δ cells.
As Btn2 appears necessary for GFP-Yif1 and RFP-Yif1 recycling, we examined whether other proteins are mistrafficked in btn2Δ cells. We first examined whether GFP-Snc1 undergoes normal recycling from early endosomes to the Golgi apparatus (47). We examined the localization of GFP-Snc1 expressed from a single-copy plasmid in btn2Δ cells (both Euroscarf and RKY5 btn2Δ strains were examined), as well as rcy1Δ and end4-1 cells, which served as positive controls for retention to early endosomes and for inhibition of internalization, respectively (28, 47). We found that GFP-Snc1 primarily labeled the bud plasma membrane as well as small internal punctate structures that correspond to early endosomes and trans Golgi structures (47, 70) in both btn2Δ cells and wild-type yeast (Fig. 6A). In contrast, GFP-Snc1 labeled the surface of both the mother and the bud in cells defective in endocytosis (e.g., end4-1 cells; Fig. 6A), as seen earlier (28, 70). Likewise, GFP-Snc1 gave punctate labeling corresponding to the early endosomes in rcy1Δ cells (Fig. 6A), also as seen earlier (28, 70). These results suggest that there is no significant deficiency in the endosomal sorting and retrieval of Snc1 in the absence of Btn2.
FIG. 6.
The localization of GFP-tagged Snc1, Fur4, and Ste2 is unaltered in btn2Δ cells. (A) GFP-Snc1 localization is unaltered in btn2Δ cells. Wild-type (Euroscarf and W303-1b strains tested; Euroscarf cells shown), btn2Δ (Euroscarf and RKY5 btn2Δ strains examined; Euroscarf shown), end4-1, and rcy1Δ cells expressing GFP-Snc1 from a single-copy plasmid (pRS315-GFP-cSNC1) were grown to mid-log phase on synthetic selective medium at 26°C prior to visualization by confocal microscopy. (B) Fur4-GFP localization is unaltered in cells lacking BTN2. Wild-type (Euroscarf and W303-1b strains tested; Euroscarf cells shown) and btn2Δ (Euroscarf and RKY4 btn2Δ strains examined; Euroscarf cells shown) cells expressing Fur4-GFP from a single-copy plasmid (pRS316-FUR4-GFP) were grown to mid-log phase on synthetic selective medium at 26°C prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C) before visualization using confocal microscopy. (C) Ste2-GFP localization is unaltered in cells lacking BTN2. Wild-type (W303-1b) and btn2Δ (RKY5 btn2Δ strain examined) cells expressing Ste2-GFP from a single-copy plasmid (pRS314-STE2-GFP) were grown to mid-log phase on synthetic selective medium at 26°C prior to pulse-labeling with FM4-64 (5.4 μM final concentration; 30 min at 26°C) and chase (30 min at 26°C), before visualization using confocal microscopy. Merge indicates the merger of GFP and FM4-64 (B and C) fluorescence microscopy images.
We next examined the localization of Fur4, a uracil transporter from the plasma membrane which recycles through late endosomes to the plasma membrane (11). In both wild-type and btn2Δ (Euroscarf and RKY4 btn2Δ strains) cells expressing GFP-Fur4 from a single-copy plasmid, we observed GFP-Fur4 labeling of the plasma membrane and the vacuole, where Fur4 is degraded (Fig. 6B). Thus, Fur4 trafficking appears to be normal in btn2Δ cells.
Next, we examined the localization of Ste2-GFP, the yeast α-mating factor receptor, which undergoes internalization and trafficking to the vacuole in a ligand-dependent fashion (77). In untreated wild-type cells, we found Ste2-GFP present on both the plasma and the vacuolar membranes (Fig. 6C), as we reported previously (34). Similar results were obtained with btn2Δ (RKY4) cells (Fig. 6C), implying that Btn2 does not alter the ability of Ste2 to reach either membrane.
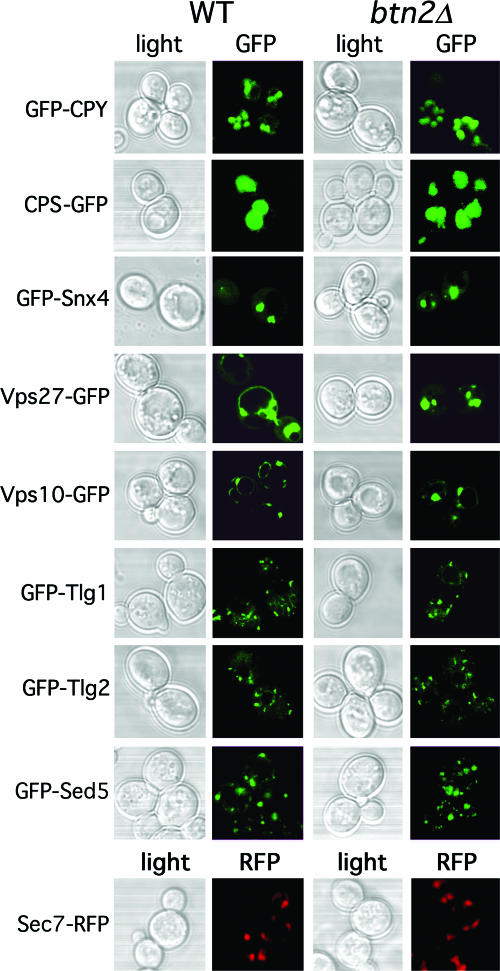
Finally, we examined the localization of other proteins that are known to reside in the Golgi apparatus and endosomal compartments or are to be trafficked to the vacuole, in btn2Δ cells. We examined GFP-tagged proteins, including Tlg1, Tlg2, Snx4, Vps27, Vps10, CPY, and CPS in btn2Δ cells (Euroscarf and RKY4 btn2Δ strains) but found no changes in their patterns of localization (Fig. 7). In addition, we examined the localization of Golgi markers, including the Sed5 t-SNARE (GFP-Sed5), which labels the cis Golgi location (36), and the Sec7 Arf exchange factor (Sec7-RFP), which labels the trans Golgi location (26). However, neither of these markers were mislocalized in btn2Δ cells (Fig. 7). Thus, the integrity of the Golgi and the endosomal and vacuolar trafficking pathways is intact in btn2Δ cells. The principle defect observed in cells lacking BTN2, therefore, is the apparent failure to retrieve Yif1 to the Golgi apparatus (Fig. 5). Together, these results imply that Btn2 is probably involved in protein export from a late endosomal compartment. This effect is specific to certain cargo proteins, as neither CPS nor CPY trafficking (trafficking through late endosomes) is affected in btn2Δ cells.
