Abstract
Transcriptional co-activator LEDGF/p75 is the major cellular interactor of HIV-1 integrase (IN), critical to efficient viral replication. In this work, a series of INs from the Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Spumavirus and Lentivirus retroviral genera were tested for interaction with the host factor. None of the non-lentiviral INs possessed detectable affinity for LEDGF in either pull-down or yeast two-hybrid assays. In contrast, all lentiviral INs examined, including those from bovine immunodeficiency virus (BIV), maedi-visna virus (MVV) and equine infectious anemia virus (EIAV) readily interacted with LEDGF. Mutation of Asp-366 to Asn in LEDGF ablated the interaction, suggesting a common mechanism of the host factor recognition by the INs. LEDGF potently stimulated strand transfer activity of divergent lentiviral INs in vitro. Unprecedentedly, in the presence of the host factor, EIAV IN almost exclusively catalyzed concerted integration, whereas HIV-1 IN promoted predominantly half-site integration, and BIV IN was equally active in both types of strand transfer. Concerted BIV and EIAV integration resulted in 5 bp duplications of the target DNA sequences. These results confirm that the interaction with LEDGF is conserved within and limited to Lentivirus and strongly argue that the host factor is intimately involved in the catalysis of lentiviral DNA integration.
INTRODUCTION
Retroviruses are distinguished by reverse transcription of their RNA genomes and integration of the resulting cDNA replicas into a host cell chromosome. Members of the Retroviridae family can be subdivided into several genera, of which five were isolated from mammals: Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Spumavirus and Lentivirus. The two remaining genera, Alpharetrovirus and Epsilonretrovirus, are retroviruses from birds and fish respectively [(1) and http://www.ncbi.nlm.nih.gov/ICTVdb]. DNA integration is essential for replication of human lentiviruses HIV-1 and HIV-2 and is fundamental to the viral persistence that leads to AIDS (2–4) (http://www.retroconference.org/2006/). Integrase (IN) is the virus-encoded enzyme responsible for the key catalytic events associated with integration [reviewed in (5)]. IN acts on the linear double stranded cDNA molecule generated by reverse transcription of the retroviral genomic RNA. In a reaction termed 3′-processing, IN trims the 3′-termini of the cDNA exposing 3′ hydroxyls of the invariant CA dinucleotides. A dinucleotide is removed from the U3 and U5 termini of HIV-1 cDNA. Next, IN inserts the processed 3′-termini of the viral cDNA into opposing strands of the chromosomal DNA. Integrated viral cDNA is initially flanked by a pair of short single-stranded gaps, which are repaired by host cell enzymes to produce a stable provirus.
Retroviral integration is not entirely random with respect to the target DNA. Weak consensus integration site sequences have been reported for HIV-1, avian sarcoma leukosis virus (ASLV) and murine leukemia virus (MLV) (6). Furthermore, on the genomic scale retroviruses display distinct, genus-specific patterns of preferred integration regions [reviewed in (7)]. The lentiviruses whose integration site preferences have been reported to-date [HIV-1, HIV-2, simian immunodeficiency virus (SIV), equine infectious anemia virus (EIAV) and feline immunodeficiency virus (FIV)] share a strong bias toward integration within transcription units (8–13), whereas a gammaretrovirus MLV favors transcription start sites and CpG islands (9). Integration of an alpharetrovirus ASLV and a spumaretrovirus prototype foamy virus (PFV) appears to correlate only weakly with gene expression (14–17). These observations collectively suggested a play of virus- or genus-specific mechanisms for integration target selection.
Of several cellular proteins implicated in retroviral integration, transcriptional co-activator p75 received considerable attention since its identification as an interactor of HIV-1 IN (18–20) [for recent reviews see (21,22)]. P75, commonly referred to as lens epithelium-derived growth factor (LEDGF), is a nuclear chromatin-associated protein, ubiquitously expressed at all stages of development (23–26). Early work has implicated LEDGF and its alternative splice form p52 in regulation of gene expression (23,27,28). Recently, LEDGF was shown to associate with JPO2, an interactor of the transcription factor and proto-oncogene c-Myc, corroborating its role in transcription regulation (29). LEDGF interacts with HIV-1 IN via the integrase binding domain (IBD) found within its C-terminal region (residues 347–429) (24,30). A solution structure of the isolated IBD and a crystal structure of the IBD in complex with the catalytic core domain (CCD) of HIV-1 IN have been recently reported (31,32). The IBD adopts a compact, α-helical fold, reminiscent of a pair of HEAT repeats (31). The tip of its finger-like structure binds at the dimer interface of the IN CCD (32). In addition to the CCD, the aminoterminal domain of HIV-1 IN required for high-affinity binding is thought to be involved in the protein–protein interaction (25). LEDGF dramatically alters biochemical properties of recombinant HIV-1 IN and potently stimulates its enzymatic activity (18,24,33,34).
