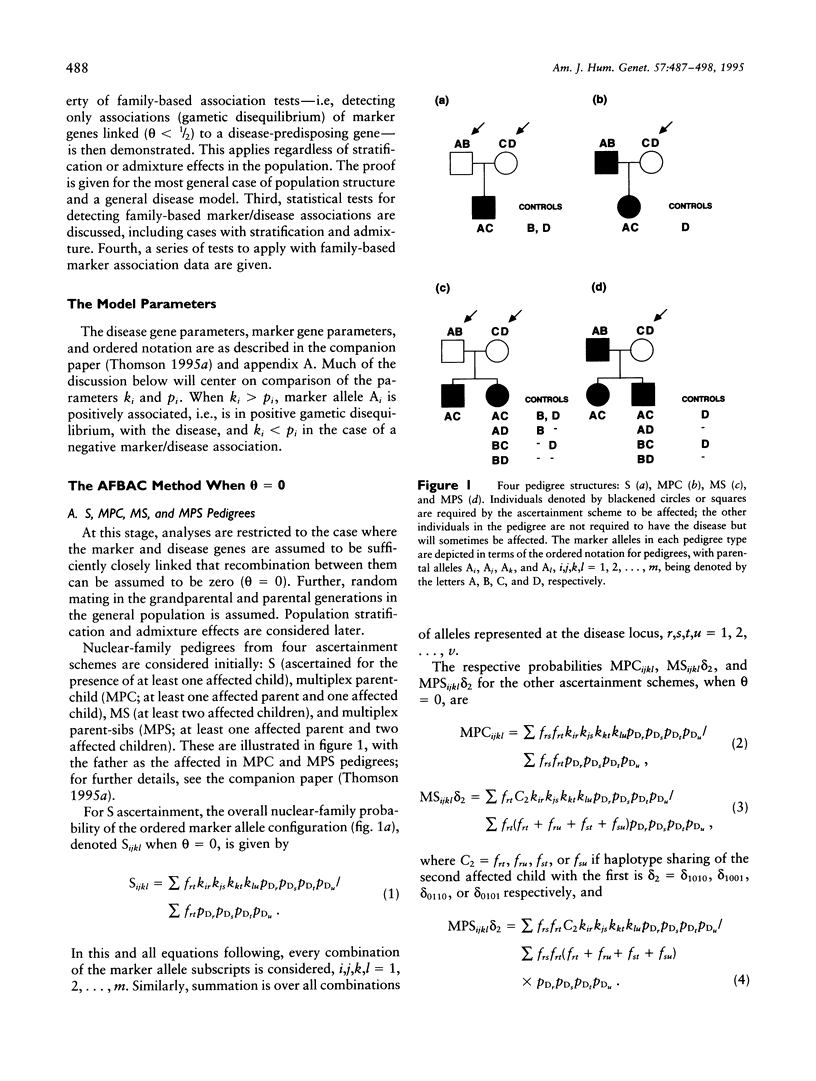
Abstract
With recent rapid advances in mapping of the human genome, including highly polymorphic and closely linked markers, studies of marker associations with disease are increasingly relevant for mapping disease genes. The use of nuclear-family data in association studies was initially developed to avoid possible ethnic mismatching between patients and randomly ascertained controls. The parental marker alleles not transmitted to an affected child or never transmitted to an affected sib pair form the so-called AFBAC (affected family-based controls) population. In this paper, the theoretical foundation of the AFBAC method is proved for any single-locus model of disease and for any nuclear family-based ascertainment scheme. In a random-mating population, when there is a marker association with disease, the AFBAC population provides an unbiased estimate of the overall population (control) marker alleles when the recombination fraction (theta) between the marker and disease genes is sufficiently small that it can be taken as zero (theta = 0). With population stratification, only marker associations present in the subpopulations will be detected with family-based analyses. Of more importance, however, is the fact that, when theta not equal to 0, differences between transmitted parental (patient) marker allele frequencies and non- or never-transmitted parental marker allele frequencies (implying a marker association with disease) can only be observed for marker genes linked to a disease gene (theta < 1/2). Thus, associations of unlinked marker loci with disease at the population level, caused by population stratification, migration, or admixture, are eliminated. This validates the use of family-based association tests as an appropriate strategy for mapping disease genes.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bengtsson B. O., Thomson G. Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens. 1981 Nov;18(5):356–363. doi: 10.1111/j.1399-0039.1981.tb01404.x. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Baker L., Spielman R. S. Insulin-gene sharing in sib pairs with insulin-dependent diabetes mellitus: no evidence for linkage. Am J Hum Genet. 1988 Jan;42(1):167–172. [PMC free article] [PubMed] [Google Scholar]
- Davies J. L., Kawaguchi Y., Bennett S. T., Copeman J. B., Cordell H. J., Pritchard L. E., Reed P. W., Gough S. C., Jenkins S. C., Palmer S. M. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994 Sep 8;371(6493):130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- Dean M., Stephens J. C., Winkler C., Lomb D. A., Ramsburg M., Boaze R., Stewart C., Charbonneau L., Goldman D., Albaugh B. J. Polymorphic admixture typing in human ethnic populations. Am J Hum Genet. 1994 Oct;55(4):788–808. [PMC free article] [PubMed] [Google Scholar]