FIG. 7.
The deletion of BTN2 does not affect the trafficking of a wide variety of other endosomal cargo proteins. Wild-type (WT) and btn2Δ cells expressing CPY-GFP [pGALΔ BglII-CPY(1-50)GFP], CPS-GFP (pGO426-CPS1-GFP), GFP-Snx4 (pAD54-GFP-SNX4), GFP-Vps27 (pGO426-GFP-VPS27), Vps10-GFP (pAD54-VPS10-GFP), GFP-Tlg1 (pRS315-GFP-TLG1), GFP-Tlg2 (pRS315-GFP-TLG2), GFP-Sed5 (pRS315-GFP-SED5), or Sec7-RFP were grown to mid-log phase on synthetic selective medium at 26°C prior to visualization by confocal microscopy. Protein expression was performed using the Euroscarf wild-type and btn2Δ cells, except in the case of CPS-GFP expression, which was performed with W303-1b and RKY4 btn2Δ cells, and Sec7-RFP expression, which was performed using the W303-1a-SEC7-RFP and RKY6 strains.
GFP-Yif1 degradation is enhanced in btn2Δ cells.
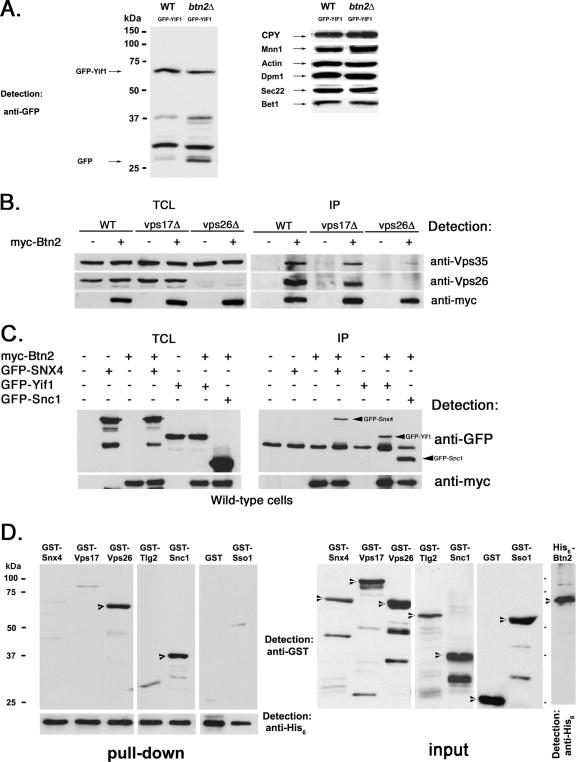
Since GFP-Yif1 appears mislocalized to the vacuole in btn2Δ cells and in cells defective in late endosome-Golgi trafficking (Fig. 5), we examined whether it is indeed degraded therein. By Western analysis, we determined that more GFP-Yif1 is found in its various smaller (degraded) forms in btn2Δ cells than in wild-type cells, as detected using anti-GFP antibodies (Fig. 8A). In the representative experiment shown, 68% of GFP-Yif1 was present as degradation products, compared to 27% in wild-type cells, after normalization for expression. In contrast, the levels of other proteins known to reside in or transit through the Golgi apparatus (i.e., Mnn1, Sec22, Bet1, and CPY) were basically unchanged in btn2Δ cells (Fig. 8A). Likewise, the levels of an ER marker, Dpm1, and a cytosolic marker, actin, were also unchanged (Fig. 8A). Thus, GFP-Yif1 that is not recycled to the Golgi apparatus is probably degraded in the vacuole in btn2Δ cells. This explains why the vacuolar lumen and not the limiting vacuolar membrane is labeled by GFP-Yif1 or RFP-Yif1 in btn2Δ cells and other cells deficient in late endosome-Golgi sorting (Fig. 5).
FIG. 8.
