Abstract
The activation of sex-specific alternative splice sites in the Drosophila melanogaster doublesex and fruitless pre-mRNAs has been well studied and depends on the serine-arginine-rich (SR) splicing factors Tra, Tra2, and Rbp1. Little is known, however, about how SR factors negatively regulate splice sites in other RNAs. Here we examine how Tra2 blocks splicing of the M1 intron from its own transcript. We identify an intronic splicing silencer (ISS) adjacent to the M1 branch point that is sufficient to confer Tra2-dependent repression on another RNA. The ISS was found to function independently of its position within the intron, arguing against the idea that bound repressors function by simply interfering with branch point accessibility to general splicing factors. Conserved subelements of the silencer include five short repeated sequences that are required for Tra2 binding but differ from repeated binding sites found in Tra2-dependent splicing enhancers. The ISS also contains a consensus binding site for Rbp1, and this protein was found to facilitate repression of M1 splicing both in vitro and in Drosophila larvae. In contrast to the cooperative binding of SR proteins observed on the doublesex splicing enhancer, we found that Rbp1 and Tra2 bind to the ISS independently through distinct sequences. Our results suggest that functionally synergistic interactions of these SR factors can cause either splicing activation or repression.
Alternative splicing is a crucial regulatory mechanism for the control of gene expression in higher eukaryotes. The ability to join pre-mRNA splice sites in different combinations allows single genes to produce multiple products with distinct functions. Alternative splicing is surprisingly prevalent in the genome. It is estimated that up to 73% of human genes undergo alternative splicing and that, in about ∼80% of these cases, the encoded protein is altered (14, 15, 29). Alternative splicing plays important roles in the regulation of a variety of physiological and developmental events, including sexual differentiation in Drosophila melanogaster (21), both neuronal (20) and immune functions (22), and apoptosis in mammalian cells (33).
Among the most important factors affecting alternative splicing are the serine-arginine-rich (SR) proteins and SR-related factors (10). These proteins are known to bind to exonic splicing enhancers (ESEs) in genes with both constitutive and alternative splicing. SR proteins bound to ESEs can activate splicing by facilitating interactions between the general splicing machinery and the adjacent splicing signals in the RNA. The Drosophila sex determination system provides a classic example of how these factors exert developmentally specific control over splicing. Expression of sex-specific products from the doublesex (dsx) and fruitless (fru) genes depends on several repeated ESE elements present in alternative exon sequences (13, 19, 31). SR proteins bind to the ESEs in cooperation with the SR-related factors Transformer (Tra) and Transformer2 (Tra2) in females and facilitate sex-specific recognition of nearby alternative splice sites (23, 24, 30, 38). Several SR proteins were found to interact with these enhancers, but only Rbp1 has been shown to functionally affect activation of both dsx and fru splicing in vivo (12, 13, 18). The mechanism by which SR factors promote assembly of spliceosomal complexes on alternative splice sites has been well studied. In vitro tethering experiments show that association of the Arg-Ser-rich effector regions (RS domains) of these proteins with the pre-mRNA is sufficient for activation (11, 19, 35). When associated with the exon, these regions are known to interact with U2AF and other prespliceosomal components to promote their binding at nearby splicing signals (9). In addition, it has been found in vitro that ESE-associated RS domains can interact directly with the pre-mRNA at the branch site to promote base pairing between the U2 snRNA and the pre-mRNA (35). Although it is not known which of these ESE functions is most important in vivo, it is clear that the binding of proteins with RS domains to ESEs activates splicing.
Given their intrinsic ability to activate splicing through ESE elements, it is perhaps surprising that SR proteins and related factors also repress splice site recognition in some pre-mRNAs (16, 36, 37). A striking example of this is the Tra2 protein, which represses the splicing of a specific intron (M1) in its own pre-mRNA (25). This intron is normally retained in about 60% of the Tra2 transcripts produced in the male germ line, provided that functional Tra2 protein is present. In the absence of Tra2 protein, the intron is efficiently spliced in vivo. Recombinant Tra2 protein can specifically block splicing of the intron in nuclear extracts derived from Schneider 2 cells via direct interactions with the RNA (4). In the studies presented here, we identify an intronic splicing silencer (ISS) from the regulated intron that is sufficient to confer Tra2-dependent repression on other introns in vitro. This silencer differs from the dsx ESE in its position, organization, and binding elements but behaves like the dsx ESE in mediating synergistic regulatory interactions between Tra2 protein and the SR protein Rbp1. These studies thus suggest that both splicing activation and repression can occur through functional interactions of similar SR factors.
MATERIALS AND METHODS
Plasmids and in vitro mutagenesis.
The ftz and M1/ftz3′ splicing substrates were transcribed from plasmid DNAs as previously described (4). Primer-derived mutagenesis was used to produce substitutions and deletions thereof by amplification of template plasmids with a pair of outwardly oriented phosphorylated primers with Pfu polymerase (Stratagene). Substitutions were produced by incorporating desired sequences or restriction sites at the 5′ ends of these primers. The PCR products were circularized with T4 DNA ligase (New England Biolabs) or digested and ligated to an insertion with the same restriction sites. DNA was transformed into the XL-1 Blue strain of Escherichia coli (Stratagene). The presence of the desired mutations were verified by DNA sequencing in all cases. Splicing substrates were transcribed from plasmid DNAs linearized by XhoI. Plasmids ftz-M1 1-208, ftz-M1 71-208, ftz-M1 120-208, ftz-M1 167-208, and ftz-M1 120-193 were generated using this strategy by substituting various PCR-amplified M1 intron sequences analogous to the deleted regions of the ftz intron. Plasmids M1/ftz3′-del ISS, +3, +10, +30, multimer, del 28, sub 28, and del 16, and plasmids with point mutations of the CAAGR repeats in the ISS, including 2mt, 3mt, and 5mt, were all generated by primer-derived mutagenesis using different templates. The deletion del28 extends from 138 to 165 nucleotides (nt) downstream of the 5′ splice site. In the sub 28 substrate, the region from nt 138 to 165 of M1 intron was replaced by a sequence located 67 to 95 nt downstream of the 5′ splice site in the ftz intron. The plasmid used to produce the 2BPS substrate was made by ligation of a blunted XhoI-PvuI vector fragment from ftz-M1 1-208 with an SspI-PvuI ftz fragment from the plasmid pG2V61. The splicing substrate was transcribed from this plasmid after being linearized with XhoI.
RNA substrate synthesis.