When expressed in human cells, HIV-1 IN stably associates with mitotic chromosomes (35,36). This property was found to be strictly dependent on the endogenous LEDGF, which appears to tether the viral protein to chromosomal DNA (20,25,37,38). These findings suggested that through its interaction with IN, LEDGF could direct HIV-1 preintegration complex (PIC) to its preferred genomic loci. Concordantly, functional HIV-1 PICs could be immunoprecipitated with antibodies to LEDGF (37). Activity of HIV-1 PICs partially disrupted by treatment in high ionic strength conditions could be rescued by recombinant LEDGF (39). Moreover, a partial knock-down of endogenous LEDGF in target cells resulted in statistically-significant reduction of HIV-1 integration into transcription units (40). In addition, HIV-1 integration could be in part re-targeted in vitro to the vicinity of a phage lambda repressor binding site by a chimeric protein containing the IBD fused to the DNA binding domain of lambda repressor (41). Current reports from several groups indicate that LEDGF plays a critical role in HIV-1 DNA integration and is required for efficient viral replication (42–45) (http://www.ascb.org).
In addition to HIV-1 IN, LEDGF was shown to bind primate lentiviral HIV-2 and SIV INs, as well as divergent FIV IN; whereas no interaction was detected between LEDGF and the INs from a gammaretrovirus MLV, deltaretrovirus human T cell lymphotropic virus type 2 (HTLV-2) and alpharetrovirus ASLV (20,34,37). Accordingly, knock-down of LEDGF expression in target cells did not affect MLV, while significantly impairing HIV-1 and FIV integration (45). Although a functional interaction with LEDGF has only been demonstrated for HIV-1 IN, the available data seem to indicate it is universal within but limited to Lentivirus. The present study was designed to test this conjecture and to clarify whether the modulation of enzymatic activities by LEDGF is a conserved feature of lentiviral INs.
MATERIALS AND METHODS
Viral DNA and PCR primers
Molecular clones KV1772 (46), pBIV127 (47), pSPeiav19 (48), pEECC-FeLV (49), pcHSRV2 (50) were sources of maedi-visna virus (MVV), bovine immunodeficiency virus (BIV), EIAV, feline leukemia virus (FeLV) and PFV DNA, respectively. Genomic DNA of Akata cells infected with Mason-Pfizer monkey virus (MPMV) served as template for amplification of the MPMV IN coding sequence (CDS). HTLV-1 DNA was from a clinical isolate.
