Abstract
Although the sequencing of the human genome has been completed, the number and identity of genes contained within it remains to be fully determined. We used LongSAGE to analyze 660,357 human transcripts from human brain mRNA and identified expression of 17,409 known genes and >15,000 different transcripts that were not annotated in genome databases. Analysis of a subset of these unannotated transcripts suggests that 85% were differentially expressed in various tissue types and that fewer than 20% would have been detected by ab initio gene predictions. These studies suggest that the human genome contains on the order of twice as many transcribed regions as are currently annotated and that experimental approaches will be required to fully elucidate the novel genes corresponding to these transcripts.
Now that the human genome project is essentially complete, efforts have turned to annotating the genes encoded within it. The recent analyses of the human genome identified ∼20,000 well-characterized protein coding genes and another ∼5,000 predicted genes (Lander et al. 2001; Venter et al. 2001; International Human Genome Sequencing Consortium 2004). Initial ab initio and homology approaches used to annotate the genome were limited in that they tended to identify protein coding genes with similarity to those already existing in DNA databases, potentially missing genes with novel motifs or which were noncoding. Additionally, these methods could not provide evidence that the identified genes were actually expressed. Subsequent computational and experimental approaches have begun to suggest that a substantially larger number of previously uncharacterized transcribed regions may be encoded in the genome (Shoemaker et al. 2001; Kapranov et al. 2002; Lim et al. 2003; Brandenberger et al. 2004; Imanishi et al. 2004; Porcel et al. 2004).
In an effort to obtain experimental evidence for such novel transcripts, we have used LongSAGE (Saha et al. 2002) to perform expression analyses on a genome-wide scale. This approach has been documented to quantitatively measure transcript levels regardless of whether such transcripts correspond to known genes (Saha et al. 2002; Shiraki et al. 2003; Hashimoto et al. 2004; Wei et al. 2004).
Results
We used LongSAGE to analyze transcripts from developing human brain, as this tissue is among the most highly complex in terms of the number of genes expressed within it (Velculescu et al. 1999). The brain RNA was treated with DNase I and doubly purified by polyA selection to ensure that no contaminating DNA fragments were present in the isolated RNA. As human cells are thought to contain ∼300,000 mRNA molecules (Lewin 1980), we aimed to obtain at least twice this number of transcripts. At this level of analysis, one would expect to detect >85% of transcripts expressed at a single copy per cell and >95% of transcripts expressed at three or more copies per cell.
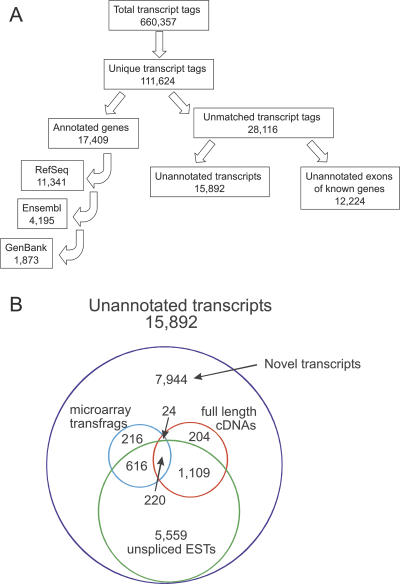
A total of 660,357 transcripts were analyzed in this manner. We first evaluated genes previously annotated in extant gene databases including RefSeq, Ensembl, and GenBank. Transcript tags were found to match annotated exons or UTR regions for 17,409 characterized genes, suggesting that most of the currently annotated genes were expressed in brain RNA (Fig. 1A). Expression levels of these genes ranged from 1 to 856 transcript copies per cell. Although expression was distributed throughout all chromosomes, certain regions contained clusters of highly expressed genes, consistent with previous descriptions of genomic regions of increased gene expression (Caron et al. 2001) (Supplemental Fig. 1). Additionally, transcripts with large introns were generally expressed at lower levels than those with smaller introns, supporting the notion of selection for short introns in highly expressed genes (Castillo-Davis et al. 2002) (Supplemental Fig. 2).