Splicing substrates and unlabeled RNA competitors were transcribed as previously described (4). For UV cross-linking experiments, each RNA was transcribed from a PCR product generated with a sense primer that included the T7 promoter and an antisense primer at the 3′ end of the ISS. In vitro transcription reactions were carried out with 5 μl of a 100-μl PCR volume for standard T7 transcription reactions as described in the Ambion Maxiscript (labeled substrates) or Megascript (unlabeled competitors) RNA transcription kits. The m7G cap analog (New England Biolabs) was included in the synthesis of both cross-linking and splicing substrates. All RNAs were purified by denaturing polyacrylamide electrophoresis before being used. Quantities of unlabeled competitors were determined by UV absorbance after gel purification. Positive- and negative-control RNAs from the doublesex gene were produced as previously described (4).
Recombinant proteins and nuclear extracts.
Recombinant six-His-Tra2 was expressed from baculovirus as described previously (4, 17). The six-His-Rbp1 protein was prepared as described previously (12) and dialyzed against D buffer before use. Glutathione S-transferase (GST) and GST-Rbp1 were expressed in the E. coli strain BL21 and purified by affinity with glutathione-agarose. Nuclear extracts were prepared from spinner cultures of Drosophila S2 cells essentially as described previously (7).
In vitro splicing assay.
Radiolabeled substrate (0.1 pmol) was incubated at 22°C for 2 h in the presence of a mixture containing 50% S2 nuclear extract, 1.2 mM MgCl2, 0.8 mM dithiothreitol, 1.2% polyethylene glycol, 40 U Protector RNase inhibitor (Roche), 20 mM phospho-l-arginine, and 2 mM ATP. For Tra2 repression studies, 3.5, 7, or 14 pmol recombinant six-His-Tra2 protein was added to the splicing reaction mixtures. For synergism experiments, 4 or 8 pmol Tra2 protein was used with 6.5, 13, 26, or 52 pmol six-His-Rbp1 protein in various combinations. Control reaction mixtures were supplemented with the same volume of buffer used for dialysis of the recombinant proteins. The RNA splicing products and intermediates were separated on 6% denaturing polyacrylamide gels and visualized by exposure to a PhosphorImager. For competition experiments, 0.5, 2.5, or 12.5 pmol of the specified unlabeled competitor RNAs were added at time zero to reactions containing either 0 or 14 picomoles Tra2 protein as indicated in each experiment. The signals produced by individual bands were quantitated using a Molecular Dynamics Storm PhosphorImager and Kodak one-dimensional quantitation software. For quantitation of splicing assays, the percentage of splicing was approximated as the sum of signals detected for all spliced products and intermediates divided by the signal from both the spliced and unspliced substrate RNA. In competition assays, splicing was carried out in the presence of 0.5, 2.5, and 12.5 pmol unlabeled ISS or 5mt RNA fragment (deriving from nt 120 to 197 nt of the M1 intron) in addition to the radiolabeled ftz-M1 1-208 splicing substrate.
RNA-protein UV cross-linking experiments.
For cross-linking assays, 0.1 pmol of labeled RNA substrate was incubated at 30°C, with 4 pmol Tra2 protein and 20% S2 nuclear extracts under splicing conditions. The mixtures were UV irradiated on ice for 10 min at a distance of 3.5 cm under a Philips TUV 15W/G15T8 UV bulb. Reaction products were digested with RNase A and RNase T1 for 20 min at 37°C. Labeled proteins were separated on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized using a phosphorimager. For reactions with Rbp1, 13, 40, and 120 pmol of recombinant protein were used under similar conditions.
Transgenic flies and crosses.
Transgenic flies were generated by injecting w1118 embryos with a P element vector carrying the pUAS-Rbp1 (Genetic Services, Inc.). Transformed lines were identified in the G1 progeny of injectees based on w+ eye pigmentation. Stable lines were crossed with a strain carrying the reporter transgene P{CZP-ORF3} (25), Clones of cells expressing Rbp1 were produced by crossing w1118/Y; P{UAS-Rbp1}, tra2b/CyO; P{CZP-ORF3} males with females of the genotype P{hs-FLP}; P{AyGAL4}, P{UAS-eGFP}. Progeny were subjected to 37°C heat shock for 30 min at between 24 and 36 h of development. This induced FLP recombinase expression and formation of random GAL4-expressing clones by excision of the FRT-flanked yellow gene which separates the actin 5C promoter from GAL4 coding sequences in P{AyGAL4}. To produce control larvae, males of the genotype w1118/Y; tra2b/CyO; P{CZP-ORF3} or w1118/Y; P{UAS-MycTra2}, tra2b/CyO; P{CZP-ORF3} were used in a parallel cross. Other strains were obtained from the Bloomington Drosophila Stock Center.
RESULTS
An intronic splicing silencer is required for M1 repression.
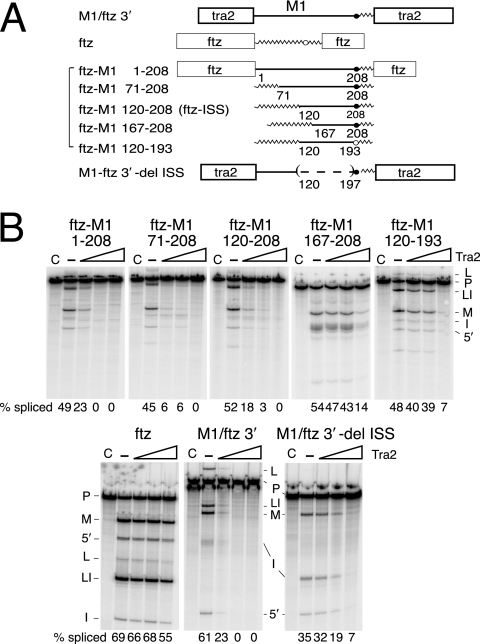
It was previously found that sequences required for repression of splicing are located within the M1 intron (4). To test if intron sequences are sufficient to confer Tra2 responsiveness on another RNA substrate, we used the first 208 nt of the M1 intron to replace analogous sequences from the intron of the fushi-tarazu (ftz) pre-mRNA, which is not a normal target of Tra2 repression in Drosophila nuclear extracts (Fig. 1B). In the hybrid substrate ftz-M1 1-208, all intron sequences located upstream of the ftz polypyrimidine are replaced by these M1 intron sequences. Splicing of this substrate in vitro was repressed by recombinant Tra2 (Fig. 1B) to the same extent as a previously studied substrate (4) flanked by native Tra2 exons (Fig. 1B, M1-ftz 3′). Thus, the M1 intron sequences upstream of the polypyrimidine tract are sufficient to confer Tra2-dependent splicing repression.
FIG. 1.