Btn2 interacts with a complex containing Yif1, Snx4, and retromer. (A) GFP-Yif1 is more degraded in cells lacking BTN2. Wild-type (WT) (W303-1b) and btn2Δ (RKY4) cells expressing GFP-Yif1 (GFP-Yif1) from a single-copy plasmid (pRS316-GFP-YIF1) were grown to mid-log phase and lysed. Equal amounts of protein from TCLs were subjected to SDS-PAGE and Western analysis with antibodies against GFP (1:1,000; left panel) to detect both GFP and GFP-Yif1, and intermediates. In the right panels, antibodies against CPY (1:1,000), Mnn1 (1:1,000), Dpm1 (1:1,000), Sec22 (1:1,000), Bet1 (1:1,000), and actin (1:10,000) were used. (B) Btn2 coimmunoprecipitates with Vps26 and Vps35. Wild-type, vps17Δ, and vps26Δ cells (all Euroscarf strains) expressing myc-Btn2-RFP from a single-copy plasmid (+) (pRS316-myc-Btn2-mRFP) or bearing a control vector (−) (pRS316) were grown to mid-log phase and processed for immunoprecipitation. Samples from the TCLs and precipitates (IP) were resolved by SDS-PAGE and detected in blots with anti-Vps26 (1:1,000), anti-Vps35 (1:1,000), and anti-myc (1:1,000) antibodies. (C) Btn2 coimmunoprecipitates with Snx4. Wild-type yeast (BY4741) expressing myc-Btn2 from a single copy plasmid (+) (pRS316-myc-Btn2-mRFP) or bearing a control vector (−) (pRS316) were transformed with plasmids expressing GFP-Snx4 (pAD54-GFP-SNX4), GFP-Yif1 (pAD54-GFP-YIF1), GFP-Snc1 (pAD54-GFP-cSNC1), or a control vector (−) (pAD54). Cells were grown to mid-log phase and processed for immunoprecipitation. Samples from the TCLs and precipitates were resolved by SDS-PAGE and detected in blots with anti-GFP (1:1,000) and anti-myc (1:1,000) antibodies. (D) His6-Btn2 binds directly to GST-Snc1 and GST-Vps26. Pull-down experiments (left panels) were performed by mixing equal amounts of protein (10 μg) of purified recombinant His6-Btn2 and GST-tagged Snx4, Vps17, Vps26, Tlg2, Snc1, and Sso1, and GST alone. After binding, nickel-charged beads were used to precipitate complexes which were resolved by SDS-PAGE and detected by anti-GST (1:500) or anti-His6 (1:1,000) antibodies. Arrowheads indicate the detection of GST-Vps26 and GST-Snc1 in the blots. Samples (10 μg) of the purified recombinant proteins (input; right panels) were also resolved by SDS-PAGE and detected with anti-GST (1:500) or anti-His6 (1:1,000) antibodies. Arrowheads indicate the appropriate full-length recombinant proteins.
btn2Δ cells do not secrete CPY or Kar2 and are calcofluor sensitive.
Although the deletion of BTN2 does not affect the trafficking (Fig. 7) of CPY-GFP or its receptor, Vps10 (19), nonetheless, a portion of CPY may be secreted outside the cell, as seen with vps mutants. To examine this, we grew wild-type, btn2Δ, and control vps cells on nitrocellulose filters and examined them for the presence of CPY by immunoblotting. However, CPY was not detected on filters at levels different than those seen with wild-type cells, while it was secreted by the control vps cells (data not shown). Next, we examined whether cells lacking BTN2 are defective for the retention of Kar2 to the ER by using immunoblotting (5). Defects in the Golgi-ER retrograde transport typically seen in COPI mutants (20, 21, 46) lead to Kar2 secretion onto filters. However, we found that Kar2 was not secreted onto nitrocellulose filters from cells lacking BTN2, unlike sec21-2 and sec33-1 control cells (data not shown). Thus, the lack of Btn2 does not lead to obvious defects in retrograde transport early in the secretory pathway, nor does it abolish proper CPY trafficking to the vacuole.
btn2Δ cells have no obvious defects in protein secretion or growth, indicating that the secretory pathway is not generally altered in the absence of this factor (58-60; our unpublished observations). Nevertheless, to examine whether protein export from early endosomes is generally affected, we determined whether btn2Δ cells are sensitive to calcofluor, a molecule which binds to chitin and inhibits growth. Cells in which Chs3 delivery to the plasma membrane is blocked are resistant to calcofluor (84). This assay has been used to identify mutants inhibited in Chs3 export from early endosomes (i.e., chs4Δ) as well as compensatory suppressor mutations (84). We examined whether btn2Δ and other mutants are sensitive to calcofluor. We found that unlike control chs4Δ cells, btn2Δ cells and other yeast cells (i.e., rhb1 cells) remain sensitive to increasing concentrations of calcofluor (data not shown). Thus, btn2Δ and rhb1Δ cells are unlikely to be significantly defective in Chs3 export from early endosomes.
Btn2 coimmunoprecipitates retromer components and Snx4.
As Btn2 is predicted to act upon the retrieval of specific cargo molecules (i.e., Yif1, but not Vps10) from the late endosome to the Golgi apparatus, we examined whether it interacts with retromer, a protein complex composed of Vps5, -17, -26, -29, and -35 that confers Vps10 endosome-Golgi recycling (73, 74). myc-tagged Btn2 was used to precipitate proteins from both wild-type cells and cells bearing mutations in retromer (e.g., vps17Δ and vps26Δ cells), using anti-myc antibodies. We employed the latter strains in case Btn2 was more tightly held in a complex with retromer under conditions where retromer disassembly might be compromised. Indeed, myc-Btn2 was able to precipitate components of retromer, including native Vps26 and Vps35 (Fig. 8B). This interaction was apparent in both wild-type cells and vps17Δ cells but was abolished in vps26Δ cells (Fig. 8B). This suggests that Btn2 is a novel interacting component with retromer and that the interaction therewith may depend upon Vps26 and/or retromer assembly.