- Falk C. T. Characteristics of a multiplex IDDM sample: unexplained differences with other samples. Genet Epidemiol. 1989;6(1):95–100. doi: 10.1002/gepi.1370060118. [DOI] [PubMed] [Google Scholar]
- Falk C. T., Rubinstein P. Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann Hum Genet. 1987 Jul;51(Pt 3):227–233. doi: 10.1111/j.1469-1809.1987.tb00875.x. [DOI] [PubMed] [Google Scholar]
- Field L. L., Fothergill-Payne C., Bertrams J., Baur M. P. HLA-DR effects in a large German IDDM dataset. Genet Epidemiol Suppl. 1986;1:323–328. doi: 10.1002/gepi.1370030749. [DOI] [PubMed] [Google Scholar]
- Field L. L. Genes predisposing to IDDM in multiplex families. Genet Epidemiol. 1989;6(1):101–106. doi: 10.1002/gepi.1370060119. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Weir B. S. Maximum-likelihood estimation of gene location by linkage disequilibrium. Am J Hum Genet. 1994 Apr;54(4):705–714. [PMC free article] [PubMed] [Google Scholar]
- Hodge S. E. Linkage analysis versus association analysis: distinguishing between two models that explain disease-marker associations. Am J Hum Genet. 1993 Aug;53(2):367–384. [PMC free article] [PubMed] [Google Scholar]
- Jin K., Speed T. P., Klitz W., Thomson G. Testing for segregation distortion in the HLA complex. Biometrics. 1994 Dec;50(4):1189–1198. [PubMed] [Google Scholar]
- Jorde L. B., Watkins W. S., Carlson M., Groden J., Albertsen H., Thliveris A., Leppert M. Linkage disequilibrium predicts physical distance in the adenomatous polyposis coli region. Am J Hum Genet. 1994 May;54(5):884–898. [PMC free article] [PubMed] [Google Scholar]
- Khoury M. J. Case-parental control method in the search for disease-susceptibility genes. Am J Hum Genet. 1994 Aug;55(2):414–415. [PMC free article] [PubMed] [Google Scholar]
- Knapp M., Seuchter S. A., Baur M. P. The haplotype-relative-risk (HRR) method for analysis of association in nuclear families. Am J Hum Genet. 1993 Jun;52(6):1085–1093. [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Thomson G. Three-allele synergistic mixed model for insulin-dependent diabetes mellitus. Diabetes. 1986 Sep;35(9):958–963. doi: 10.2337/diab.35.9.958. [DOI] [PubMed] [Google Scholar]
- Maxwell A. E. Comparing the classification of subjects by two independent judges. Br J Psychiatry. 1970 Jun;116(535):651–655. doi: 10.1192/bjp.116.535.651. [DOI] [PubMed] [Google Scholar]
- Motro U., Thomson G. The affected sib method. I. Statistical features of the affected sib-pair method. Genetics. 1985 Jul;110(3):525–538. doi: 10.1093/genetics/110.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Linkage disequilibrium in subdivided populations. Genetics. 1973 Sep;75(1):213–219. doi: 10.1093/genetics/75.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom G. T., Erlich H. MHC class-II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- Ott J. Statistical properties of the haplotype relative risk. Genet Epidemiol. 1989;6(1):127–130. doi: 10.1002/gepi.1370060124. [DOI] [PubMed] [Google Scholar]