Mapping the intronic splicing silencer. (A) A schematic of the transcripts that were used in the in vitro splicing assays. The boxes indicate exons from the tra2 or ftz gene as labeled. The straight lines indicate M1 intron sequences, and jagged lines indicate intron sequences from the ftz gene. The positions of endpoints for M1 intron sequences are indicated as the number of nucleotides from the M1 5′ splice site. Branch point sequences deriving from the M1 intron (solid circle) and ftz intron (open circle) are indicated. The parentheses and the dashed line indicate deleted sequences. (B) A set of five in vitro splicing reactions is shown for each transcript. Lanes C are control reactions with no ATP; —, reactions supplemented with ATP but not recombinant Tra2 protein. Lanes under the triangles are reactions supplemented with increasing amounts of recombinant Tra2 protein. Products from in vitro splicing reactions are indicated to the side of each gel, including pre-mRNA (P), mRNA (M), lariat intermediate (L), lariat intron (LI), 5′ exon (5′), and linear intron (I). The total percentage of splicing products and intermediates is given below each gel lane (% splicing).
It is worth noting here that in previous studies on M1 splicing, strong repression was observed only when splicing occurred under the influence of an ESE in the upstream exon (4). Although ftz-M1 1-208 lacks the natural A/C-rich ESE found in the preceding exon, we found that the ftz 5′ exon contains an ESE that functionally substitutes for it (data not shown). This is consistent with a previous finding that the Tra2 A/C-rich ESE plays only a generic role in repression and can be replaced by exogenous ESEs (4).
To map the specific elements in the intron that are responsible for repression, we substituted progressively smaller segments of M1 sequences into the ftz gene-derived substrate (Fig. 1A). All fusions containing the region 120 to 208 nt of the M1 intron were found to undergo repression (Fig. 1B). For example, splicing of the substrates ftz-M1 71-208 and ftz-M1 120-208 was repressed by Tra2 at a level similar to that of ftz-M1 1-208. Splicing of other substrates (ftz-M1 167-208 and ftz 120-193) with shorter segments of the M1 intron were repressed only by highest levels of Tra2 protein tested, where we also began to observe nonspecific effects on the ftz control substrate. These results indicate that M1 intron sequences from 120 to 208 nt downstream of the 5′ splice site are sufficient to confer Tra2-dependent splicing repression on another RNA. We therefore define this region as an ISS and refer to the construct ftz-M1 120-208 as “ftz-ISS.”
The ISS defined above includes the predicted Tra2 branch point at its 3′ end. However, when M1 sequences in ftz-ISS are further reduced by substituting the branch point with ftz RNA, there was little or no effect on repression by Tra2, indicating that the exact branch point sequences are not important (data not shown).
We next tested whether there are additional elements in the tra2 RNA that function redundantly with the ISS to repress splicing. The construct M1/ftz 3′-del ISS (Fig. 1A) contains all M1 intron sequences except the ISS and both of the tra2 exons that normally flank it. This substrate supported a much lower level of Tra2-dependent repression than did substrates with the ISS, but the repression observed was nonetheless significantly above that obtained with a nonspecific control (Fig. 1B). This suggests that although the ISS defined above is the major element mediating Tra2-dependent repression, there also exist other sequences in the RNA through which Tra2 can interfere with splicing.
Conserved short repeats in the ISS are required for Tra2 binding and repression.
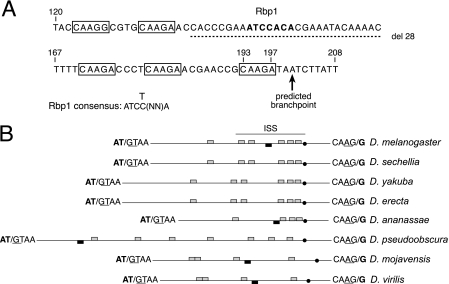
Inspection of sequences in the ISS revealed the presence of five copies of the short repeat CAAGR (R = G or A). These short repeats are interspersed with nonrepetitive sequences in the ISS and are clustered with two copies located very near the silencer's 5′ end and three near its 3′ end adjacent to the predicted M1 branch point (Fig. 2A). Previous studies on transgenic flies have shown that signals in the pre-mRNA needed for M1 repression are conserved between the Tra2 genes of Drosophila virilis and D. melanogaster (2). We therefore examined whether CAAGR repeats or any other elements of the ISS are conserved in analogous introns of other Drosophila species. Notably, in each of seven species examined, an intron was identified that splits a translation initiation codon and is thus positioned to allow autoregulation of Tra2 protein expression in a manner like the D. melanogaster M1 intron. Repeats of the sequence CAAG were found in each case (Fig. 2B). Each intron has more than 4 copies of the repeats. Although their exact positions vary, in all cases, at least one CAAG element was found in close proximity to the branch point. These observations are consistent with the idea that CAAG elements are needed for M1 repression.
FIG. 2.
Repeated sequences in the ISS are conserved between Drosophila species. (A) The sequence of the ISS element is shown with CAAGR repeats boxed. The predicted branch point for the M1 intron is indicated by an arrow. The 28-nt deletion of nonrepeat sequences is indicated by the dashed line. The numbers above the sequence indicate distance to the 5′ end of the intron. The predicted Rbp1 binding site in the ISS is indicated in boldface type, and the consensus sequence for type B Rbp1 binding sites (12) is shown below. (B) Schematic showing the relative position of CAAG repeats (gray boxes) and potential Rbp1 binding sites (black boxes) in the M1 introns of all Drosophila species examined. All introns are represented to scale (the D. melanogaster M1 intron is 232 nt in length), with the exception of the intron from Drosophila pseudoobscura, which is 758 nt long. Sequences at exon-intron junctions are shown. Splice sites are indicated by a slash mark, and canonical splicing signals are underlined. The translation initiation codon split by M1 is indicated in boldface type. The solid dots indicate the predicted branch point of each intron. The position of the ISS sequences defined by experiments in the D. melanogaster M1 intron is indicated by a line above the diagram.
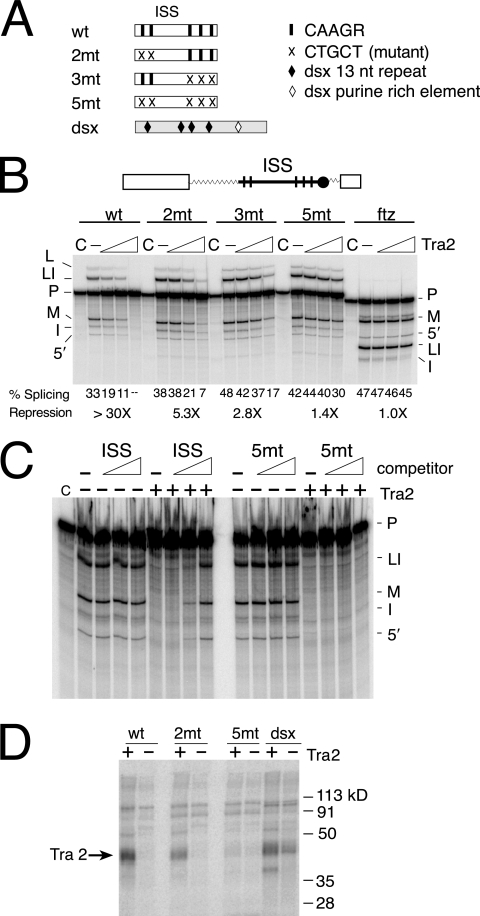
To functionally test whether the short repeats found in the D. melanogaster ISS are required for regulation of splicing, we generated point mutations that change each repeat from CAAGR to CTGCT. Substrates containing various combinations of these mutated repeats (Fig. 3A) were then assayed for their ability to support Tra2 repression in splicing assays. The results shown in Fig. 3B demonstrate that mutation of either the first two (2mt) or the last three (3mt) repeat copies significantly reduced repression. The largest effect was observed when all five copies of the repeat were removed. Repression in this case was only 1.4-fold compared to >30-fold repression observed with the wild-type ISS. This confirmed our hypothesis that the short repeats are required for repression of M1 splicing.