In parallel, we examined immunoprecipitates for the presence of GFP-tagged Vps10, the CPY receptor known to interact with retromer (19), in lysates derived from cells overexpressing Vps10-GFP but observed no physical interaction with myc-Btn2 (data not shown). Since the deletion of BTN2 does not alter Vps10 or CPY trafficking (Fig. 7) and Btn2 does not colocalize with Vps10 in wild-type cells (Fig. 3C), we suggest that Btn2 may be part of a retromer-mediated trafficking complex independent of Vps10.
Next, we examined whether Snx4, which interacts with retromer components and is involved in endosome-Golgi recycling of Snc1 (38), binds to Btn2. We found that myc-Btn2 was able to precipitate GFP-tagged Snx4 from wild-type cells, along with GFP-Yif1 and GFP-Snc1, which served as controls (Fig. 8C). Thus, Btn2 may form a complex with a sorting nexin and the retromer coat in order to retrieve proteins (i.e., Yif1) to the Golgi apparatus.
His6-Btn2 interacts directly with GST-Snc1 and GST-Vps26.
To test whether Btn2 interacts directly with individual SNAREs, retromer components, and Snx4, we produced recombinant His6-tagged Btn2 in E. coli and determined whether it could bind to recombinant GST-tagged SNAREs (i.e., GST-Snc1, GST-Tlg2, and GST-Sso1), retromer components (i.e., GST-Vps17, and GST-Vps26), and GST-Snx4 in vitro. His6-Btn2 purified from bacteria ran as a single band of ∼60 kDa with SDS-PAGE (Fig. 8D, far right panel). This indicates that recombinant His6-Btn2 is not modified, in contrast to HA-Btn2 which appears as a doublet in yeast extracts (Fig. 2B). We note that similar results were found for another Snc v-SNARE-interacting protein, Vsm1 (49), though the nature of these modifications is unknown. Purified GST-tagged proteins of the correct molecular mass were also produced, although some degradation products were apparent in the different preparations (Fig. 8D, right panels). All recombinant SNAREs contained their soluble amino-terminal portions and SNARE binding motifs but lacked their transmembrane domains.
In an in vitro binding assay, we mixed equal amounts of His6-Btn2 and the individual GST-tagged proteins, performed pull-downs with nickel beads, and separated the bound products with SDS-PAGE. We found that His6-Btn2 did not bind to GST alone but bound to both GST-Snc1 and GST-Vps26 specifically (Fig. 8D, left panels). Densitometric analysis of His6-Btn2 binding to GST-Snc1 or GST-Vps26 showed that it bound equally well to both and suggests that the affinity of Btn2 for either the v-SNARE or the retromer component is fairly similar. In contrast, His6-Btn2 did not bind to other SNAREs (i.e., GST-Tlg2 or GST-Sso1), GST-Vps17, or GST-Snx4 (Fig. 8D, left panels) individually. Together with the immunoprecipitation data (Fig. 2B and 8B and C), this suggests that Btn2 interacts with specific components of a complex involved in Yif1 sorting from late endosomes to the Golgi apparatus but not with all members of the complex.
DISCUSSION
Batten disease or JNCL is a severe neurodegenerative disorder characterized by the accumulation of autofluorescent material in the lysosomes of cells derived from afflicted individuals. This phenotype implies defects in the sorting of material away from the lysosome or the faulty degradation of normally trafficked materials or both. In the course of our work, we have found that a Batten disease-related protein in yeast, Btn2 (18, 59), acts upon protein trafficking from late endosomes. In particular, cells lacking BTN2 accumulate a fluorescent protein-tagged Golgi marker (Yif1) in the vacuole (Fig. 5A and B; Table 3), as shown earlier (14), where it undergoes degradation (Fig. 8A). Yif1 mislocalization also occurs in mutants preferentially defective in late endosome-Golgi trafficking (Fig. 5A; Table 3); thus, Btn2 is likely to act upon this same transport step. Defects in late endosome-Golgi protein recycling may, therefore, be the mechanism underlying Batten disease/NCL pathogenesis in mammals.
BTN2 encodes a Hook1 ortholog (Fig. 1) that is a late component of the endocytic pathway (Fig. 3A) and which localizes adjacent to the vacuole but is clearly nonnuclear (Fig. 3A and B). Btn2, along with the Rhb1 small GTPase, appears to play a prominent role in the recycling of Yif1, as the deletion of either gene results in Yif1 missorting to the vacuole (Fig. 5; Table 3). Thus, we propose that both Btn2 and Rhb1 mediate late endosome-Golgi recycling. This is supported by several lines of evidence. First, known late endosome-Golgi trafficking mutants, such as the vps26Δ strain, have a similar effect upon Yif1 sorting (Fig. 5A; Table 3), and GST-Vps26 binds directly to His6-Btn2 in in vitro binding assays (Fig. 8D). VPS26 encodes a component of retromer, a pentameric complex composed of Vps5, Vps17, Vps26, Vps29, and Vps35 (73, 74). Retromer regulates the trafficking of the CPY receptor, Vps10, and retrieves it from the late endosome to the Golgi apparatus. Vps26 has been proposed to link both cargo selection and retromer assembly, while other retromer components, Vps5 and Vps17, form a membrane-associated subcomplex and are the yeast equivalents of sorting nexins 1 and 2 (Snx1 and -2) (73, 74). These and other retromer components have all been shown to localize to the late endosome (73, 74). Second, Btn2 localizes to large perivacuolar structures that are consistent with late endosomes (Fig. 3 and 4). Btn2 colocalizes in part with the sorting nexin Snx4, the Vps27 late endosome/MVB marker, and the Tlg2 endosomal t-SNARE to these same structures located adjacent to the vacuole (Fig. 3C). Third, Btn2 coimmunoprecipitates with Yif1, Snx4, and retromer (Fig. 8B,C), along with an endosomal SNARE complex (Fig. 2B), suggesting that it may be a component of an assembled retromer-mediated Yif1 sorting complex. Finally, in vitro binding studies (Fig. 8D) show that recombinant Btn2 binds to specific components of this putative complex, including the recycling v-SNARE, Snc1, which recycles from early endosomes to the Golgi apparatus (47) and, by inference from this work, from late endosomes to the Golgi apparatus. In addition, recombinant Btn2 binds to Vps26, which is involved in retromer assembly and cargo sorting (73). Thus, we propose that Btn2 confers cargo retrieval (i.e., Yif1 and, possibly, SNAREs) from late endosomes to the Golgi apparatus along with retromer.