The following oligodeoxynucleotides were synthesized by Invitrogen: PC122, GCGCGTCGACTTCTTTTTGTATTTCTTTTGGTGG; PC123, TGTGGATAGAAAATATTCCC; PC124, GCGCGTCGACATTTGGTTTTATTAGTAACTTAGTCC; PC125, TGTGGGTAGAAAATATTCAGG; PC126, GCGCGTCGACCGAACTCCCATCTTGGATATC; PC127, TGTTCCTAGAAAATATCCCCTC; PC138, CCCCACAGTACTGTTGGGTGTTCTTCACC; PC139, GGCGGGTACCTTCAGCTCGTGTAGCTCA; PC140, CACCCAACAGTACTGTGGGGCAGGGTGG; PC141, GGCGGAATTCGTACAGAGTGAAGATATGG; PC142, TCAGGGACCCGGGTTCCTAGAAAATATCCCCT; PC143, GCCTTGAAGTCCTCTTTCAGGGACCCGGGTTCC; PC144, GGCCCTCGAGTCACGAACTCCCATCTTGG; PC145, GCGCGTCGACTTCATTTTTTTCCAAATGATCCATTG; PC146, TGAAAGGATATCCCAAACAATATAC; PC155, TGTTCCTGGAAAAAATAGAGCC; PC156, GCGCGTCGACTGCCATTTCTCCATCCTC; PC160, GCGCGTCGACATTGAACAAGATGGATTGCACG; PC161, TGAGTAACATAAACACAAACCTCG; PC169, GGCGGGTACCGGCACTCAGATTCTGCGGTC; PC170, GGCGGAATTCCAATTGTCAGAATACAAGCAC; PC171, CCCACAGTACTGTAGGATCTCGAACAG; PC172, GATCCTACAGTACTGTGGGGTTTTTATGAGG; PC173, GAGCCCCGGGTGGGTAGAAAATATTCAGG; PC174, GCGCGGATCCTCAATTTGGTTTTATTAGTAACTTAG; PC175, TGCAGCTCTCTCCTGCAGACC; PC176, GCGCGTCGACCCCATGGTGTTGGTGGTCTTTTTCTTTGG; PC186, GCGCGTCGACAGTTCCACGAGAGAGTC; PC187x, TGCCCACTGAACTTATAGAGG; PC188, TACCTGGTACCTGTACTGGGTCTCTCTGG; PC192, GGCGGGTACCAGAAAGCAGGTAGCTTGCAGTGG; PC193, GGCGGGTACCCGAAGAACTCCAGCATGAGATCC; PC200, CGCACCTCCTCCCTGGGCTTCGGAC; PC203, TTTTCCTTATTTTCTGAGTAAGG; PC204, GAAAGGACAACATGAGAAAGAGGC; PC206, CTATCAATTACACATTAACATACACAC; PC210, GGACTGAGGGGCCTGAAATGAGC; PC214, CCATTACAAACTTCTCAAATGTTCTTTATATTCCAGG; PC217, GTAAGCCCACTGCAAGCTACC; PC218, GGATCTCATGCTGGAGTTCTTCG; T3P, GCAATTAACCCTCACTAAAGG.
DNA constructs for protein expression
To make constructs for production of C-terminally His6-tagged INs, DNA fragments encoding HIV-2, MVV, BIV, EIAV, MPMV, FeLV, HTLV-1 and PFV INs were PCR-amplified using Pfu DNA polymerase (Stratagene) and the primer pairs PC155/PC156, PC123/PC122, PC127/PC126, PC125/PC124, PC161/PC160, PC187x/PC186, PC175/PC176 and PC146/PC145, respectively. Resulting amplicons digested with SalI were ligated between NdeI and SalI sites of pET20-b(+) (Novagen) (NdeI site of the vector was filled-in using Pfu polymerase to allow blunt-end ligation). The plasmid pKB-IN6H used for bacterial expression of HIV-1 IN with a C-terminal His6-tag has been described (25).
To make pCPH6P-BIV-IN, for production of BIV IN with N-terminal His6-tag and a human rhinovirus (HRV) 3C protease cleavage site, a PCR fragment obtained using primers PC143 and PC144 and re-amplified using primers PC142 and PC144, was digested with XhoI and ligated between NdeI and XhoI sites of pET15b (Novagen) (NdeI site of the vector was filled-in to allow blunt-end ligation). To make pCPH6P-EIAV-IN for expression of EIAV IN with a removable His6-tag, a PCR amplicon obtained with primers PC173 and PC174 was digested with XmaI and BamHI and subcloned into pCPH6P-BIV-IN, to replace the BIV IN CDS. The plasmids pRP1012, pCP-Nat75 and pCP-Nat75(D366N) were used for bacterial expression of HIV-1 IN with an N-terminal His6-tag and a thrombin cleavage site, non-tagged human LEDGF and LEDGF(D366N), respectively (25,31,51).