FIG. 3.
The CAAGR repeats in the ISS are required for Tra2 repression and binding. (A) A schematic showing the positions of point mutations affecting repeats in the ISS is shown. The black bars indicate CAAGR repeats, and the crosses indicate mutated repeats. Sequences from the dsx splicing enhancer were used as a positive control in UV cross-linking assays. (B) Splicing assays on substrates with ISS mutations are shown. Above the gel is a diagram of ftz-ISS, the base substrate used in all assays. Within this substrate, different forms of ISS including wt, 2mt, 3mt, and 5mt are tested for response to recombinant Tra2 protein. The ftz RNA substrate (ftz) served as a negative control. Splicing percentages and maximal relative (n-fold) repression are listed below the gel. Products and intermediates are labeled as described for Fig. 1. (C) Unlabeled ISS or 5mt RNA fragments (120 to 197 nt) were used as competitors in reactions containing repressive levels of Tra2 protein and the radiolabeled substrate ftz-M1 1-208. Increasing height of the triangle indicates increasing amount of RNA competitors. Note that splicing is restored by the wild-type ISS but not by the 5mt RNA with altered repeat elements. Reactions without Tra2 are shown as controls for nonspecific repression of splicing. (D) Labeled wt, 2mt, and 5mt ISS RNA fragments (120 to 197 nt) were UV cross-linked in Drosophila S2 nuclear extract under splicing conditions both with (+) and without (−) recombinant Tra2 protein present. Cross-linking reactions carried out on RNA from the dsx splicing enhancer are shown for comparison. The position of cross-linked recombinant Tra2 protein is indicated by the arrow.
To test if the ISS binds factors needed for M1 repression, both wild-type and mutant ISS RNAs were used as competitors in splicing assays that were supplemented with recombinant Tra2 protein at levels giving strong repression. To avoid titrating essential splicing factors, only ISS sequences upstream of the branch point (120 to 197 nt) were used for these binding assays. Addition of unlabeled wild-type ISS RNA reversed Tra2-dependent repression, while competitors with mutations of CAAGR repeats did not (Fig. 3C). These results indicate that the ISS RNA can bind Tra2 or other factors required for Tra2-dependent repression and that the binding of these factors is dependent on the repeat elements.
To examine Tra2 binding more directly, we performed UV cross-linking assays on the same ISS sequences after incubation in S2 nuclear extracts under splicing conditions with and without recombinant Tra2. As shown in Fig. 3D, the ISS RNAs bind to recombinant Tra2 proteins in a manner dependent on the CAAGR repeats. Point mutations in the first two copies of the repeats (2mt) reduce Tra2 binding only partially, but mutations in all five copies of repeats (5mt) eliminated Tra2 cross-linking. In summary, our splicing and binding data indicate that the conserved repeats in the ISS are required for the Tra2 protein to recognize and repress splicing of the M1 intron.
The ISS can function independently of its proximity to the branch point.
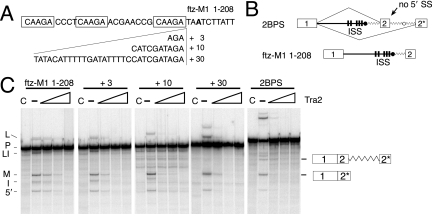
Three CAAGR repeats are located less than 29 nt upstream of the predicted M1 branch point (Fig. 2). The proximity of these elements suggested the possibility that Tra2 bound at these positions might block access of general splicing factors to sequences near the branch point. In particular, non-sequence-specific interactions have been observed between components of the U2 snRNP and intronic sequences immediately upstream of the branch point (8). To test if this location of the ISS is important for its function, we produced a series of modified splicing substrates in which spacer sequences of 3 nt, 10 nt, or 30 nt in length were inserted to separate the CAAGR repeats from the branch point (Fig. 4A). We then assayed the ability of these substrates to undergo Tra2-dependent splicing repression in vitro. As shown in Fig. 4C, the ISS functioned normally in each case, indicating that it does not depend acutely on local position.
FIG. 4.
The ISS can function at a distance from the branch point. (A) A diagram of spacer sequences of 3 nt, 10 nt, or 30 nt inserted between the last CAAGR repeat and the predicted M1 branchpoint in the ftz-M1 1-208 substrate is shown. Spacer sequences were derived from the ftz intron. The branch point adenosine is indicated in boldface type. (B) The structure of predicted splice products from the 2BPS substrate is shown and compared to ftz-M1 1-208. It is derived by adding ftz branch point and duplicated 3′ exon (2*) over 100 nt downstream of the ISS. The wavy line indicates ftz intron sequences. (C) The ftz-M1 1-208 substrate as well as +3, +10, and +30 RNAs and 2BPS were spliced in Drosophila S2 nuclear extract in the presence and absence of recombinant Tra2 protein. Products from ftz-M1 1-208 are indicated on the left side of the gel using symbols as in previous figures. The positions of spliced products using each 3′ splice site of 2BPS are diagrammed on the right side of the gel.
To more stringently test if the ISS functions independently of position in the intron, we moved it further away from the branch point by another strategy illustrated in Fig. 4B. In this case, we duplicated the branch point and polypyrimidine tract, 3′ splice site and downstream exon sequences at a position over 100 nt downstream of the native ISS. We then asked whether splicing to the downstream exon could be repressed by Tra2 through the ISS. Splicing products from these two acceptor sites of the 2BPS substrate were detected in the absence of Tra2, and notably, both were significantly repressed in its presence (Fig. 4C). Together, the above data suggest that the ISS functions independently of its position with respect to the branch point and other 3′ elements. Thus, Tra2-ISS complexes are unlikely to simply block required splicing signals. These results suggest instead that such complexes interfere with the function of splicing activators or general splicing factors (see Discussion).
Nonrepeat ISS sequences are needed for repression of M1 splicing.