Interestingly, neither CPY nor Vps10 trafficking was affected by the deletion of BTN2 (Fig. 7), although both should access the late endosome on their way to the vacuole in a retromer-dependent fashion (19, 73, 74, 78). This may imply that there are multiple retromer-mediated cargo sorting events occurring at the late endosome or perhaps that there are multiple late endosomal compartments serviced by retromer (7, 8). In either case, some may be mediated by Btn2 and others may not, although at this point we cannot distinguish between these two distinct possibilities. Despite this, we were able to show that Btn2 forms complexes with Yif1, retromer, and Snx4 but not with Vps10 (Fig. 8B and C and data not shown). This is consistent with the idea that there are distinct nexin-retromer sorting complexes operating in yeast. That neither Snc1 recycling from early endosomes to the Golgi apparatus (Fig. 6A) nor protein export to the plasma membrane (as assayed by calcofluor sensitivity and growth assays [data not shown]) are affected in btn2Δ cells suggests that the role of Btn2 is limited to regulating the trafficking of specific cargo molecules from a late endosome to the Golgi apparatus. This is supported by the other physical interaction, colocalization, and protein trafficking data presented here. Models outlining the trafficking functions mediated by Btn2 in wild-type cells and the consequences of BTN2 disruption therein are shown (Fig. 9).
FIG. 9.
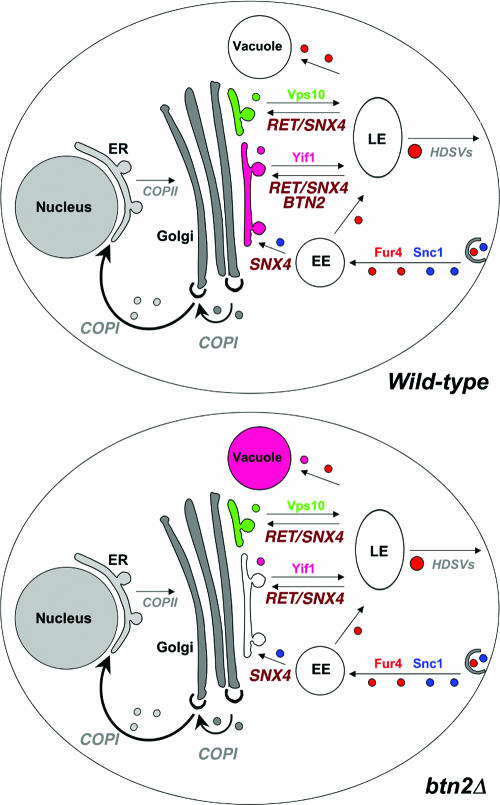
A model for Btn2 function in late endosome-Golgi protein sorting. Wild-type cells: the Snc1 exo/endocytic v-SNARE (blue) and Fur4 (red) are both endocytosed and delivered by endocytic vesicles to the early endosome (EE). Snc1 is delivered to the trans Golgi location in a manner dependent upon Snx4, Rcy1, Ric1, and Ypt31/32, etc. (later components are not listed). Fur4 is trafficked to the late endosome (LE) for either recycling to the plasma membrane or delivery to the vacuole for degradation. Retrieval to the plasma membrane is presumably mediated by high-density secretory vesicles (HDSVs), which originate from late endosomes (35, 37). A Golgi marker, Yif1 (purple), exits the Golgi apparatus to the late endosome but is retrieved in a retromer-, Snx4-, and Btn2-dependent fashion back to the Golgi apparatus. Vps10 (green), the CPY receptor, is also recycled to the Golgi apparatus from a late endosome via retromer and Snx4 but in a manner independent of Btn2. btn2Δ cells: in the absence of BTN2, the recycling of Yif1 (but not the other tested Golgi markers, e.g., Sed5 and Sec7) to the Golgi apparatus is blocked and the protein is sent to the vacuole for degradation. The endosomal sorting of Vps10, Snc1, and Fur4 is unaffected in the absence of Btn2. Thus, we predict that Btn2 acts, along with retromer and Snx4, to mediate the retrieval of Yif1 from a sorting late endosome back to the Golgi apparatus.