DNA substrates for integration assays
To make pBIV-U3U5, RU5 and U3 fragments of the BIV long terminal repeat (LTR) were PCR-amplified using primers pairs PC138/PC139 and PC140/PC141 respectively and spliced by re-amplification with primers PC139 and PC141. The resulting amplicon digested with KpnI and EcoRI was ligated between KpnI and EcoRI sites of pBK-RSV (Stratagene). To make pEIAV-U3U5, RU5 and U3 fragments of the EIAV LTR PCR-amplified using primer pairs PC169/PC171 and PC170/PC172 were spliced in a PCR with primers PC169 and PC170. The resulting amplicon was digested with KpnI and EcoRI and subcloned into pBK-RSV. Mini-viral substrates for HIV-1, BIV and EIAV INs were prepared by digestion of pU3U5 (51), pBIV-U3U5 and pEIAV-U3U5 with ScaI, respectively.
To obtain RU5 substrates for HIV-1, BIV and EIAV INs, PCR fragments obtained using Taq DNA polymerase and T3P primer in combination with PC188, PC139, and PC169, respectively, and pU3U5, pBIV-U3U5 and pEIAV-U3U5 as templates, respectively, were digested with ScaI and the RU5-containing fragments were purified by electrophoresis through 2% agarose gels. The longer RU5+300 donors were obtained in a similar fashion, substituting PC188, PC139 or PC169 for PC210 primer that anneals 300 bp upstream of the KpnI site of the mini-viral constructs. Where indicated, the reactive strands of the RU5 substrates were 5′ end labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (GE Healthcare).
Recombinant proteins
Endonuclease A-deficient Escherichia coli strain PC2 [BL21(DE3), endA::TetR, T1R, pLysS], recovered as a spontaneous T1 phage-resistant mutant of PC1 (51,52), was used for protein production. Shake flask cultures of PC2 transformed with various expression constructs were grown in LB medium at 30°C to an A600 of 0.9–1.0 prior to induction with 0.25 mM isopropyl-thio-β-d-galactopyranoside. Following 4 h induction at 25–28°C, bacteria were harvested and stored at −70°C.
To isolate His6-tagged retroviral IN proteins, thawed bacterial paste was sonicated in buffer B (1 M NaCl, 7.5 mM CHAPS, 50 mM Tris–HCl, pH 7.4) containing 0.5 mM phenylmethlysulfonyl fluoride and 15 mM imidazole. Crude extracts pre-cleared by centrifugation were incubated with Ni NTA agarose (Qiagen). The resin was extensively washed in buffer B containing 15 mM imidazole, and His6-tagged proteins were eluted with 200 mM imidazole in buffer B. IN-containing fractions diluted with 3 volumes of 50 mM Tris–HCl, pH 7.4 were injected into a 5 ml HiTrap Heparin column (GE Healthcare), and bound proteins were eluted with a linear gradient of 0.25–1 M NaCl in 50 mM Tris–HCl, pH 7.4. Immediately after elution, 10 mM DTT was added to each fraction and NaCl concentration was adjusted to 1 M. HIV-1, BIV and EIAV INs produced to carry cleavable N-terminal His6-tags were digested with thrombin (3 NIH units of thrombin per mg of His6-tagged IN) for 3 h at 25°C in the presence of 20 mM β-mercaptoethanol or HRV14 3C protease (20 μg of protease per mg of His6-tagged IN) for 6–12 h at 4°C in the presence of 20 mM DTT; the tag-free INs were then purified by chromatography on Heparin sepharose as above.
Wild type and D366N LEDGF were made as previously described (24,25,31). Purified proteins supplemented with 10% glycerol were flash-frozen in liquid nitrogen and stored at −70°C. Protein concentration was determined using Bradford assay (Bio-Rad Laboratories) with a BSA standard. Where indicated, molar concentrations refer to monomer protein forms.
His6-tag pull-down assays
His6-tag pull-down assays were done as previously described (25,31). Briefly, 4 μg wild type or D366N LEDGF was incubated with 6.7 μg C-terminally His6-tagged retroviral IN, 10 μg BSA and 15 μl Ni-NTA agarose (settled bead volume) in 650 μl pull-down buffer [PB, 150 mM NaCl, 2 mM MgCl2, 25 mM imidazole, 0.1% Nonidet P40 and 50 mM Tris–HCl (pH 7.4)] for 4–5 h at 4°C with gentle rocking. The beads washed trice in ice-cold PB were boiled in 16 μl 2× Laemmli sample buffer containing 50 mM ethylenediaminetetraacetic acid (EDTA) and 50 mM DTT. Eluted proteins were separated by electrophoresis by SDS–PAGE and detected by staining with Coomassie-R250.