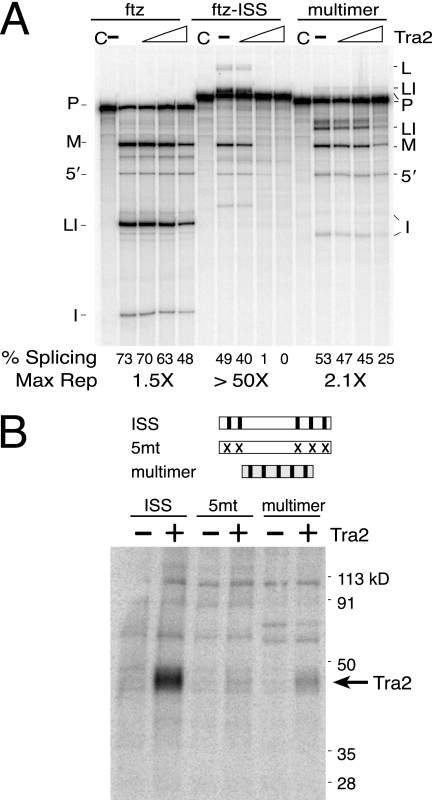
As the CAAG repeats are required for Tra2 binding and splicing repression, we next tested whether they alone are sufficient for these functions. A 50-nt-long repeat multimer consisting of 5 copies of the sequence CAAGA separated by 5 nt of spacer sequences was used to replace the entire ISS in the ftz-ISS splicing substrate. When assayed in vitro, this multimer supported only twofold repression (Fig. 5A), a level only slightly higher than the nonspecific repression measured on the ftz substrate in parallel control assays. To investigate why the repeat multimer does not support better repression, we compared its ability to bind Tra2 with that of the ISS in cross-linking assays. As shown in Fig. 5B, binding of Tra2 to the repeat multimer was significantly reduced in relation to the intact ISS. Binding of the multimer was only slightly stronger than a copy of the ISS with mutant repeats (5mt). We conclude that additional sequences outside the repeats are needed for Tra2 binding and function.
FIG. 5.
The repeat multimer is not sufficient to support Tra2 repression and binds Tra2 weakly. (A) Parallel splicing assays are shown for the ftz control substrate (ftz), a similar substrate with the ISS inserted (ftz-ISS), and a substrate in which a multimer containing 5 copies of the sequence CAAGA was substituted for the ISS in ftz-ISS (multimer). Splicing percentages and maximal relative (n-fold) repression are shown below the gel. (B) Cross-linking of Tra2 to the same CAAGA RNA multimer is compared to that with the ISS (ISS) and ISS sequences with mutated repeats (5mt). Reactions were carried out in Drosophila S2 nuclear extracts both in the presence (+) and absence (−) of recombinant Tra2 protein. The position of cross-linked recombinant Tra2 is indicated. The cross-linked RNAs did not include splice junction or branch point sequences. Mutant repeats and symbols are like those described for Fig. 3.
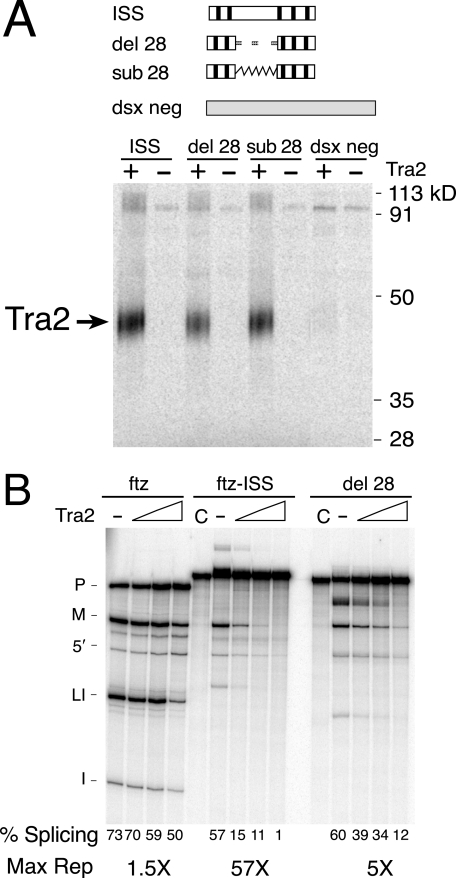
To further determine if nonrepeat sequences are required for repression, we examined mutations altering the region between the two clusters of repeats in the ISS (see Fig. 2A and Fig. 6A). Alteration of the central 28 nt in this region had only a small effect on Tra2 binding (Fig. 6A, del 28 and sub 28), but the level of Tra2-dependent repression was reduced from 50-fold to 5-fold when these sequences were deleted (Fig. 6B). These results indicate that, in addition to the CAAGR repeat, nonrepeat sequences are required for ISS function.
FIG. 6.
Nonrepeat ISS sequences are needed for repression of M1 splicing. (A) A deletion (del 28) and a substitution (sub 28) of the 28-nt spacer region were compared to the wild-type ISS for their ability to cross-link with Tra2. The negative control RNA “dsx neg” is derived from sequences in exon 4 of the dsx gene outside the splicing enhancer and is included as a control for nonspecific cross-linking. The dashed line indicates deleted sequences, and the jagged line indicates substitution with sequences from ftz intron. (B) A ftz-ISS-derived substrate with the del 28 mutation was tested in splicing assays for its response to Tra2. The ftz and ftz-ISS substrates were used as controls. Splicing percentages and maximum repression levels are indicated below the gel.
Rbp1 binds the ISS and represses M1 splicing in vitro.
The above results raised the possibility that a previously unidentified factor involved in M1 repression may bind the ISS through sequences in the 28-nt region between the repeat clusters. Examination of the sequences in this region revealed a predicted binding site for the splicing factor Rbp1 (ATCCACA), a Drosophila homolog of mammalian SRp20 protein (Fig. 2A). This site is a perfect match to the type B Rbp1 target sequence that was identified previously using an in vitro selection approach (12).
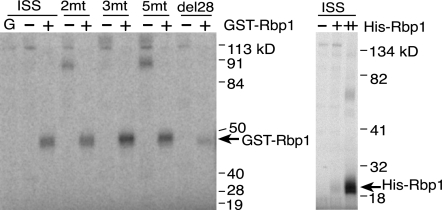
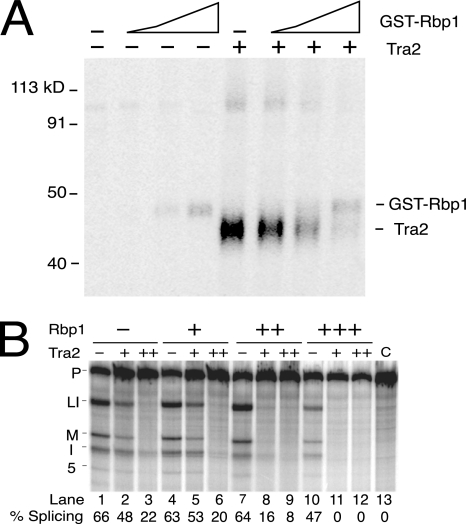
To determine if Rbp1 binds to this region of the M1 ISS, we carried out UV cross-linking assays on labeled ISS RNAs in the presence of nuclear extracts supplemented with recombinant Rbp1 proteins. Both six-His-Rbp1 and GST-Rbp1 cross-linked to the ISS, but GST protein alone did not (Fig. 7). Importantly Rbp1 cross-linking occurred independently of the CAAGR repeat elements and was significantly reduced when the region containing the putative consensus Rbp1 binding site was deleted (Fig. 7, del 28). We found, however, that significant residual binding of recombinant protein was consistently detected in the absence of this site, suggesting that interactions with Rbp1 also occur at other positions in the ISS.