Figure 1.
Categorization of LongSAGE transcript tags. (A) Flowchart of transcript tag matches against genome databases. The 660,357 transcript tags obtained resulted in 111,624 unique transcript tags that matched single loci in the genome. The positions of transcript tags in the genome were sequentially compared to the RefSeq, Ensembl, and GenBank tracks of the UCSC human genome database and the number of annotated genes that was identified is listed in each of the corresponding boxes (see Methods for details). The positions of unmatched transcript tags were compared with the intron–exon structures of annotated genes to determine whether these tags corresponded to unannotated exons of known genes or novel transcripts. The number of transcript tags matching novel exons or unannotated transcripts is indicated in the corresponding boxes. (B) Venn diagram of unannotated transcript tags. The numbers of transcript tags for which independent evidence of expression exists are indicated. These include transcript tag matches to unspliced ESTs, or to full-length cDNAs and microarray transcriptional fragments that were described during the course of this study (Ota et al. 2004; Cheng et al. 2005).
The remaining transcript tags corresponded to either previously uncharacterized transcripts or to transcripts containing unannotated exons of known genes. To discriminate between these two possibilities, transcript tags were compared with intron/exon structures of all annotated genes. Once the tags matching the exon sequences noted above were removed, a total of 12,224 transcript tags were found to be derived from intronic regions of known genes (Fig. 1A; Supplemental Table 1). As the transcript databases used already contained annotated alternative splice forms of known genes, such intron-derived transcripts may be the result of previously uncharacterized exons within these genes.
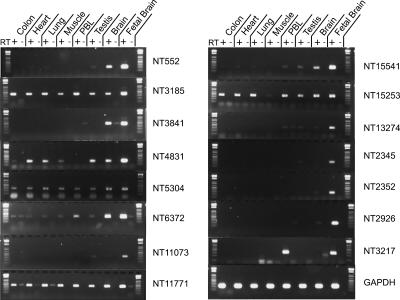
The remaining 15,892 tags were derived from transcripts located in intergenic regions (Fig. 1A; Supplemental Table 2). Seven levels of evidence suggested that many of these transcripts were generated from previously uncharacterized genes. First, these transcript fragments were located on average 186 kb (median 40 kb) from previously annotated genes and were at least 5 kb from annotated gene transcription stop sites and 500 bp from gene transcription start sites. Such distances are substantially longer than the average 5′ and 3′ UTR sequences of known genes (Lander et al. 2001). Second, analysis of the transcript tags and adjacent genomic regions showed that nearly half were located in areas that were evolutionarily conserved among vertebrate genomes (Supplemental Table 2). Third, comparison of these transcripts with databases of unspliced ESTs (not included in current gene annotations) identified 7504 matching ESTs (Fig. 1B). These provide a separate measure of expression of these genes from a variety of different tissue types. Fourth, we used RT-PCR to independently evaluate expression of 18 arbitrarily selected candidate novel genes. The template for these analyses was cDNA produced by reverse transcription (RT) of mRNA from brain tissue. As intron–exon boundaries of these genes were not known, primers were designed to flank the transcript tag and produce a ∼200 bp product. Of the 18 candidate genes analyzed, 15 (83%) were shown to be expressed in an RT-dependent manner. Fifth, to determine whether these candidate genes were differentially expressed in various tissue types, we evaluated their expression in RNA derived from colon, heart, lung, skeletal muscle, peripheral blood leukocytes, testes, and fetal and adult brain. All of the candidate genes were expressed in at least one tissue type, and 13 of 15 displayed expression profiles that varied between the different tissues (Fig. 2). This pattern of differential expression suggests that most of these unannotated genes are tissue-regulated and may have specific physiologic roles. Sixth, to demonstrate experimentally that these differentially expressed candidate genes were derived from previously uncharacterized mRNAs, we screened four of the candidate genes in brain cDNA libraries using rapid amplification of cDNA ends (RACE). Sequence analysis of the resulting RACE products identified two novel full-length transcripts, a single exon transcript with alternative transcriptional start sites for the