In this study, we found that Btn2 interacts physically and genetically with SNAREs (Fig. 2 and 8C and D) involved in endocytosis and endosomal sorting (e.g., Snc, Tlg1, Tlg2, and Vti1) (2, 10, 15, 25, 34, 40). In vitro binding studies reveal a direct interaction with Snc1, which may help recruit Btn2 to the endosomal SNARE complex. This may facilitate interactions with Yif1 (14) (Fig. 8C) and retromer (Fig. 8B and D) and might be dependent on the Rhb1 small GTPase, which is known to interact with Btn2 (13) and whose deletion phenocopies btn2Δ cells (Fig. 5A). More work is clearly necessary to determine the temporal order of events leading to the assembly of the putative retrieval complex in which Btn2 functions, along with retromer, Snx4, and SNAREs. Finally, there is a connection to COPI which bears future exploration. First, systematic interaction studies have demonstrated that Sec27, a COPI component, and Btn2 interact physically (39). Second, a role for COPI in late endosome-MVB sorting events has been described for mammalian cells (3, 24, 33, 87). Third, we recently demonstrated a potential role for specific COPI subunits (COPI B) (52), including Sec27, in the delivery of CPS, Ste2, Ste3, and Fur4 to multivesicular bodies in yeast (27). Fourth, retromer components cosediment with COPI-coated membranes when separated by size exclusion chromatography (74). Finally, work in progress suggests that Btn2 interacts genetically and physically with COPI B components (R. Kama and J. E. Gerst, unpublished results). Thus, Btn2 might function in conjunction with COPI, as well as with retromer, in mediating specific post-Golgi trafficking events.
Previous studies have shown that Btn2 interacts with a wide variety of proteins (i.e., Rhb1, Yif1, and Ist2) (13, 14, 43) purportedly involved in numerous processes relevant to cell growth and homeostasis (i.e., ion, pH, or amino acid balance). However, the precise role of Btn2 in intracellular membrane transport has remained obscure. Here we reveal that role to be the regulation of cargo protein recycling from the late endosome. Thus, we predict that changes in ion, pH, or amino acid homeostasis, seen in the absence of Btn2, result from the improper recycling of cargo proteins within the endomembrane transport system. If so, this would preclude a general role in the regulation of cell homeostasis, as predicted by Kim et al. (43). Furthermore, we predict that Batten disease/JNCL in humans may originate from the failed recycling of cargo proteins to their proper sites of action. This might result in the accumulation of nonrecycled cargo in other intracellular compartments (i.e., lysosome) and lead to cellular dysfunction. Interestingly, a connection between the defects in retromer function and the production of the amyloid-β peptide, which is involved in the pathogenesis of Alzheimer's disease, has been noted (76). Along with this study, it suggests that alterations in late endosome-Golgi protein recycling may play a significant role in the control of cell metabolism and viability.
Finally, an analysis of a role for Btn1 in endomembrane trafficking would seem to be a high priority, in light of the results obtained with Btn2. BTN1 encodes the yeast ortholog of CLN3 (18, 59), a gene known to be mutated in NCL patients (30, 31, 53). Btn1 has been proposed to play a role in vacuolar pH homeostasis (57) and the import of basic amino acids therein (44), although the mechanism is unknown. Our results may suggest a potential role for Btn1/Cln3 in endosomal protein sorting, which could account for its effects upon vacuolar/lysosomal homeostasis.
Acknowledgments
We thank Hagai Abeliovich, Pat Brennwald, Olivier Deloche, Rainer Duden, Scott Emr, Susan Ferro-Novick, Gabrielle Fischer von Mollard, Won-Ki Huh, Sirkka Keranen, Andreas Mayer, Hugh Pelham, Howard Riezman, Randy Schekman, Matthew Seaman, Anne Spang, Chris Stefan, Fuyu Tamanoi, and Michael Wigler for the generous gifts of reagents. We also thank Michael Marash for providing the unpublished yeast and bacterial expression plasmids used in this study.
This study was supported by grants to J.E.G. from the Batten Disease Research and Support Association (United States), the Josef Cohn Minerva Center for Biomembrane Research, Weizmann Institute (Israel), and the Kekst Family Family Center for Medical Genetics, Weizmann Institute (Israel). J.E.G. holds the Henry Kaplan Chair in cancer research.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Aalto, M. K., H. Ronne, and S. Keranen. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeliovich, H., E. Grote, P. Novick, and S. Ferro-Novick. 1998. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273:11719-11727. [DOI] [PubMed] [Google Scholar]
- 3.Aniento, F., F. Gu, R. G. Parton, and J. Gruenberg. 1996. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J. Cell Biol. 133:29-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becherer, K. A., S. E. Rieder, S. D. Emr, and E. W. Jones. 1996. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7:579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belden, W. J., and C. Barlowe. 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell 12:957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensen, E. S., B. G. Yeung, and G. S. Payne. 2001. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell 12:13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonifacino, J. S., and R. Rojas. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7:568-579. [DOI] [PubMed] [Google Scholar]
- 8.Bowers, K., and T. H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744:438-454. [DOI] [PubMed] [Google Scholar]
- 9.Brennwald, P., B. Kearns, K. Campion, S. Keranen, V. Bankaitis, and P. Novick. 1994. Sec9 Is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79:245-258. [DOI] [PubMed] [Google Scholar]