FIG. 7.
Rbp1 binds the ISS independently of the CAAGR repeats. In the left panel, UV cross-linking assays carried out on ISS RNAs after incubation in S2 nuclear extracts both with (+) and without (−) added recombinant GST-Rbp1 fusion protein are shown. The labeled RNAs are the wild-type ISS, ISS RNAs with mutations in two repeats (2mt), three repeats (3mt), or all 5 repeats (5mt), and ISS RNA with the spacer region deleted (del28). The lane at the far left labeled “G” shows results from cross-linking with the GST protein without fused Rbp1 sequences. In the right panel, an identical cross-linking assay on labeled wild-type ISS RNA using increasing amounts (indicated by + and ++) of a six-His-Rbp1 fusion protein to supplement S2 nuclear extracts.
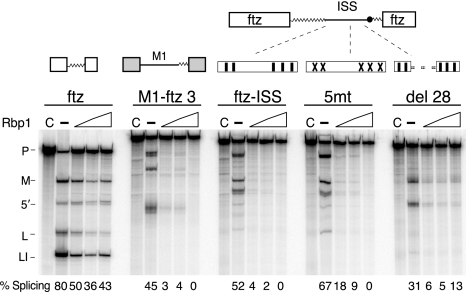
We next tested whether recombinant Rbp1 plays a role in the repression of M1 splicing in vitro. Surprisingly, recombinant Rbp1 protein strongly repressed splicing of M1-ftz 3′ substrate in extracts that were not supplemented with recombinant Tra2. Repression by Rbp1 was specific to the M1 intron and over 20-fold greater than the nonspecific repression observed with the ftz intron control substrate (Fig. 8). The ISS is sufficient to direct Rbp1 repression, as the ftz-ISS substrate supported the same level of repression as did a substrate with more extensive M1 sequences (M1-ftz 3′).
FIG. 8.
Rbp1 represses M1 splicing in vitro. RNA substrates ftz, M1-ftz 3′, ftz-ISS, 5mt, and del 28 were tested in the splicing reactions for their response to recombinant six-His-Rbp1 proteins. The diagram illustrates the ISS regions in ftz-ISS, 5mt, and del 28 using the symbols described for Fig. 3. Note that repression is stimulated by the ISS alone when it is inserted into the ftz intron but does not depend on the CAAGR repeat elements. Splicing percentages are shown below each lane.
Consistent with binding assays, repression was independent of the CAAGR repeat elements and was reduced by deletion of its consensus binding site. However, significant residual repression was observed in the absence of this site (Fig. 8, del28). This is consistent with the finding that a point mutation affecting only the predicted Rbp1 binding site did not alter observed ISS function (data not shown). Thus, our data indicate that the predicted Rbp1 site identified above is not required to account for the interaction of this protein with the ISS. Taken together, the above results demonstrate that Rbp1 can physically interact with the ISS and facilitates repression of M1 splicing but that this protein must interact with additional sites within the ISS.
Tra2 and Rbp1 function synergistically but bind to the ISS independently.
The observations that supplementation of S2 extracts with either Tra2 or Rbp1 repressed M1 splicing through the ISS led us to test whether Tra2 and Rbp1 synergize in binding and/or function. To examine binding, we carried out a titration experiment in which a constant amount of Tra2 was cross-linked to the ISS in the presence of increasing amounts of Rbp1. Both proteins were present in the same concentration range, where they cause measurable repression of splicing in vitro. As can be seen in Fig. 9A, increasing amounts of Rbp1 did not augment Tra2 binding, nor did the presence of Tra2 increase Rbp1 binding. Notably, at high Rbp1 concentrations, the proteins appear to compete for binding. The latter effect may be due to artificially high levels of Rbp1 (the same levels yield some nonspecific inhibition of RNA splicing); however, binding of the two proteins to the ISS is clearly not cooperative.
FIG. 9.
Tra2 and Rbp1 bind the ISS independently but function synergistically. (A) Wild-type ISS RNA was incubated in the Drosophila S2 nuclear extract and UV cross-linked with increasing amounts of GST-Rbp1 and Tra2 protein. The concentration of Tra2 used (4 picomoles) is at the lower end of the range where repression is observed but has no effect on Rbp1 binding. (B) Limiting amounts of recombinant Rbp1 and Tra2 proteins were used in splicing assays to investigate the combined effect on the repression of M1 splicing. The substrate used in these assays is ftz-ISS. Symbols are as described for Fig. 1.
Although their binding appears to be independent of one another, titrations of Rbp1 and Tra2 indicate that the proteins can function synergistically to repress M1 splicing. Using amounts of Rbp1 and Tra2 proteins that are limiting for repression, we examined their combined effect. Up to 100 ng of Rbp1 did not repress ftz-ISS splicing by itself (Fig. 9B, compare lane 1 to lanes 4 and 7); however, when combined with an amount of Tra2 that by itself results in less than 0.5× repression (Fig. 9B, compare lanes 1 and 2), splicing of the substrate was repressed by fourfold (Fig. 9B, compare lanes 7 and 8). When an amount of Rbp1 causing modest repression was used (Fig. 9B, lane 10), the addition of the same limiting amount of Tra2 eliminated splicing (Fig. 9B, lane 11). Together, the above results suggest that, when combined, Tra2 and Rbp1 bind to the ISS independently but function synergistically to more effectively repress M1 splicing.
Increased Rbp1 expression in vivo induces M1 retention.