NT2926 transcript, and a spliced two exon transcript for the NT552 transcripts, as well as two novel partial transcripts (NT5192 and NT12163) (Supplemental Fig. 3). These results suggest that LongSAGE transcript tags can be directly used to obtain either full-length or partial sequences of previously unannotated transcripts. Finally, we compared these transcript tags with a large collection of full-length cDNAs, microarray expression data, and ENCODE gene annotations that were published during the course of our study (Ota et al. 2004; Cheng et al. 2005; International HapMap Consortium 2005; Harrow et al. 2006). A total of 1557 of the transcript tags matched these full-length cDNAs, 1076 transcript tags matched transcription fragments (transfrags) detected by microarrays, and 129 matched annotated genes within the ENCODE regions (Fig. 1B; Supplemental Table 2). Most of the matching cDNAs contained multiple exons and >450 of them had open reading frames >300 bp in length (Supplemental Table 2; examples in Supplemental Figure 4). The expression levels of transcript tags matching full-length cDNAs in silico varied widely, with over half of the matching transcripts expressed at very low levels (≤1 copy per cell). These results provide further evidence of expression of the identified unannotated transcript tags and suggest that at least some correspond to multi-exon protein coding genes. In total, 10,986 of the 15,892 unannotated transcript tags had at least one of the types of functional evidence mentioned above, thereby suggesting that most of these transcripts were derived from unannotated genes.
Figure 2.
Expression analysis of novel transcripts in different tissue types. RT-PCR was performed on each novel transcript using equal amounts of cDNA from total RNA of human colon, heart, lung, skeletal muscle, peripheral blood leukocytes, testis, and brain, and poly(A+) RNA from human fetal brain. Representative pictures of experiments performed in triplicate are shown. GAPDH was used as a control.
Discussion
In summary, our data suggest that the human genome encodes a substantial number of unannotated transcripts. Although it is possible that some of these could be the result of spurious transcription in intergenic regions, the observation that many of the analyzed transcripts are differentially expressed and spliced, suggests that a substantial fraction correspond to bona fide genes. At least two questions emerge from these results.
First, how many uncharacterized genes are actually present in the genome? An upper boundary for the number of transcribed regions detected in our analyses would be the 15,892 identified transcripts from unannotated intergenic regions. This is likely to be an overestimate, as multiple transcript tags may be derived from the same gene because of splice variants, alternative sites of polyadenylation, and other mechanisms. An estimation that includes this possibility may be obtained by evaluating the relative positions of intergenic transcripts within the genome and considering clusters of nearby transcripts to be derived from the same gene. Using a window size of 15,000 bp (corresponding to the median genomic length of characterized genes; Lander et al. 2001), the intergenic transcripts can be grouped into 11,257 clusters. Using an even larger window size of 30,000 bp, there would still remain 10,699 clusters for the intergenic transcripts. Finally, if one considers the average number of transcript tags obtained for each characterized gene and uses this ratio to interpret the unannotated transcripts observed, one would obtain 3310 clusters of putative genes as a lower boundary. This latter figure is likely to be too drastic a correction as many uncharacterized transcripts are expressed at low levels and we therefore would not have expected to detect all tags associated with these transcripts in our analyses. Because a substantial fraction of the transcripts we identified appeared to be expressed in a tissue-specific manner, additional human cell types will have to be evaluated by LongSAGE or other experimental approaches to completely identify the compendium of transcripts that are encoded in the human genome. The recent availability of massively parallel sequencing approaches that can be used to sequence transcript tags (Rogers and Venter 2005) may provide a cost-effective method of achieving such comprehensive expression analyses.
