Abstract
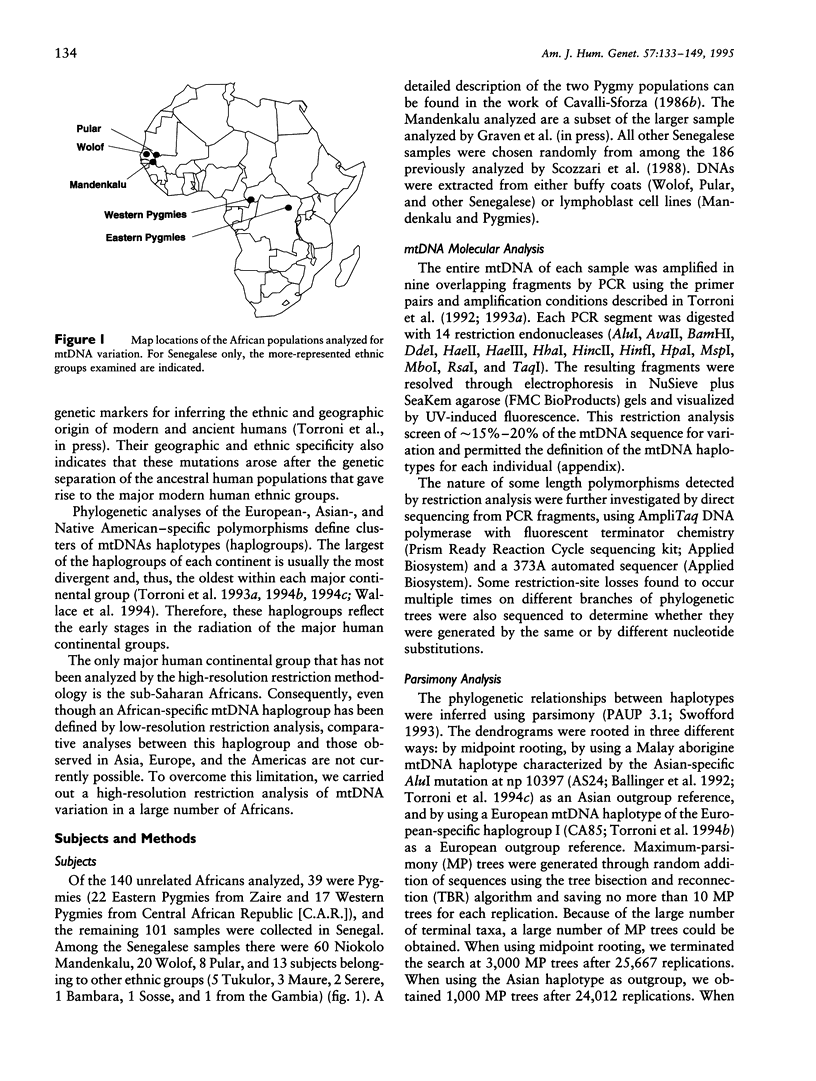
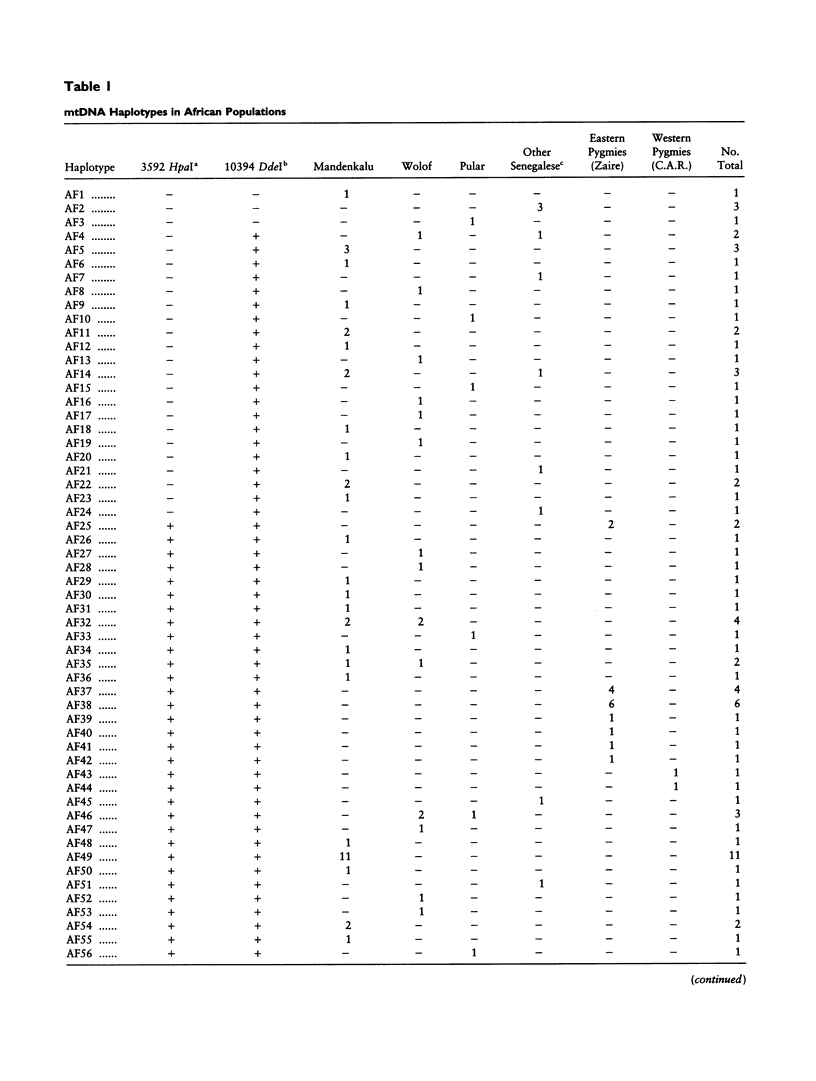
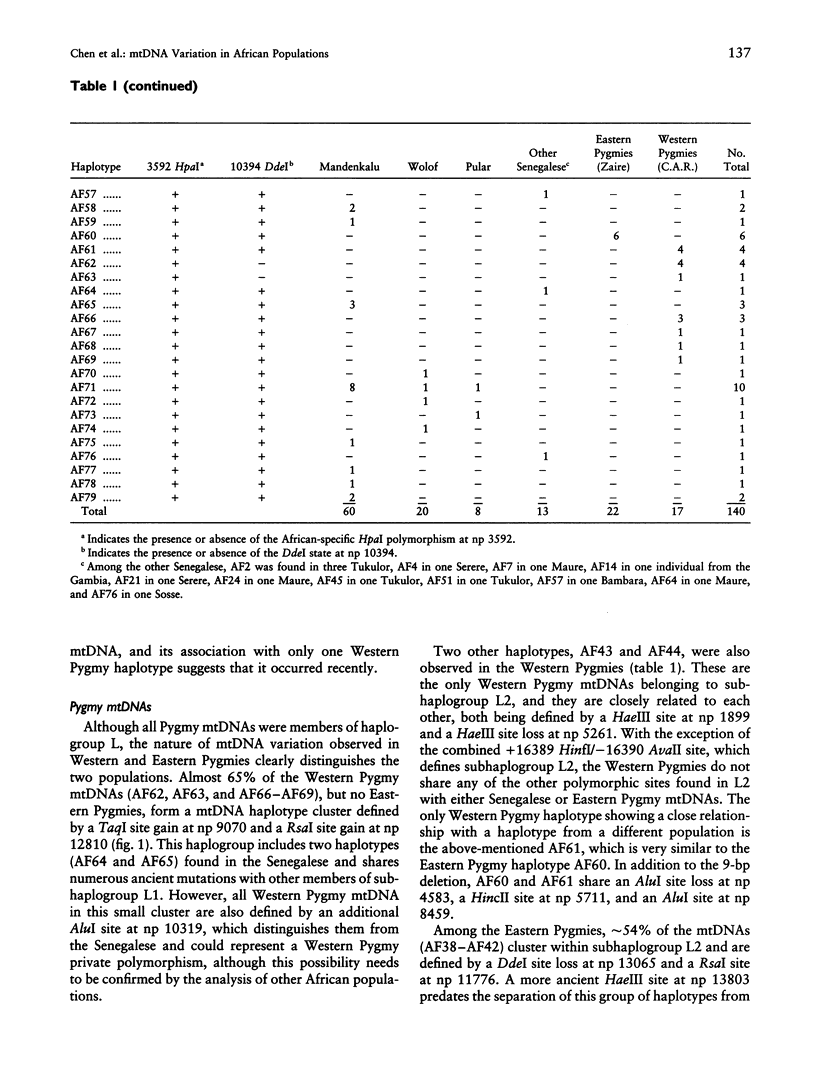
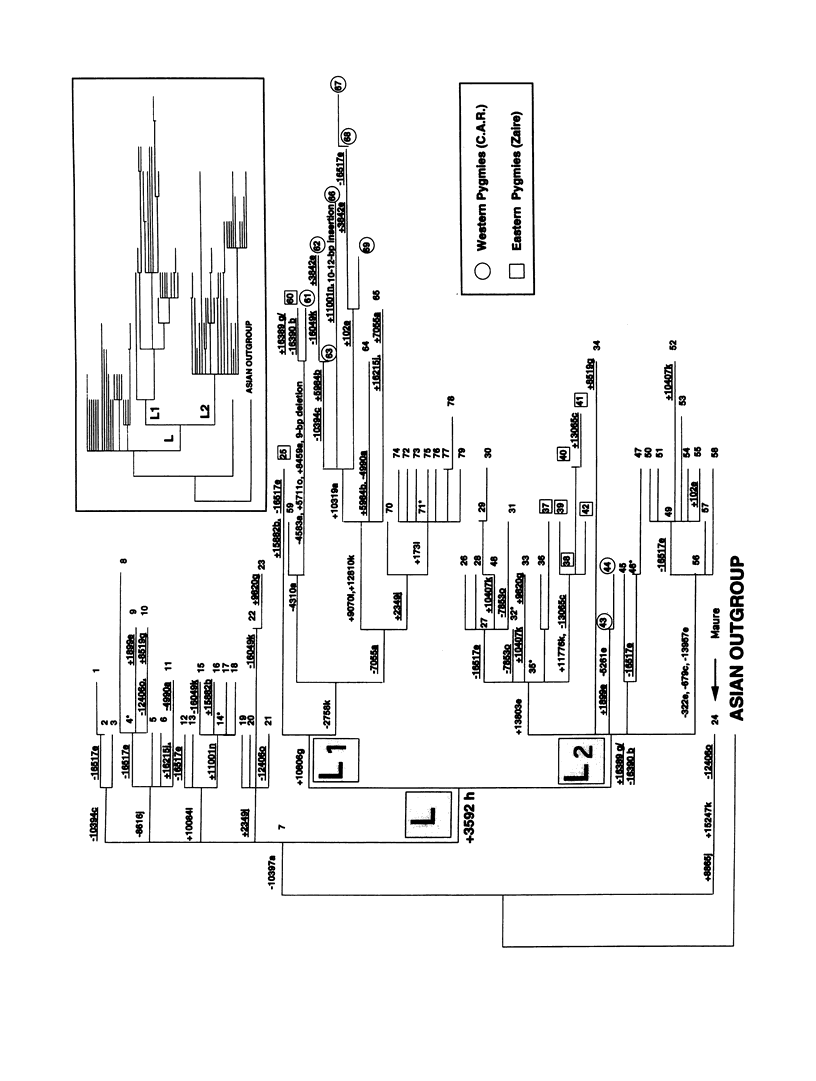
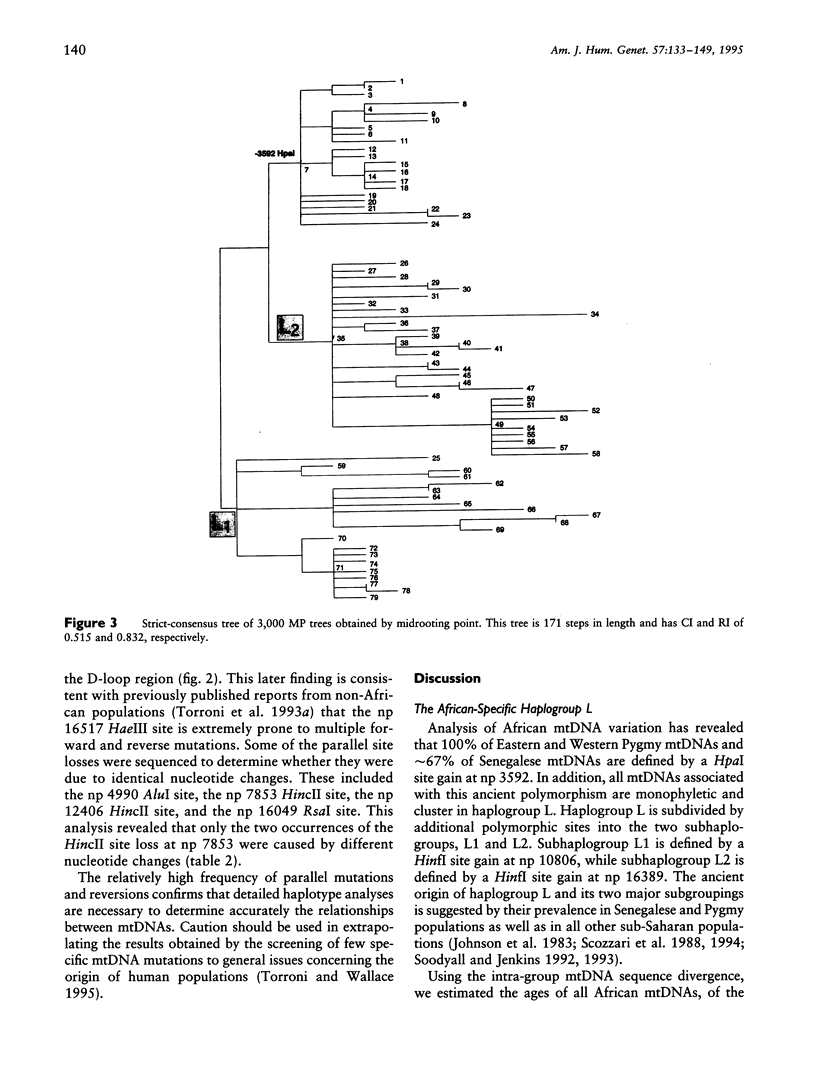
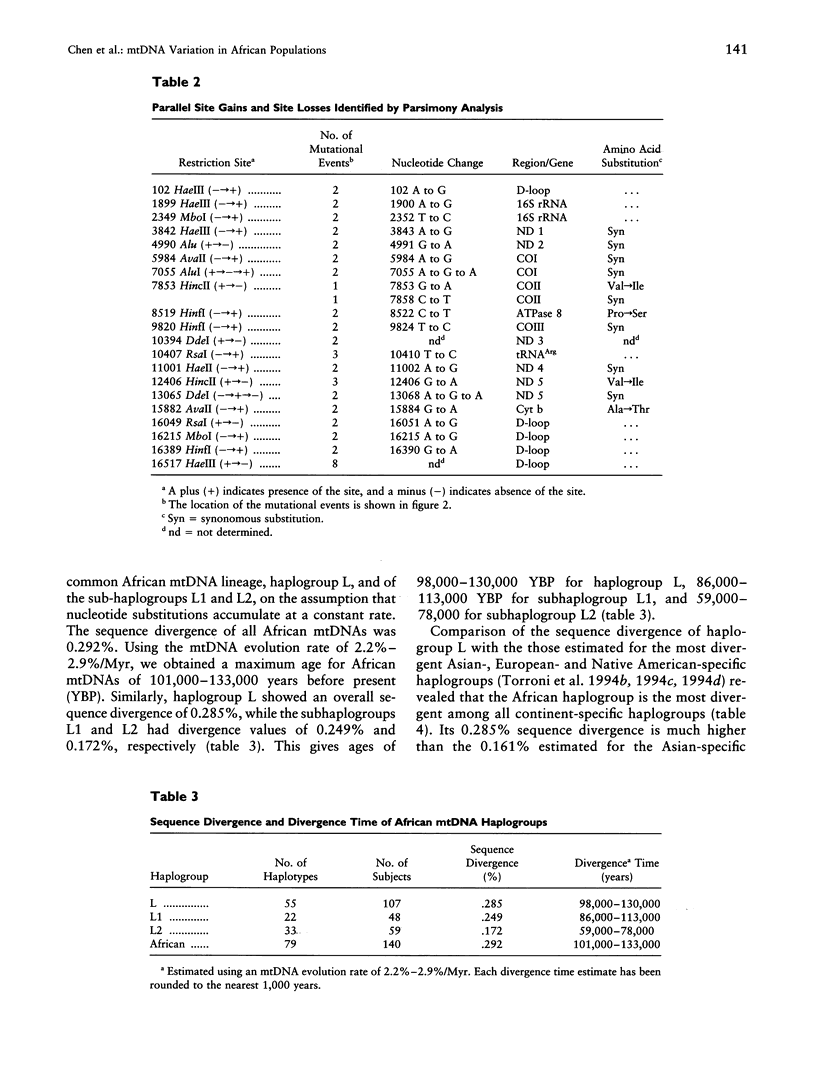
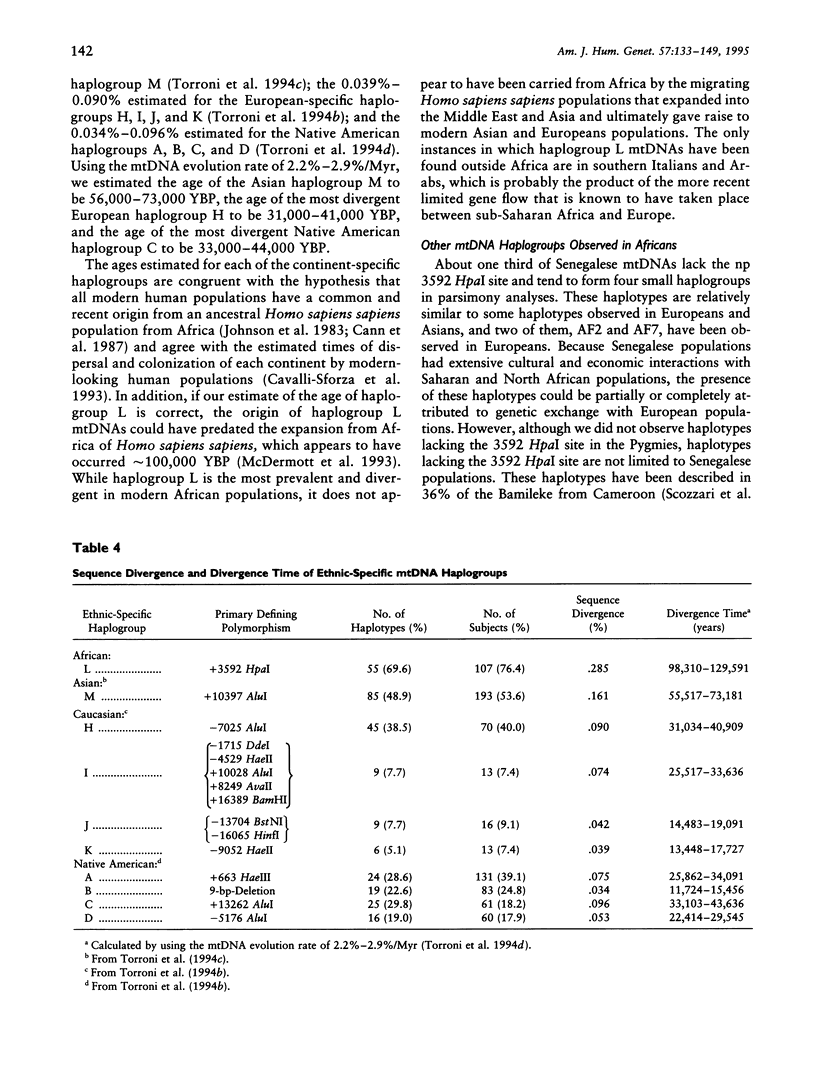
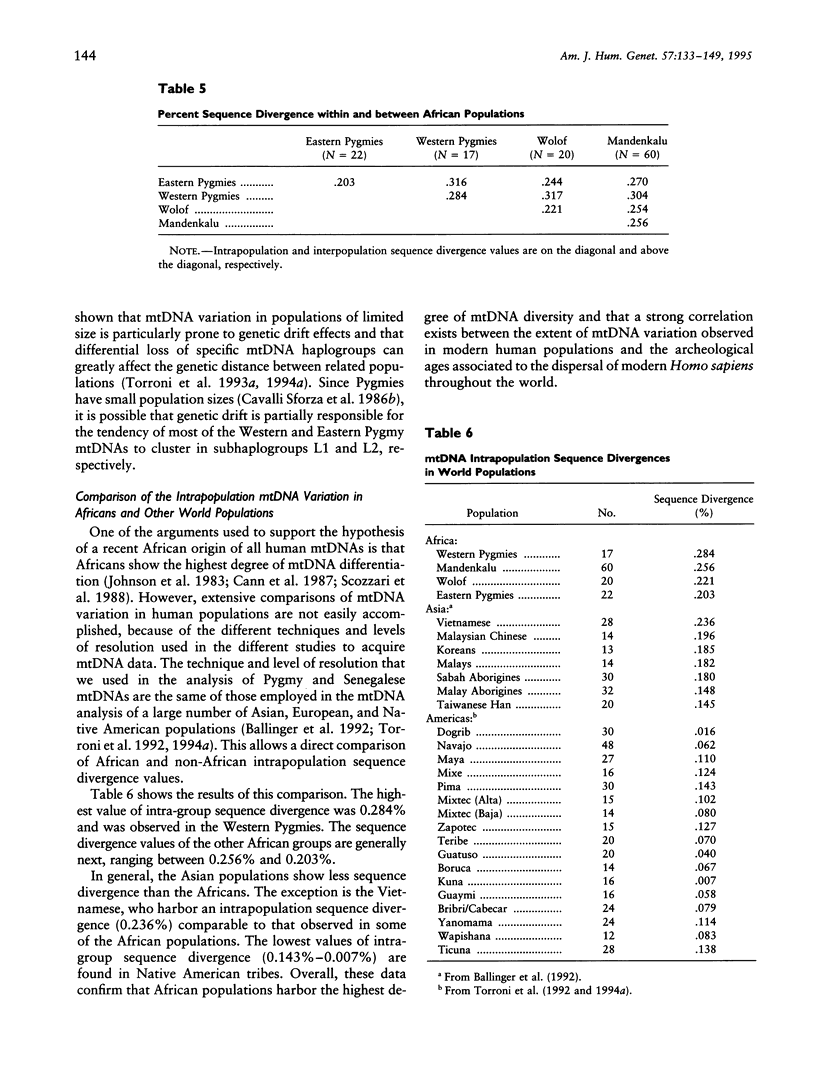
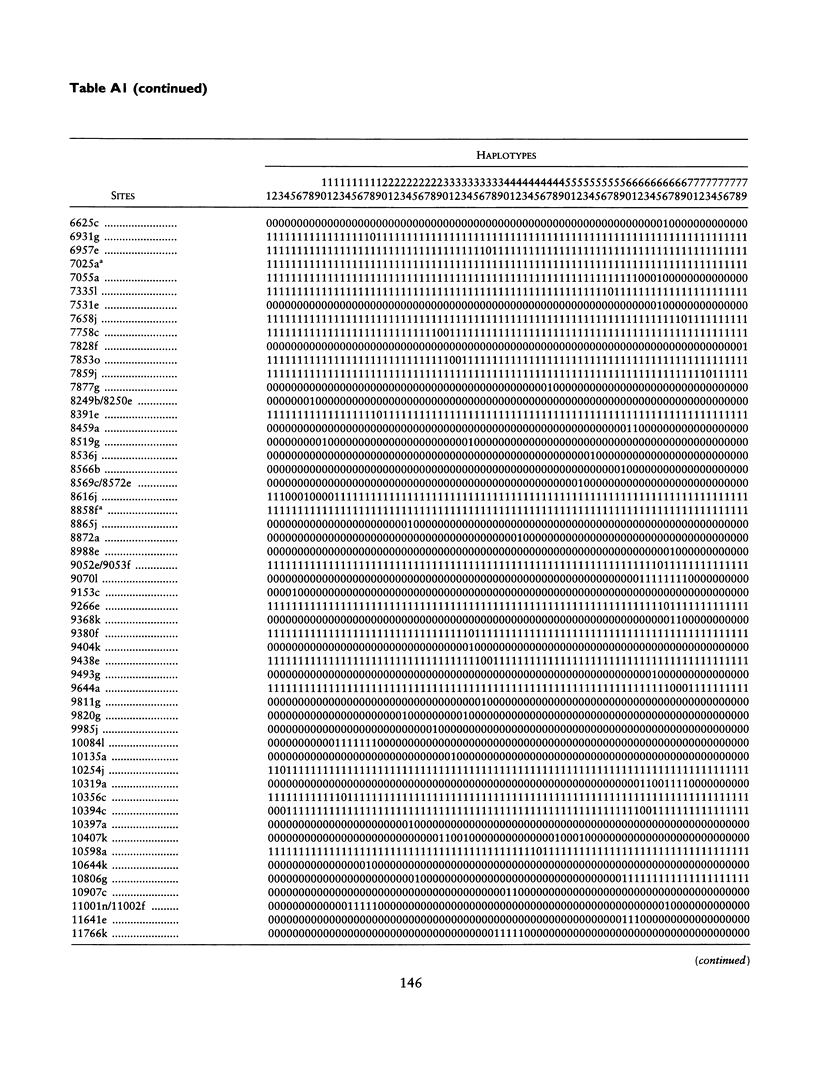
mtDNA sequence variation was examined in 140 Africans, including Pygmies from Zaire and Central African Republic (C.A.R.) and Mandenkalu, Wolof, and Pular from Senegal. More than 76% of the African mtDNAs (100% of the Pygmies and 67.3% of the Senegalese) formed one major mtDNA cluster (haplogroup L) defined by an African-specific HpaI site gain at nucleotide pair (np) 3592. Additional mutations subdivided haplogroup L into two subhaplogroups, each encompassing both Pygmy and Senegalese mtDNAs. A novel 12-bp homoplasmic insertion in the intergenic region between tRNA(Tyr) and cytochrome oxidase I (COI) genes was also observed in 17.6% of the Pygmies from C.A.R. This insertion is one of the largest observed in human mtDNAs. Another 25% of the Pygmy mtDNAs harbored a 9-bp deletion between the cytochrome oxidase II (COII) and