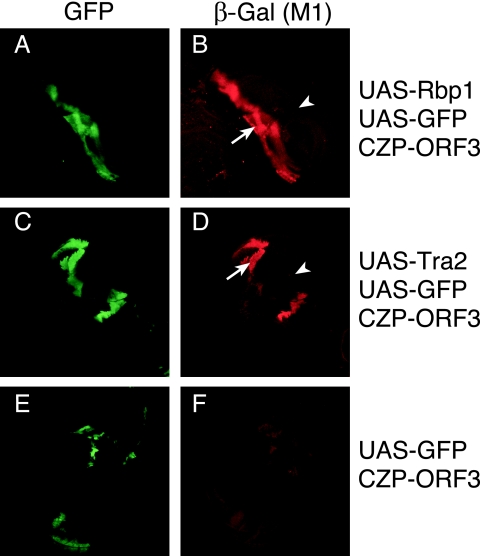
The above experiments show that the Rbp1 protein stimulates M1 repression in vitro. To test whether, like Tra2, Rbp1 can cause M1 retention in the fly, we employed the Gal4-UAS system to increase expression of Rbp1 in specific tissues of Drosophila larvae. In these experiments, an established reporter transgene that expresses β-galactosidase from M1-containing RNAs was used to monitor M1 retention (25, 27). Shown in Fig. 10A is a clone of imaginal disk cells in which a recombination-activated Gal4 transgene simultaneously drives expression of UAS-GFP and UAS-Rbp1. Staining of the same cells with a β-galactosidase antibody shows dramatically enhanced expression of the M1 reporter relative to surrounding GFP-negative cells where UAS-Rbp1 expression is not induced (Fig. 10B). The enhancement in reporter expression caused by Rbp1 is similar to that obtained in a parallel experiment where Tra2 expression is activated in a similar manner (Fig. 10C and D). No such enhancement was observed in GFP-positive clones lacking the UAS-Rbp1 transgene (Fig. 10E and F). These results are consistent with our in vitro findings and indicate that Rbp1 can function in vivo to increase M1 retention.
FIG. 10.
Clones of imaginal disk cells with increased Rbp1 expression have increased M1 retention. Results from whole-mount immunostaining of Drosophila larval imaginal disks are shown. An FLP-induced Gal4-positive clone expresses UAS-Rbp1 and is marked by the activity of a UAS-GFP transgene (A). The same cells (arrow) show enhanced expression of β-galactosidase from the M1 splicing reporter P{CZP-ORF3} relative to cells neighboring the clone (arrowhead) (B). For comparison, a GFP-positive clone expressing UAS-MycTra2 is shown (C) that similarly induces the reporter (D). In a parallel cross that omits the UAS-Rbp1 transgene, the GFP-expressing clones do not increase expression of the same reporter (E and F).
DISCUSSION
The splicing of the M1 intron has previously been shown to depend on multiple distinct sequences (3, 4). These include an AC-rich exonic splicing enhancer in the 5′ exon, a conserved intronic splicing enhancer immediately downstream of the 5′ splice site, and a variant 3′ splice site/polypyrimidine tract. Together, these elements ensure that the intron's basal recognition by the general splicing machinery is permissive for repression by Tra2 in the male germ line. However, both experiments in transgenic flies and in nuclear extracts indicate that these sequences function only as “context elements” and do not mediate specific interactions with Tra2. In this respect, the ISS element differs. It interacts specifically with Tra2 and can confer Tra2-dependent repression on the ftz intron. Thus, the ISS is sufficient for repression provided it is placed in an appropriate context.
A series of repeats of the sequence CAAGR within the ISS are critical for both binding and repression by Tra2. Several lines of evidence suggest, however, that the interaction between Tra2 and these repeats is not sufficient to cause repression. Multimers of the repeats alone were not able to support splicing repression in vitro and supported only weak cross-linking of Tra2. In addition, we have observed that cross-linking of recombinant Tra2 with the ISS depends on the presence of nuclear extract (J. Qi and W. Mattox, unpublished), suggesting that additional factors are involved. Finally, we found that the SR protein Rbp1 binds to the ISS independently of CAAGR repeats and is capable of repressing M1 splicing both in vitro and in vivo. Thus, it appears that the ISS interacts with multiple factors that act to repress M1 splicing.
The ability of Rbp1 to repress M1 splicing in nuclear extracts and in larval cells is consistent with a recent report that cell strains overexpressing Rbp1 or the related Rbp1-like protein had increased levels of M1 retention in endogenous Tra2 RNAs originating from its somatic promoter (18). In the same study, it was found that extensive cross-regulatory interactions occur at the level of splicing between Rbp1 and several other SR protein genes. The repression of M1 splicing by Rbp1 is likely to represent yet another extension of this regulatory network. The role that Rbp1 might play in regulating Tra2 expression is not known. However, M1 splicing plays a critical role in regulating levels of Tra2 during spermatogenesis. Splicing of M1 is necessary for expression of functional Tra2 protein and thus male fertility (26), but failure to limit Tra2 expression through M1 retention also results in sterility due to spermatogenesis defects (27). Whether Rbp1 and Rbp1-like play roles in male fertility is not yet known and awaits the identification of viable strains deficient in these factors.
Functional interaction between Tra2 and Rbp1 also occur at the splicing enhancers in dsx and fru pre-mRNAs, where these proteins bind in an interdependent manner to repeated elements and activate female-specific alternative splice sites (23). Although similar, there are some significant differences in the way these proteins interact at the ISS. First, the conserved CAAG element required for Tra2 binding in the ISS is not found within the 13-nt repeated elements of fru and dsx RNA; thus, the sequence specificity of Tra2 binding may differ depending on the context. The formation of activator complexes on the dsx enhancer repeats involves cooperative binding of Tra2 and Rbp1 or other SR proteins at adjacent positions (23, 24). In contrast, binding of Tra2 and Rbp1 within the ISS appears to occur independently at distinct sequences. It is significant that, in splicing assays on ISS-containing substrates, we observed some functional synergism between Tra2 and Rbp1. Presumably, this reflects cooperation at the level of their effector functions or in facilitating interactions of other repressors with the ISS complex. A final difference is that the formation of Tra2/Rbp1 complexes on the dsx/fru repeats depends on the presence of the female-specific Tra protein, which is not present in the male germ line where M1 repression occurs (23, 24, 30, 38, 39). No protein similar to Tra has been associated with complexes on the ISS; however, binding of both Tra2 and Rbp1 depends on the presence of nuclear extract, suggesting the involvement of other factors.