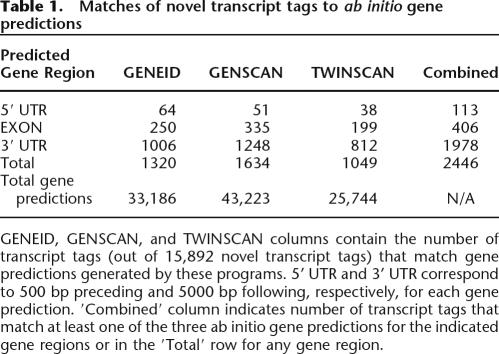
Second, what is the biologic function of these novel transcripts? While additional work will be needed to fully identify and characterize these transcripts, it is already clear that they are different in at least several ways from previously annotated genes. In contrast to known genes, which computational approaches can predict with >70% sensitivity (Rogic et al. 2001), <20% of the uncharacterized transcript tags were detected by gene predictions from three different ab initio programs (GENSCAN [Burge and Karlin 1997], GENEID [Parra et al. 2000], and TWINSCAN [Korf et al. 2001]) (Table 1). In addition, the average GC content of transcript tags derived from unannotated transcripts was significantly lower than for those derived from annotated transcripts (42% for identified novel transcripts vs. 47% for previously annotated transcripts). Interestingly, both of these features are similar to those of nonprotein coding genes (Ota et al. 2004), suggesting that many of these transcripts may represent noncoding RNAs. This is consistent with the observation that of the transcript tags matching full-length cDNAs noted above, more than half had no obvious open reading frames. Of the transcript tags matching cDNAs that did contain open reading frames, some had homology with proteins implicated in a variety of different cellular processes, including those involved in transcriptional activation, signal transduction, cytoskeletal structure, metabolism, and intracellular transport (Supplemental Table 2). Additionally, 160 transcript tags matched cDNAs containing tetratricopeptide repeat regions, a degenerate 34-amino-acid motif believed to mediate protein interactions in a wide range of proteins.
Table 1.
Matches of novel transcript tags to ab initio gene predictions
GENEID, GENSCAN, and TWINSCAN columns contain the number of transcript tags (out of 15,892 novel transcript tags) that match gene predictions generated by these programs. 5' UTR and 3' UTR correspond to 500 bp preceding and 5000 bp following, respectively, for each gene prediction. ′Combined′ column indicates number of transcript tags that match at least one of the three ab initio gene predictions for the indicated gene regions or in the ′Total′ row for any gene region.
Finally, it appears that the expression level of these novel transcripts is substantially lower than that of well-characterized genes (average of 0.84 transcript copies per cell for uncharacterized transcripts vs. 2.3 transcript copies per cell for known genes). This may explain why such genes have historically been more difficult to detect experimentally and suggests that these transcripts may be involved in specialized cellular functions that do not require high transcript levels or that are present only in certain cell subpopulations. Given the significant role of many noncoding and newly characterized RNAs in a variety of cellular processes (Morey and Avner 2004), it will be important to evaluate the function of these genes in the years to come.
Methods
LongSAGE library construction
LongSAGE libraries were generated from 500 ng of human fetal brain poly(A+) selected RNA (BD Biosciences) following the LongSAGE protocol (Saha et al. 2002) with the following modifications. Poly (A+) RNA was treated with 0.5 units of RNase-free DNase I for 15 min at room temperature following manufacturer’s protocol (Invitrogen). Three different LongSAGE libraries were generated, using NlaIII, Sau3A, and XspI as anchoring enzymes. The NlaIII library cDNA was digested in a 200 μL reaction for 1 h at 37°C with 60 units of NlaIII in 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM DTT, and 100 μg/mL BSA. The Sau3A library cDNA was digested