Abstract
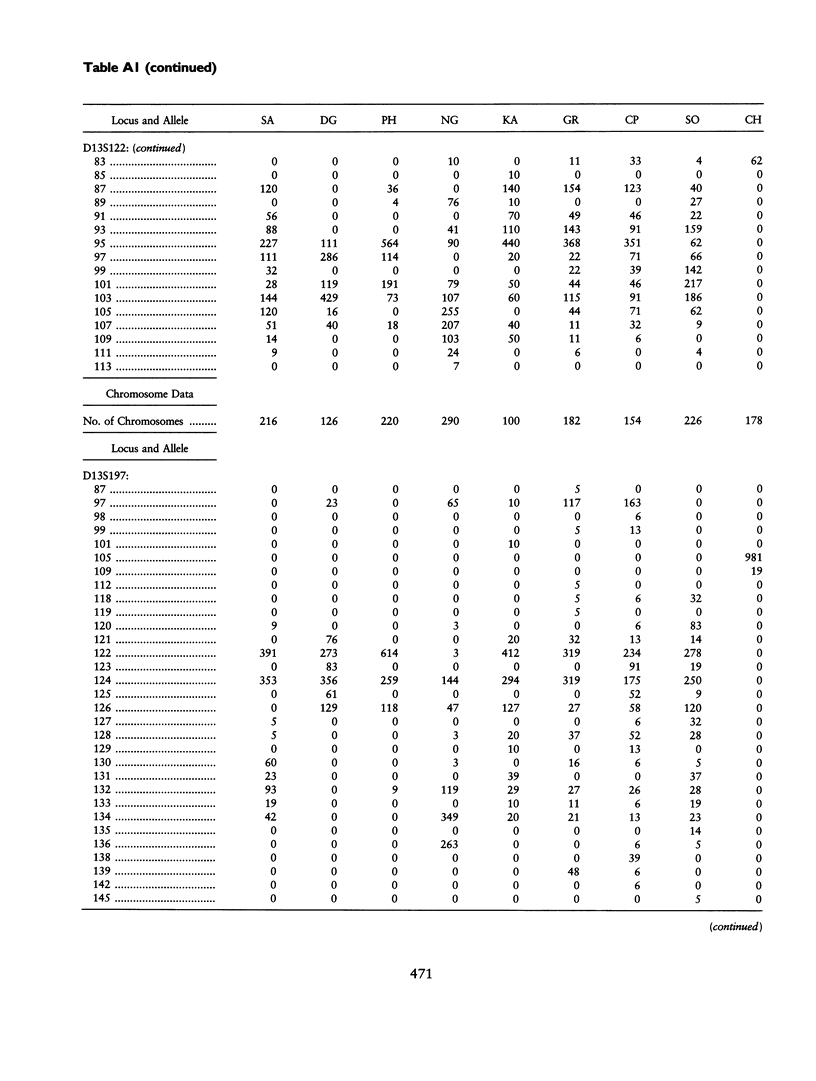
We have characterized eight dinucleotide (dC-dA)n.(dG-dT)n repeat loci located on human chromosome 13q in eight human populations and in a sample of chimpanzees. Even though there is substantial variation in allele frequencies at each locus, at a given locus the most frequent alleles are shared by all human populations. The level of heterozygosity is reduced in isolated or small populations, such as the Pehuenche Indians of Chile, the Dogrib of Canada, and the New Guinea highlanders. On the other hand, larger average heterozygosities are observed in large and cosmopolitan populations, such as the Sokoto population from Nigeria and German Caucasians. Conformity with Hardy-Weinberg equilibrium is generally observed at these loci, unless (a) a population is isolated or small or (b) the repeat motif of the locus is not perfect (e.g., D13S197). Multilocus genotype probabilities at these microsatellite loci do not show departure from the independence rule, unless the loci are closely linked. The allele size distributions at these (CA)n loci do not follow a strict single-step stepwise-mutation model. However, this features does not compromise the ability to detect population affinities, when these loci are used simultaneously. The microsatellite loci examined here are present and, with the exception of the locus D13S197, are polymorphic in the chimpanzees, showing an overlapping distribution of allele sizes with those observed in human populations.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowcock A. M., Ruiz-Linares A., Tomfohrde J., Minch E., Kidd J. R., Cavalli-Sforza L. L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994 Mar 31;368(6470):455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Budowle B., Giusti A. M., Waye J. S., Baechtel F. S., Fourney R. M., Adams D. E., Presley L. A., Deadman H. A., Monson K. L. Fixed-bin analysis for statistical evaluation of continuous distributions of allelic data from VNTR loci, for use in forensic comparisons. Am J Hum Genet. 1991 May;48(5):841–855. [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Fuerst P. A., Nei M. Statistical Studies on Protein Polymorphism in Natural Populations. III. Distribution of Allele Frequencies and the Number of Alleles per Locus. Genetics. 1980 Apr;94(4):1039–1063. doi: 10.1093/genetics/94.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R. Sample size requirements for addressing the population genetic issues of forensic use of DNA typing. Hum Biol. 1992 Apr;64(2):141–159. [PubMed] [Google Scholar]
- Chakraborty R., Weiss K. M. Genetic variation of the mitochondrial DNA genome in American Indians is at mutation-drift equilibrium. Am J Phys Anthropol. 1991 Dec;86(4):497–506. doi: 10.1002/ajpa.1330860405. [DOI] [PubMed] [Google Scholar]
- Deka R., Chakraborty R., DeCroo S., Rothhammer F., Barton S. A., Ferrell R. E. Characteristics of polymorphism at a VNTR locus 3' to the apolipoprotein B gene in five human populations. Am J Hum Genet. 1992 Dec;51(6):1325–1333. [PMC free article] [PubMed] [Google Scholar]
- Deka R., Chakroborty R., Ferrell R. E. A population genetic study of six VNTR loci in three ethnically defined populations. Genomics. 1991 Sep;11(1):83–92. doi: 10.1016/0888-7543(91)90104-m. [DOI] [PubMed] [Google Scholar]