tRNA(Lys) genes, a length polymorphism previously reported in non-African populations. In addition to haplogroup L, other haplogroups were observed in the Senegalese. These haplogroups were more similar to those observed in Europeans and Asians than to haplogroup L mtDNAs, suggesting that the African mtDNAs without the HpaI np 3592 site could be the ancestral types from which European and Asian mtDNAs were derived. Comparison of the intrapopulation sequence divergence in African and non-African populations confirms that African populations exhibit the largest extent of mtDNA variation, a result that further supports the hypothesis that Africans represent the most ancient human group and that all modern humans have a common and recent African origin. The age of the total African variation was estimated to be 101,000-133,000 years before present (YBP), while the age of haplogroup L was estimated at 98,000-130,000 YBP. These values substantially exceed the ages of all Asian- and European-specific mtDNA haplogroups.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Ballinger S. W., Schurr T. G., Torroni A., Gan Y. Y., Hodge J. A., Hassan K., Chen K. H., Wallace D. C. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992 Jan;130(1):139–152. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonné-Tamir B., Johnson M. J., Natali A., Wallace D. C., Cavalli-Sforza L. L. Human mitochondrial DNA types in two Israeli populations--a comparative study at the DNA level. Am J Hum Genet. 1986 Mar;38(3):341–351. [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Torroni A., Huoponen K., Chen Y. S., Lott M. T., Wallace D. C. Pathological significance of the mtDNA COX III mutation at nucleotide pair 9438 in Leber hereditary optic neuropathy. Am J Hum Genet. 1994 Aug;55(2):410–412. [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Wilson A. C. Length mutations in human mitochondrial DNA. Genetics. 1983 Aug;104(4):699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Menozzi P., Piazza A. Demic expansions and human evolution. Science. 1993 Jan 29;259(5095):639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- Denaro M., Blanc H., Johnson M. J., Chen K. H., Wilmsen E., Cavalli-Sforza L. L., Wallace D. C. Ethnic variation in Hpa 1 endonuclease cleavage patterns of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5768–5772. doi: 10.1073/pnas.78.9.