in a 200 μL reaction for 1 h at 37°C with 60 units of Sau3A in 100 mM NaCl, 10 mM Bis Tris Propane-HCl at pH 7.0, 10 mM MgCl2, 1 mM DTT, and 100 μg/mL BSA. The XspI library cDNA was digested in a 200 μL reaction for 1 h at 37°C with 50 units of XspI in 20 mM Tris-HCl at pH 8.5, 10 mM MgCl2, 1 mM DTT, and 100 mM KCl. Linkers containing the MmeI recognition site were ligated to 3′ cDNA ends after NlaIII, Sau3A, or XspI digestion. The following linkers were used for the three libraries: NlaIII-LS linker 1A, 5′-TTTGGATTTGCTGGTGCAGTACAACTAGGCTTAATATC CGACATG-3′; NlaIII-LS linker 1B, 5′-phosphate-TCGGATATTAA GCCTAGTTGTACTGCACCAGCAAATCC-amino modified C7–3′; NlaIII-LS linker 2A, 5′-TTTCTGCTCGAATTCAAGCTTCTAAC GATGTACGTCCGACATG-3′; NlaIII-LS linker 2B, 5′-phosphate-TCGGACGTACATCGTTAGAAGCTTGAATTCGAGCAGamino modified C7–3′; SAU3A-LS Linker 1A, 5′-TTTGGATTTG CTGGTGCAGTACAACTAGGCTTAATATCCGAC-3′; SAU3A-LS Linker 1B, 5′-phoshphate-GATCGTCGGATATTAAGCCTAGTTG TACTGCACCAGCAAATCC-amino modified C7–3′; SAU3A-LS Linker 2A, 5′-TTTCTGCTCGAATTCAAGCTTCTAACGATGTACG TCCGAC-3′; SAU3A-LS Linker 2B, 5′-phoshphate-GATCGTCGG ACGTACATCGTTAGAAGCTTGAATTCGAGCAG-amino modified C7–3′; XspI-LS Linker 1A, 5′-TTTGGATTTGCTGGTGCAG TACAACATGGCTTAATATCCGAC-3′; XspI-LS Linker 1B, 5′-phoshphate-TAGTCGGATATTAAGCCATGTTGTACTGCACCAG CAAATCC-amino modified C7–3′; XspI-LS Linker 2A, 5′-TTTCT GCTCGAATTCAAGCTTCTAACGATGTACGTCCGAC-3′; XspI-LS Linker 2B, 5′-phoshphate-TAGTCGGACGTACATCGTTAGAAGC TTGAATTCGAGCAG-amino modified C7–3′.
Linker tag molecules were released from the cDNA using the MmeI type IIS restriction endonuclease (University of Gdansk Center for Technology Transfer, Gdansk, Poland). Digestion was performed at 37°C for 1 h using 4 units MmeI in 250 μL of 10 mM HEPES at pH 8.0, 2.5 mM potassium acetate, 5 mM magnesium acetate, 2 mM DTT, and 40 M S-adenosylmethionine. The linker-1-tag and linker-2-tag molecules were not polished and were directly ligated together in a 6-μL reaction containing 4 units T4 DNA ligase (Invitrogen) in the supplied buffer for 2.5 h at 16°C. The SAGE software (Velculescu et al. 1995) parameters were modified to allow extraction of 20- or 21-bp tags from sequences of concatemer clones. Detailed protocols for performing SAGE and LongSAGE and software for extraction of LongSAGE data are available at http://www.sagenet.org/sage_protocol.htm.
RT-PCR analysis of candidate genes
Single-stranded cDNA was synthesized from 800 ng of human fetal brain poly(A+) selected RNA (BD Biosciences), 4 μg of total human brain RNA (BD Biosciences), 4 μg of total human peripheral blood leukocyte RNA (BD Biosciences), 4 μg of total human colon RNA (BD Biosciences), 4 μg of total human heart RNA (BD Biosciences), 4 μg of total human lung RNA (BD Biosciences), 4 μg of total human skeletal muscle RNA (BD Biosciences), and 4 μg of total human testis RNA (BD Biosciences) using Superscript II reverse transcriptase (Invitrogen) following the manufacturer's protocol, and mock template preparations were prepared in parallel without the addition of reverse transcriptase. All RNA samples were DNase I treated as described above before reverse transcription. Regions surrounding 18 transcript tags with no matches to annotated genes were arbitrarily selected for RT-PCR analysis. Nine of the 18 transcript tags had matches to ESTs and expression of the transcript tags was low (range, 5–60 transcript copies per cell). Primers were designed using Primer 3 interface (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to span a 200-bp region that included the transcript tag, and were synthesized by Integrated DNA Technologies. Reactions were performed in triplicate. Products were separated on 2% TBE gels and stained with ethidium bromide.