- Payami H., Joe S., Farid N. R., Stenszky V., Chan S. H., Yeo P. P., Cheah J. S., Thomson G. Relative predispositional effects (RPEs) of marker alleles with disease: HLA-DR alleles and Graves disease. Am J Hum Genet. 1989 Oct;45(4):541–546. [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., Sommer S. S. Genotype relative risks: methods for design and analysis of candidate-gene association studies. Am J Hum Genet. 1993 Nov;53(5):1114–1126. [PMC free article] [PubMed] [Google Scholar]
- Sham P. C., Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995 Jan;59(Pt 1):97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- Spielman R. S., Baur M. P., Clerget-Darpoux F. Genetic analysis of IDDM: summary of GAW5 IDDM results. Genet Epidemiol. 1989;6(1):43–58. doi: 10.1002/gepi.1370060111. [DOI] [PubMed] [Google Scholar]
- Spielman R. S., McGinnis R. E., Ewens W. J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993 Mar;52(3):506–516. [PMC free article] [PubMed] [Google Scholar]
- Stephens J. C., Briscoe D., O'Brien S. J. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet. 1994 Oct;55(4):809–824. [PMC free article] [PubMed] [Google Scholar]
- Suarez B. K., Hampe C. L. Linkage and association. Am J Hum Genet. 1994 Mar;54(3):554–563. [PMC free article] [PubMed] [Google Scholar]
- Suarez B. K., Hampe C. L. Linkage and association. Am J Hum Genet. 1994 Mar;54(3):554–563. [PMC free article] [PubMed] [Google Scholar]
- Svejgaard A., Jersild C., Nielsen L. S., Bodmer W. F. HL-A antigens and disease. Statistical and genetical considerations. Tissue Antigens. 1974;4(2):95–105. doi: 10.1111/j.1399-0039.1974.tb00230.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger J. D., Ott J. A haplotype-based 'haplotype relative risk' approach to detecting allelic associations. Hum Hered. 1992;42(6):337–346. doi: 10.1159/000154096. [DOI] [PubMed] [Google Scholar]
- Thomson G., Robinson W. P., Kuhner M. K., Joe S., Klitz W. HLA and insulin gene associations with IDDM. Genet Epidemiol. 1989;6(1):155–160. doi: 10.1002/gepi.1370060129. [DOI] [PubMed] [Google Scholar]
- Thomson G., Robinson W. P., Kuhner M. K., Joe S., MacDonald M. J., Gottschall J. L., Barbosa J., Rich S. S., Bertrams J., Baur M. P. Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes mellitus. Am J Hum Genet. 1988 Dec;43(6):799–816. [PMC free article] [PubMed] [Google Scholar]
- Thomson G. The effect of a selected locus on linked neutral loci. Genetics. 1977 Apr;85(4):753–788. doi: 10.1093/genetics/85.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiret L., Nicaud V., Ehnholm C., Havekes L., Menzel H. J., Ducimetiere P., Cambien F. Inference of the strength of genotype-disease association from studies comparing offspring with and without parental history of disease. Ann Hum Genet. 1993 May;57(Pt 2):141–149. doi: 10.1111/j.1469-1809.1993.tb00895.x. [DOI] [PubMed] [Google Scholar]