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Langaney A. Origin and differentiation of human mitochondrial DNA. Am J Hum Genet. 1989 Jan;44(1):73–85. [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M., Mickleson K. N., Serjeantson S. W., Prior J. F., Trent R. J. An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. Am J Hum Genet. 1989 Apr;44(4):504–510. [PMC free article] [PubMed] [Google Scholar]
- Johnson M. J., Wallace D. C., Ferris S. D., Rattazzi M. C., Cavalli-Sforza L. L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19(3-4):255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- McDermott F., Grün R., Stringer C. B., Hawkesworth C. J. Mass-spectrometric U-series dates for Israeli Neanderthal/early modern hominid sites. Nature. 1993 May 20;363(6426):252–255. doi: 10.1038/363252a0. [DOI] [PubMed] [Google Scholar]
- Nei M., Tajima F. Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics. 1983 Sep;105(1):207–217. doi: 10.1093/genetics/105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman N. J., Torroni A., Brown M. D., Lott M. T., Fernandez M. M., Wallace D. C. Epidemic neuropathy in Cuba not associated with mitochondrial DNA mutations found in Leber's hereditary optic neuropathy patients. Cuba Neuropathy Field Investigation Team. Am J Ophthalmol. 1994 Aug 15;118(2):158–168. doi: 10.1016/s0002-9394(14)72895-8. [DOI] [PubMed] [Google Scholar]