RACE analysis of candidate genes
Full-length cDNA obtained from 5′ end selected human brain mRNA was used for RACE analysis following the manufacturer’s protocol (FirstChoice RACE-Ready Kit, Ambion). Analysis of low-expression transcripts was also performed by circularization of cDNA and inverse PCR (Ye and Connor 2000). Human fetal brain poly(A+) selected RNA (1 μg) (BD Biosciences) was used to make cDNA using Superscript III reverse transcriptase (Invitrogen) following the manufacturer's protocol. cDNA was purified with a Qiagen PCR column and circularized with 15 units of T4 ligase (Invitrogen) for 16 h at 8°C followed by 2 h at 15°C. PCR primers were designed using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to anneal within a 100-bp region of DNA surrounding the LongSAGE tag to be analyzed and were synthesized by Invitrogen. PCR products were run on 1% TBE gels. DNA bands were excised, purified, and cloned into TOPO vectors (Invitrogen) for sequencing. Transcript sequences corresponding to previously uncharacterized genes have been submitted to GenBank.
Matching transcript tags to the genome
All 17-bp tags adjacent to the NlaIII anchoring enzyme site (CATG) and XspI anchoring enzyme site (CTAG) and 16-bp tags adjacent to the Sau3A anchoring enzyme site (GATC), along with corresponding position information were computationally extracted from the June 2002 human genome assembly (http://genome-archive.cse.ucsc.edu/goldenPath/28jun2002/chromosomes/) and were considered to be LongSAGE “virtual tags.” The 660,357 experimentally derived transcript tags were directly compared with these virtual tags to identify the precise location of experimental tag matches in the genome. Of the 660,357 transcript tags, a total of 512,419 (78%) representing 137,756 different transcript tags perfectly matched the genome sequence. The remaining nonmatching transcript tags were likely due to sequence polymorphisms in the tag region, transcribed sequences not represented in the genome databases, or sequencing errors in the tags or human genomic sequences. Of the 137,756 different transcript tags matching the genome, 111,624 (81%) matched unique loci in the genome. Transcript tags matching multiple locations in the genome were considered to be duplicated genes or repeat sequences and were not included in further analyses. All transcript tags used in these analyses can be obtained from http://cgap.nci.nih.gov/SAGE.
Comparison of transcript tags to annotated genes, ab initio tracks, ESTs, full-length cDNAs, microarray transfrags, and ENCODE regions
Exon coordinates of RefSeq, Ensembl, and GenBank known genes, along with any annotated alternative splice forms, were obtained from refGene.txt, ensGene.txt, and knownGene.txt tracks of the June 2002 genome assembly (http://genome-archive.cse.ucsc.edu/goldenPath/28jun2002/database/ for these and subsequent tracks), respectively. Transcript tags were considered to match their corresponding genes included in these databases when they identically matched annotated exonic sequences, 3′ UTRs < 5 kb from the terminal exon, or 5′ UTRs < 500 bp from the first exon. Transcript tags were sequentially matched to the different annotation databases in the following order: RefSeq exon, Ensembl exon, GenBank exon, RefSeq 3′ UTR, Ensembl 3′ UTR, GenBank 3′ UTR, RefSeq 5′ UTR, Ensembl 5′ UTR, GenBank 5′ UTR. Once a transcript tag matched an entry in any database, it was no longer analyzed against the remaining databases to ensure that the highest quality annotation was provided for each transcript tag. When transcript tags matched alternative splice or polyadenylation forms of the same gene in an annotation database, all transcript matches were included. Only tags matching in the sense orientation were considered for these analyses, as we could not distinguish whether antisense tags were the result of an undiscovered overlapping gene on the opposite strand or of internal oligo-dT priming during cDNA synthesis.