- 10.Bryant, N. J., and D. E. James. 2003. The Sec1p/Munc18 (SM) protein, Vps45p, cycles on and off membranes during vesicle transport. J. Cell Biol. 161:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugnicourt, A., M. Froissard, K. Sereti, H. D. Ulrich, R. Haguenauer-Tsapis, and J. M. Galan. 2004. Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell 15:4203-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay, S., N. E. Muzaffar, F. Sherman, and D. A. Pearce. 2000. The yeast model for Batten disease: mutations in btn1, btn2, and hsp30 alter pH homeostasis. J. Bacteriol. 182:6418-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chattopadhyay, S., and D. A. Pearce. 2002. Interaction with Btn2p is required for localization of Rsglp: Btn2p-mediated changes in arginine uptake in Saccharomyces cerevisiae. Eukaryot. Cell 1:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chattopadhyay, S., P. M. Roberts, and D. A. Pearce. 2003. The yeast model for Batten disease: a role for Btn2p in the trafficking of the Golgi-associated vesicular targeting protein, Yif1p. Biochem. Biophys. Res. Commun. 302:534-538. [DOI] [PubMed] [Google Scholar]
- 15.Coe, J. G., A. C. Lim, J. Xu, and W. Hong. 1999. A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 10:2407-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conibear, E., J. N. Cleck, and T. H. Stevens. 2003. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14:1610-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couve, A., and J. E. Gerst. 1994. Yeast Snc proteins complex with Sec9. Functional interactions between putative SNARE proteins. J. Biol. Chem. 269:23391-23394. [PubMed] [Google Scholar]
- 18.Croopnick, J. B., H. C. Choi, and D. M. Mueller. 1998. The subcellular location of the yeast Saccharomyces cerevisiae homologue of the protein defective in the juvenile form of Batten disease. Biochem. Biophys. Res. Commun. 250:335-341. [DOI] [PubMed] [Google Scholar]
- 19.Deloche, O., and R. W. Schekman. 2002. Vps10p cycles between the TGN and the late endosome via the plasma membrane in clathrin mutants. Mol. Biol. Cell 13:4296-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duden, R., M. Hosobuchi, S. Hamamoto, M. Winey, B. Byers, and R. Schekman. 1994. Yeast beta- and beta′-coat proteins (COP). Two coatomer subunits essential for endoplasmic reticulum-to-Golgi protein traffic. J. Biol. Chem. 269:24486-24495. [PubMed] [Google Scholar]
- 21.Duden, R., L. Kajikawa, L. Wuestehube, and R. Schekman. 1998. Epsilon-COP is a structural component of coatomer that functions to stabilize alpha-COP. EMBO J. 17:985-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durfee, T., K. Becherer, P.-L. Chen, S.-H. Yeh, Y. Yang, A. E. Kilburn, W.-H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 23.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faure, J., R. Stalder, C. Borel, K. Sobo, V. Piguet, N. Demaurex, J. Gruenberg, and D. Trono. 2004. ARF1 regulates Nef-induced CD4 degradation. Curr. Biol. 14:1056-1064. [DOI] [PubMed] [Google Scholar]
- 25.Fischer von Mollard, G., and T. H. Stevens. 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 10:1719-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzusoff, A., K. Redding, J. Crosby, R. S. Fuller, and R. Schekman. 1991. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriely, G., R. Kama, and J. E. Gerst. 2007. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell. Biol. 27:526-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan, J.-M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerst, J. E., L. Rodgers, M. Riggs, and M. Wigler. 1992. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc. Natl. Acad. Sci. USA 89:4338-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goebel, H. H. 1995. The neuronal ceroid-lipofuscinoses. J. Child Neurol. 10:424-437. [DOI] [PubMed] [Google Scholar]
- 31.Goebel, H. H., and K. E. Wisniewski. 2004. Current state of clinical and morphological features in human NCL. Brain Pathol. 14:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruenberg, J. 2001. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2:721-730. [DOI] [PubMed] [Google Scholar]
- 33.Gu, F., F. Aniento, R. G. Parton, and J. Gruenberg. 1997. Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes. J. Cell Biol. 139:1183-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurunathan, S., D. Chapman-Shimshoni, S. Trajkovic, and J. E. Gerst. 2000. Yeast exocytic v-SNAREs confer endocytosis. Mol. Biol. Cell 11:3629-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurunathan, S., D. David, and J. E. Gerst. 2002. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 21:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardwick, K. G., and H. R. Pelham. 1992. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J. Cell Biol. 119:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harsay, E., and R. Schekman. 2002. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 156:271-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hettema, E. H., M. J. Lewis, M. W. Black, and H. R. Pelham. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 40.Holthuis, J. C., B. J. Nichols, S. Dhruvakumar, and H. R. Pelham. 1998. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17:113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 42.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 43.Kim, Y., S. Chattopadhyay, S. Locke, and D. A. Pearce. 2005. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 4:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, Y., D. Ramirez-Montealegre, and D. A. Pearce. 2003. A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl. Acad. Sci. USA 100:15458-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer, H., and M. Phistry. 1996. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell Biol. 133:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letourneur, F., E. C. Gaynor, S. Hennecke, C. Demolliere, R. Duden, S. D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79:1199-1207. [DOI] [PubMed] [Google Scholar]