- Passarino G., Semino O., Modiano G., Santachiara-Benerecetti A. S. COII/tRNA(Lys) intergenic 9-bp deletion and other mtDNA markers clearly reveal that the Tharus (southern Nepal) have Oriental affinities. Am J Hum Genet. 1993 Sep;53(3):609–618. [PMC free article] [PubMed] [Google Scholar]
- Ritte U., Neufeld E., Prager E. M., Gross M., Hakim I., Khatib A., Bonné-Tamir B. Mitochondrial DNA affinity of several Jewish communities. Hum Biol. 1993 Jun;65(3):359–385. [PubMed] [Google Scholar]
- Schurr T. G., Ballinger S. W., Gan Y. Y., Hodge J. A., Merriwether D. A., Lawrence D. N., Knowler W. C., Weiss K. M., Wallace D. C. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet. 1990 Mar;46(3):613–623. [PMC free article] [PubMed] [Google Scholar]
- Scozzari R., Torroni A., Semino O., Cruciani F., Spedini G., Santachiara Benerecetti S. A. Genetic studies in Cameroon: mitochondrial DNA polymorphisms in Bamileke. Hum Biol. 1994 Feb;66(1):1–12. [PubMed] [Google Scholar]
- Scozzari R., Torroni A., Semino O., Sirugo G., Brega A., Santachiara-Benerecetti A. S. Genetic studies on the Senegal population. I. Mitochondrial DNA polymorphisms. Am J Hum Genet. 1988 Oct;43(4):534–544. [PMC free article] [PubMed] [Google Scholar]
- Semino O., Torroni A., Scozzari R., Brega A., De Benedictis G., Santachiara Benerecetti A. S. Mitochondrial DNA polymorphisms in Italy. III. Population data from Sicily: a possible quantitation of maternal African ancestry. Ann Hum Genet. 1989 May;53(Pt 2):193–202. doi: 10.1111/j.1469-1809.1989.tb01784.x. [DOI] [PubMed] [Google Scholar]
- Shields G. F., Hecker K., Voevoda M. I., Reed J. K. Absence of the Asian-specific region V mitochondrial marker in Native Beringians. Am J Hum Genet. 1992 Apr;50(4):758–765. [PMC free article] [PubMed] [Google Scholar]
- Soodyall H., Jenkins T. Mitochondrial DNA polymorphisms in Khoisan populations from southern Africa. Ann Hum Genet. 1992 Oct;56(Pt 4):315–324. doi: 10.1111/j.1469-1809.1992.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Jorde L. B., Bhatia K., Wilson A. C. Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics. 1990 Mar;124(3):717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. R. Human origins and analysis of mitochondrial DNA sequences. Science. 1992 Feb 7;255(5045):737–737. doi: 10.1126/science.1590849. [DOI] [PubMed] [Google Scholar]