Tags were considered to match novel internal exons of annotated genes when the tags matched intronic regions between two exons of the same gene in the appropriate orientation. Alternatively, tags were considered to match previously undiscovered genes when they matched regions >5 kb from the 3′ terminal exon of an annotated gene, or >500 bp from the 5′ UTR of an annotated gene. Unannotated transcript tags were considered to match unspliced ESTs or ab initio gene predictions when they identically matched sequences present in the all_est.txt track or the gene prediction tracks (genescan.txt, geneid.txt, twinscan.txt) of the June 2002 genome assembly, respectively. Although the all_est.txt track also contains spliced ESTs, tags matching spliced ESTs were previously removed by matching to gene databases described above that included gene predictions derived from spliced ESTs (e.g., Ensembl). Unannotated transcript tags were compared with recently described full-length cDNAs (Ota et al. 2004) using local BLAST. Unannotated transcript tag genome coordinates were converted to the April 2003 and May 2004 genome assemblies using the UCSC hgLiftOver utility (http://genome.ucsc.edu/cgi-bin/hgLiftOver) and were compared with genome coordinates of microarray transfrags (Cheng et al. 2005) and GENCODE annotations (Harrow et al. 2006). Only perfect matches of the transcript tags to these databases were considered. Analyses of clusters of novel transcript tags were performed by considering tags on the same strand that were within windows of 15,000 and 30,000 bp.
Evolutionary conservation and open reading frame detection and analysis
The evolutionary conservation scores for the unannotated transcript tags and 500 bp upstream of each tag were obtained from the UCSC July 2003 assembly of the genome (http://genome.ucsc.edu/cgi-bin/hgGateway), as no such scores were available from the June 2002 assembly. Tag matches to the July 2003 assembly were performed using local BLAST and only perfect matches were considered. The conservation score represents a measure of conservation in human, chimp, mouse, rat, and chicken based on a phylogenetic hidden Markov model (HMM) (phylo-HMM) (Siepel and Haussler 2004). A conservation score of 2200 for the tag sequence indicated a region that was conserved in at least two additional organisms. Open reading frames in full-length cDNAs were detected using the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Similarity searches of full-length cDNAs were obtained through the FLJ-DB Web site (http://fldb.hgc.jp/cgi-bin/cDNA3/public/publication/CloneName.cgi?PUB=publication&inhouse=on&flj=on&genbank=on).
Acknowledgments
We thank Kurt Bachman for help with RT-PCR analysis, Ivy Riffkin for help with LongSAGE library preparation, and Bert Vogelstein for careful review of the manuscript. This work was supported by the NCI Cancer Genome Anatomy Project (CGAP), the Ludwig Trust, the Pew Charitable Trusts, and NIH grants CA121113, CA57345, CA62924. Under a licensing agreement between Genzyme and the Johns Hopkins University, K.W.K. and V.E.V. are entitled to a share of royalty received by the University on sales of products described in this article. K.W.K., V.E.V., and the University own Genzyme stock, which is subject to certain restrictions under University policy. K.W.K. and V.E.V. also receive research funding from Genzyme and are paid consultants to Genzyme. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
[Supplemental material is available online at www.genome.org]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5486607
References
- Brandenberger R., Wei H., Zhang S., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Wei H., Zhang S., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Zhang S., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Fisk G.J., Li Y., Xu C., Fang R., Guegler K., Li Y., Xu C., Fang R., Guegler K., Xu C., Fang R., Guegler K., Fang R., Guegler K., Guegler K., et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Caron H., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Asperen R., Boon K., Voute P.A., Boon K., Voute P.A., Voute P.A., et al. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis C.I., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A., Hartl D.L., Koonin E.V., Kondrashov F.A., Koonin E.V., Kondrashov F.A., Kondrashov F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Sese J., Morishita S., Sugano S., Matsushima K., Morishita S., Sugano S., Matsushima K., Sugano S., Matsushima K., Matsushima K. 5′-End SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- Imanishi T.T., Itoh Y., Suzuki C., O'Donovan S., Fukuchi K.O., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Itoh Y., Suzuki C., O'Donovan S., Fukuchi K.O., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Suzuki C., O'Donovan S., Fukuchi K.O., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., O'Donovan S., Fukuchi K.O., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Fukuchi K.O., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Koyanagi R.A., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Barrero T., Tamura Y., Yamaguchi-Kabata M., Tanino K., Tamura Y., Yamaguchi-Kabata M., Tanino K., Yamaguchi-Kabata M., Tanino K., Tanino K., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:E162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M.R., Flicek P., Duan D., Brent M.R., Duan D., Brent M.R., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17 (Suppl 1):S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lewin B. Gene expression: Eukaryotic chromosomes. Vol. 2. Hoboken, NJ: 1980. pp. 694–727. [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Yekta S., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Morey C., Avner P., Avner P. Employment opportunities for non-coding RNAs. FEBS Lett. 2004;567:27–34. doi: 10.1016/j.febslet.2004.03.117. [DOI] [PubMed] [Google Scholar]
- Ota T.Y., Suzuki T., Nishikawa T., Otsuki T., Sugiyama R., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Suzuki T., Nishikawa T., Otsuki T., Sugiyama R., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Nishikawa T., Otsuki T., Sugiyama R., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Otsuki T., Sugiyama R., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Sugiyama R., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Irie A., Wakamatsu K., Hayashi H., Sato K., Nagai K., Wakamatsu K., Hayashi H., Sato K., Nagai K., Hayashi H., Sato K., Nagai K., Sato K., Nagai K., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Parra G., Blanco E., Guigo R., Blanco E., Guigo R., Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel B.M., Delfour O., Castelli V., De Berardinis V., Friedlander L., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Delfour O., Castelli V., De Berardinis V., Friedlander L., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Castelli V., De Berardinis V., Friedlander L., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., De Berardinis V., Friedlander L., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Friedlander L., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Cruaud C., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Ureta-Vidal A., Scarpelli C., Wincker P., Schachter V., Scarpelli C., Wincker P., Schachter V., Wincker P., Schachter V., Schachter V., et al. Numerous novel annotations of the human genome sequence supported by a 5′-end-enriched cDNA collection. Genome Res. 2004;14:463–471. doi: 10.1101/gr.1481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers Y.H., Venter J.C., Venter J.C. Genomics: Massively parallel sequencing. Nature. 2005;437:326. doi: 10.1038/437326a. [DOI] [PubMed] [Google Scholar]
- Rogic S., Mackworth A.K., Ouellette F.B., Mackworth A.K., Ouellette F.B., Ouellette F.B. Evaluation of gene-finding programs on mammalian sequences. Genome Res. 2001;11:817–832. doi: 10.1101/gr.147901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Kinzler K.W., Velculescu V.E., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Kodzius R., Watahiki A., Nakamura M., Arakawa T., Watahiki A., Nakamura M., Arakawa T., Nakamura M., Arakawa T., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D.D., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Leonardson A., Lum P.Y., Cavet G., Lum P.Y., Cavet G., Cavet G., et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W., Zhang L., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Madden S.L., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Madden S.L., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., Wang C.J., Beaudry G.A., Ciriello K.M., Beaudry G.A., Ciriello K.M., Ciriello K.M., et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Ng P., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Ng P., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Lipovich L., Liu E.T., Ruan Y., Liu E.T., Ruan Y., Ruan Y. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl. Acad. Sci. 2004;101:11701–11706. doi: 10.1073/pnas.0403514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Connor J.R., Connor J.R. cDNA cloning by amplification of circularized first strand cDNAs reveals non-IRE-regulated iron-responsive mRNAs. Biochem. Biophys. Res. Commun. 2000;275:223–227. doi: 10.1006/bbrc.2000.3282. [DOI] [PubMed] [Google Scholar]