- 47.Lewis, M. J., B. J. Nichols, C. Prescianotto-Baschong, H. Riezman, and H. R. Pelham. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 49.Lustgarten, V., and J. E. Gerst. 1999. Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol. Cell. Biol. 19:4480-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marash, M., and J. E. Gerst. 2003. Phosphorylation of the autoinhibitory domain of the Sso t-SNAREs promotes binding of the Vsm1 SNARE regulator in yeast. Mol. Biol. Cell 14:3114-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matern, H., X. Yang, E. Andrulis, R. Sternglanz, H. H. Trepte, and D. Gallwitz. 2000. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19:4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon, H. T., and I. G. Mills. 2004. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16:379-391. [DOI] [PubMed] [Google Scholar]
- 53.Mole, S. 2004. Neuronal ceroid lipofuscinoses (NCL). Eur. J. Paediatr. Neurol. 8:101-103. [DOI] [PubMed] [Google Scholar]
- 54.Mole, S. E. 2004. The genetic spectrum of human neuronal ceroid-lipofuscinoses. Brain Pathol. 14:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura, N., A. Hirata, Y. Ohsumi, and Y. Wada. 1997. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272:11344-11349. [DOI] [PubMed] [Google Scholar]
- 56.Narayanan, R., H. Kramer, and M. Ramaswami. 2000. Drosophila endosomal proteins hook and deep orange regulate synapse size but not synaptic vesicle recycling. J. Neurobiol. 45:105-119. [DOI] [PubMed] [Google Scholar]
- 57.Pearce, D. A., T. Ferea, S. A. Nosel, B. Das, and F. Sherman. 1999. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 22:55-58. [DOI] [PubMed] [Google Scholar]
- 58.Pearce, D. A., S. A. Nosel, and F. Sherman. 1999. Studies of pH regulation by Btn1p, the yeast homolog of human Cln3p. Mol. Genet. Metab. 66:320-323. [DOI] [PubMed] [Google Scholar]
- 59.Pearce, D. A., and F. Sherman. 1997. BTN1, a yeast gene corresponding to the human gene responsible for Batten's disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast 13:691-697. [DOI] [PubMed] [Google Scholar]
- 60.Pearce, D. A., and F. Sherman. 1998. A yeast model for the study of Batten disease. Proc. Natl. Acad. Sci. USA 95:6915-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson, M. R., and S. D. Emr. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2:476-486. [DOI] [PubMed] [Google Scholar]
- 62.Piper, R. C., A. A. Cooper, H. Yang, and T. H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piper, R. C., and J. P. Luzio. 2001. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic 2:612-621. [DOI] [PubMed] [Google Scholar]
- 64.Poupon, V., A. Stewart, S. R. Gray, R. C. Piper, and J. P. Luzio. 2003. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell 14:4015-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price, A., D. Seals, W. Wickner, and C. Ungermann. 2000. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 148:1231-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Protopopov, V., B. Govindan, P. Novick, and J. E. Gerst. 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74:855-861. [DOI] [PubMed] [Google Scholar]
- 67.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reggiori, F., C. W. Wang, P. E. Stromhaug, T. Shintani, and D. J. Klionsky. 2003. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278:5009-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richardson, S. C., S. C. Winistorfer, V. Poupon, J. P. Luzio, and R. C. Piper. 2004. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell 15:1197-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson, M., P. P. Poon, C. Schindler, L. E. Murray, R. Kama, G. Gabriely, R. A. Singer, A. Spang, G. C. Johnston, and J. E. Gerst. 2006. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell 17:1845-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 72.Sato, T. K., P. Rehling, M. R. Peterson, and S. D. Emr. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6:661-671. [DOI] [PubMed] [Google Scholar]
- 73.Seaman, M. N. 2005. Recycle your receptors with retromer. Trends Cell Biol. 15:68-75. [DOI] [PubMed] [Google Scholar]
- 74.Seaman, M. N., J. M. McCaffery, and S. D. Emr. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siniossoglou, S., and H. R. Pelham. 2002. Vps51p links the VFT complex to the SNARE Tlg1p. J. Biol. Chem. 277:48318-48324. [DOI] [PubMed] [Google Scholar]
- 76.Small, S. A., K. Kent, A. Pierce, C. Leung, M. S. Kang, H. Okada, L. Honig, J. P. Vonsattel, and T. W. Kim. 2005. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann. Neurol. 58:909-919. [DOI] [PubMed] [Google Scholar]
- 77.Stefan, C. J., and K. J. Blumer. 1999. A syntaxin homolog encoded by VAM3 mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 274:1835-1841. [DOI] [PubMed] [Google Scholar]
- 78.Stevens, T., B. Esmon, and R. Schekman. 1982. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 30:439-448. [DOI] [PubMed] [Google Scholar]
- 79.Sunio, A., A. B. Metcalf, and H. Kramer. 1999. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell 10:847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Søgaard, M., K. Tani, R. R. Ye, S. Geromanos, P. Tempst, T. Kirchhausen, J. E. Rothman, and T. Söllner. 1994. A rab protein is required for the assembly of SNARE complex in the docking of transport vesicles. Cell 78:937-948. [DOI] [PubMed] [Google Scholar]
- 81.Tsukada, M., and D. Gallwitz. 1996. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J. Cell Sci. 109:2471-2481. [DOI] [PubMed] [Google Scholar]
- 82.Ungermann, C., and W. Wickner. 1998. Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 17:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urano, J., A. P. Tabancay, W. Yang, and F. Tamanoi. 2000. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 275:11198-11206. [DOI] [PubMed] [Google Scholar]
- 84.Valdivia, R. H., D. Baggott, J. S. Chuang, and R. W. Schekman. 2002. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell 2:283-294. [DOI] [PubMed] [Google Scholar]
- 85.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walenta, J. H., A. J. Didier, X. Liu, and H. Kramer. 2001. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whitney, J. A., M. Gomez, D. Sheff, T. E. Kreis, and I. Mellman. 1995. Cytoplasmic coat proteins involved in endosome function. Cell 83:703-713. [DOI] [PubMed] [Google Scholar]