- Torroni A., Chen Y. S., Semino O., Santachiara-Beneceretti A. S., Scott C. R., Lott M. T., Winter M., Wallace D. C. mtDNA and Y-chromosome polymorphisms in four Native American populations from southern Mexico. Am J Hum Genet. 1994 Feb;54(2):303–318. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Lott M. T., Cabell M. F., Chen Y. S., Lavergne L., Wallace D. C. mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet. 1994 Oct;55(4):760–776. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Miller J. A., Moore L. G., Zamudio S., Zhuang J., Droma T., Wallace D. C. Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol. 1994 Feb;93(2):189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- Torroni A., Schurr T. G., Cabell M. F., Brown M. D., Neel J. V., Larsen M., Smith D. G., Vullo C. M., Wallace D. C. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993 Sep;53(3):563–590. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Schurr T. G., Yang C. C., Szathmary E. J., Williams R. C., Schanfield M. S., Troup G. A., Knowler W. C., Lawrence D. N., Weiss K. M. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992 Jan;130(1):153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Sukernik R. I., Schurr T. G., Starikorskaya Y. B., Cabell M. F., Crawford M. H., Comuzzie A. G., Wallace D. C. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet. 1993 Sep;53(3):591–608. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Wallace D. C. Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance. J Bioenerg Biomembr. 1994 Jun;26(3):261–271. doi: 10.1007/BF00763098. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A. C. African populations and the evolution of human mitochondrial DNA. Science. 1991 Sep 27;253(5027):1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- Wrischnik L. A., Higuchi R. G., Stoneking M., Erlich H. A., Arnheim N., Wilson A. C. Length mutations in human mitochondrial DNA: direct sequencing of enzymatically amplified DNA. Nucleic Acids Res. 1987 Jan 26;15(2):529–542. doi: 10.1093/nar/15.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Benedictis G., Rose G., Passarino G., Quagliariello C. Restriction fragment length polymorphism of human mitochondrial DNA in a sample population from Apulia (southern Italy). Ann Hum Genet. 1989 Oct;53(Pt 4):311–318. doi: 10.1111/j.1469-1809.1989.tb01800.x. [DOI] [PubMed] [Google Scholar]