- Deka R., Shriver M. D., Yu L. M., Jin L., Aston C. E., Chakraborty R., Ferrell R. E. Conservation of human chromosome 13 polymorphic microsatellite (CA)n repeats in chimpanzees. Genomics. 1994 Jul 1;22(1):226–230. doi: 10.1006/geno.1994.1369. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Peterson A. C., Garza J. C., Valdes A. M., Slatkin M., Freimer N. B. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W. F., Miller J. C., Steen R. G., Merchant M., Damron D., Nahf R., Gross A., Joyce D. C., Wessel M., Dredge R. D. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat Genet. 1994 Jun;7(2 Spec No):220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Edwards A., Hammond H. A., Jin L., Caskey C. T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992 Feb;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Guo S. W., Thompson E. A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992 Jun;48(2):361–372. [PubMed] [Google Scholar]
- Gyapay G., Morissette J., Vignal A., Dib C., Fizames C., Millasseau P., Marc S., Bernardi G., Lathrop M., Weissenbach J. The 1993-94 Généthon human genetic linkage map. Nat Genet. 1994 Jun;7(2 Spec No):246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- Hong H. K., Giorda R., Yu L. M., Trucco M., Chakravarti A. Microsatellite repeat polymorphism at the D13S197 locus. Hum Mol Genet. 1993 Mar;2(3):337–337. doi: 10.1093/hmg/2.3.337-a. [DOI] [PubMed] [Google Scholar]
- Kamino K., Nakura J., Kihara K., Ye L., Nagano K., Ohta T., Jinno Y., Niikawa N., Miki T., Ogihara T. Population variation in the dinucleotide repeat polymorphism at the D8S360 locus. Hum Mol Genet. 1993 Oct;2(10):1751–1751. doi: 10.1093/hmg/2.10.1751. [DOI] [PubMed] [Google Scholar]
- Li W. H. A Mixed Model of Mutation for Electrophoretic Identity of Proteins within and between Populations. Genetics. 1976 Jun;83(2):423–432. doi: 10.1093/genetics/83.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Long J. C., Naidu J. M., Mohrenweiser H. W., Gershowitz H., Johnson P. L., Wood J. W., Smouse P. E. Genetic characterization of Gainj- and Kalam-speaking peoples of Papua New Guinea. Am J Phys Anthropol. 1986 May;70(1):75–96. doi: 10.1002/ajpa.1330700113. [DOI] [PubMed] [Google Scholar]
- Matise T. C., Perlin M., Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994 Apr;6(4):384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Morton N. E., Collins A., Balazs I. Kinship bioassay on hypervariable loci in blacks and Caucasians. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1892–1896. doi: 10.1073/pnas.90.5.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Tajima F., Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol. 1983;19(2):153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Pena S. D., de Souza K. T., de Andrade M., Chakraborty R. Allelic associations of two polymorphic microsatellites in intron 40 of the human von Willebrand factor gene. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):723–727. doi: 10.1073/pnas.91.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N. J., Devlin B. On the probability of matching DNA fingerprints. Science. 1992 Feb 7;255(5045):717–720. doi: 10.1126/science.1738844. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shriver M. D., Jin L., Chakraborty R., Boerwinkle E. VNTR allele frequency distributions under the stepwise mutation model: a computer simulation approach. Genetics. 1993 Jul;134(3):983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary E. J., Ferrell R. E., Gershowitz H. Genetic differentiation in Dogrib Indians: serum protein and erythrocyte enzyme variation. Am J Phys Anthropol. 1983 Nov;62(3):249–254. doi: 10.1002/ajpa.1330620304. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]