These observations raise the issue of how Tra2 and Rbp1 repress splicing on one pre-mRNA while activating splicing when bound to other transcripts. SR factors bound to exons generally enhance splicing. In vitro tethering of their Arg-Ser-rich domains to an exon is sufficient to activate nearby splice sites. These domains improve splice site recognition by recruiting general splicing factors such as U2AF or by direct interactions with the branch point that facilitate its base pairing with U2 snRNA (34, 35). Although, a priori, it would seem that SR proteins might have similar effects when bound to intron sequences, in most cases, they either repress splicing or have no effect (5, 16, 28, 37). One well-studied example of this is the adenovirus “3RE,” an intronic element through which the SR protein SF2/ASF prevents the use of the IIIa mRNA alternative 3′ splice site during early stages in infection (16). Like the ISS, this silencer features a set of three repeated 5-nt repressor binding sites immediately upstream of the affected branch point. Tethering experiments show that the presence of SF2/ASF at this position in either adenovirus or β-globin introns was sufficient to cause repression (6). The effect of the tethered protein is not mediated by its Arg-Ser-rich region but rather by its second RNA binding domain (RBD2), which is sufficient to block use of the splice site when tethered alone to the RNA. Notably, RBD2 is a variant RBD that is also found in other SR proteins with two RBDs. A conserved SWQDLKD motif characteristic of these variant RBDs is required for its repressive function and is thought to mediate protein-protein interactions. Although Rbp1 and Tra2 repress splicing and bind at a similar position, each contains only a single RBD which lacks the SWQDLKD motif and is not of the same variant class as the SF2/ASF RBD2 domain. In the case of Tra2, its RBD domain is known to be required for M1 intron retention in vivo (1), but it is not known whether the Tra2 RBD has an effector role or simply binds the pre-mRNA. Given the interdependent interactions of SR proteins on splicing enhancers, it seems plausible that Drosophila SF2/ASF (or another protein with an RBD2 domain) interacts with Tra2 and/or Rbp1 to cause repression by a mechanism similar to adenovirus.
A common strategy utilized by splicing repressors to affect specific RNA targets is to recognize regulatory signals that are in close proximity to general factor binding sites (e.g., the 5′ splice site, branch point, polypyrimidine tract) (21). This allows them to competitively inhibit binding of the general factor. The position of Tra2 binding sites in the ISS of both D. melanogaster and other Drosophila species suggests such a function. In each case, one or more sites lie very close to the intron's branch point within a region where non-sequence-specific interactions by proteins associated with U2 snRNP have been observed in mammals (8). However, we found that repositioning the ISS at locations more distant from the branch point had no effect on its function. Moreover, we found that the ISS itself did not promote splicing in the absence of Tra2, indicating that it does not bind to splicing activators. We therefore favor the idea that repressor proteins associated with the ISS actively interfere with splicing through their effector domains rather than by passively obstructing splicing signals adjacent to their binding sites.
In addition to their roles in alternative splicing, SR proteins and related factors have now been implicated in a wide range of functions, including various steps in spliceosome assembly, mRNA export, translation, and RNA surveillance (32). The ability of Tra2 and Rbp1 to participate in splicing repression as well as activation of both donor and acceptor splice sites suggests that SR proteins play versatile roles in the assembly of regulatory complexes that affect splicing.
Acknowledgments
We thank Dawn Chandler and Manli Shen for helpful comments on the manuscript. We are grateful to Kristen Lynch and Volker Heinrichs for recombinant Rbp1 protein and constructs, Ronald Richman and Mitzi Kuroda for providing the S2 cell line adapted to spinner culture, Hank Adams and Yi Du for assistance with confocal microscopy, and Supriya Kumar, Javier Lopez, Tom Cooper, Michelle Wilson, Andrew McCullough, and Susan Berget for providing useful advice. Reta Haynes and Rong Dong provided excellent technical support with Drosophila cultures and media.
This work was supported by NIH grant R01GM070892 to W.M. DNA sequencing was carried out at the U.T. M.D. Anderson DNA Analysis Facility and supported by a grant (no. CA16672, DAF) from the National Cancer Institute.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Amrein, H., M. L. Hedley, and T. Maniatis. 1994. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell 76:735-746. [DOI] [PubMed] [Google Scholar]
- 2.Chandler, D., M. E. McGuffin, J. Piskur, J. Yao, B. S. Baker, and W. Mattox. 1997. Evolutionary conservation of regulatory strategies for the sex determination factor transformer-2. Mol. Cell. Biol. 17:2908-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler, D. S., M. E. McGuffin, and W. Mattox. 2001. Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing. Nucleic Acids Res. 29:3012-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler, D. S., J. Qi, and W. Mattox. 2003. Direct repression of splicing by transformer-2. Mol. Cell. Biol. 23:5174-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, C. R., and M. T. McNally. 1998. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology 242:211-220. [DOI] [PubMed] [Google Scholar]
- 6.Dauksaite, V., and G. Akusjarvi. 2002. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 277:12579-12586. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gozani, O., R. Feld, and R. Reed. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 9.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 10.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs, V., and B. S. Baker. 1995. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 14:3987-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinrichs, V., L. C. Ryner, and B. S. Baker. 1998. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 18:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 15.Kan, Z., E. C. Rouchka, W. R. Gish, and D. J. States. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. J., P. Zuo, J. L. Manley, and B. S. Baker. 1992. The Drosophila RNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes Dev. 6:2569-2579. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., and A. J. Lopez. 2005. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 24:2646-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam, B. J., A. Bakshi, F. Y. Ekinci, J. Webb, B. R. Graveley, and K. J. Hertel. 2003. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 278:22740-22747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipscombe, D. 2005. Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 22.Lynch, K. W. 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 4:931-940. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, K. W., and T. Maniatis. 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 10:2089-2101. [DOI] [PubMed] [Google Scholar]
- 24.Lynch, K. W., and T. Maniatis. 1995. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 9:284-293. [DOI] [PubMed] [Google Scholar]
- 25.Mattox, W., and B. S. Baker. 1991. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 5:786-796. [DOI] [PubMed] [Google Scholar]
- 26.Mattox, W., M. E. McGuffin, and B. S. Baker. 1996. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics 143:303-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGuffin, M. E., D. Chandler, D. Somaiya, B. Dauwalder, and W. Mattox. 1998. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics 149:1477-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNally, L. M., and M. T. McNally. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 30:13-19.11753382 [Google Scholar]
- 30.Nagoshi, R. N., and B. S. Baker. 1990. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 4:89-97. [DOI] [PubMed] [Google Scholar]
- 31.Ryner, L. C., and B. S. Baker. 1991. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 5:2071-2085. [DOI] [PubMed] [Google Scholar]
- 32.Sanford, J. R., J. Ellis, and J. F. Caceres. 2005. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 33:443-446. [DOI] [PubMed] [Google Scholar]
- 33.Schwerk, C., and K. Schulze-Osthoff. 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19:1-13. [DOI] [PubMed] [Google Scholar]
- 34.Shen, H., and M. R. Green. 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16:363-373. [DOI] [PubMed] [Google Scholar]
- 35.Shen, H., J. L. Kan, and M. R. Green. 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13:367-376. [DOI] [PubMed] [Google Scholar]
- 36.Shin, C., and J. L. Manley. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407-417. [DOI] [PubMed] [Google Scholar]
- 37.Simard, M. J., and B. Chabot. 2002. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 22:4001-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian, M., and T. Maniatis. 1993. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74:105-114. [DOI] [PubMed] [Google Scholar]
- 39.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]