Sequence analysis of equine and bovine LEDGF cDNAs
Two partial cDNA sequences (GenBank accession nos. CX598499 and CD528249) spanning ∼900 bp of the equine LEDGF CDS were identified using translated BLAST. PC204, PC200 and PC206 primers were designed based on this known sequence and conserved portions of 5′- and 3′-untranslated regions (3′-UTRs) of mammalian LEDGF cDNA sequences. Total RNA extracted from Equus caballus-derived cell line NBL-6 was reverse-transcribed using random-primed Superscript III (Invitrogen). Sequencing of a 1 kb PCR fragment obtained using Easy-A DNA polymerase (Stratagene) primed by PC204 and PC206 revealed ∼300 bp of the 3′-UTR sequence of the equine cDNA. Finally, a 1.6 kb PCR fragment obtained using Easy-A with PC200 and PC214 revealed complete and unbiased CDS of the equine LEDGF cDNA. The sequences determined in this work were submitted to GenBank (accession codes DQ829781 and DQ873682). The entire CDS of the bovine LEDGF cDNA could be reconstructed from the available partial sequences (GenBank accession codes AF474175, CR382879, DN537967, DN536615, DT857195, DN533664, DT885845, DV915861, DT896093, DN535088, DV916069, DV913569 and DV827864).
In vitro integration assays
Tag-free preparations of HIV-1, BIV or EIAV INs were used in all enzymatic assays. Reaction conditions were essentially as previously described for the HIV-1 enzyme (18,24,31). Briefly, 2 μl IN [in dilution buffer (DB), 750 mM NaCl, 10 mM DTT and 10 mM Tris–HCl (pH 7.8)] or 2 μl DB was added to 36 μl master mixture containing appropriate substrate and target DNA in 42 mM NaCl, 6 mM MgSO4, 4.8 μM ZnCl2, 12.1 mM DTT, and 24.2 mM HEPES, pH 7.4. Samples were incubated at 25°C for 5–7 min before addition of 2 μl LEDGF (0–0.54 mg/ml in DB) and then at 37°C for 90 min. Mini-HIV, -BIV, and -EIAV donor DNA substrates were used at 2.5 nM (300 ng per 40 μl reaction), RU5 and RU5+300 were used at 10 nM. Supercoiled pGEM-9Zf(−) (300 ng) served as target DNA in the assays with RU5 and RU5+300 donors.
Integration reactions were stopped by addition of 0.5% SDS and 25 mM EDTA and deproteinized by digestion with 30 μg proteinase K (Roche Applied Science) for 45–60 min at 37°C followed by ethanol precipitation. DNA products separated in 0.8 or 1.5% agarose gels in Tris–acetate–EDTA buffer were visualized by staining with ethidium bromide. Singly-nicked pGEM-9Zf(−) DNA, used as migration standard for agarose gel electrophoresis, was prepared by digestion of the supercoiled plasmid with DNase I (Roche Applied Science) in the presence of saturating amounts of ethidium bromide (53). Radioactively-labeled reaction products were detected and quantified by phosphor autoradiography using a Storm-860 scanner (GE Healthcare).
Sequence analysis of BIV and EIAV integration products
Strand transfer products from upscaled reactions of BIV or EIAV INs with the respective RU5 donors and supercoiled pGEM-9Zf(−) target DNA were separated by electrophoresis in 1.5% agarose gels. The linear concerted integration products isolated from the gels were treated with phi29 DNA polymerase (New England Biolabs) in the presence of 200 μM of each deoxynucleotide triphosphate, digested with KpnI, and ligated with KpnI-digested kanamycin-resistance cassette. The cassette, containing the Tn5 aminoglycoside-3′-O-phosphotransferase gene, was obtained in a PCR using Easy-A DNA polymerase, PC192 and PC193 primers, and pCP15 (52) template. E.coli XL1-Blue cells transformed with the ligation products were selected with 35 μg/ml kanamycin. Plasmids isolated from individual colonies were sequenced using primers PC217 and PC218 annealing to the flanking regions of the kanamycin-resistance cassette.
RESULTS
Interaction with LEDGF is limited to and conserved among lentiviral INs
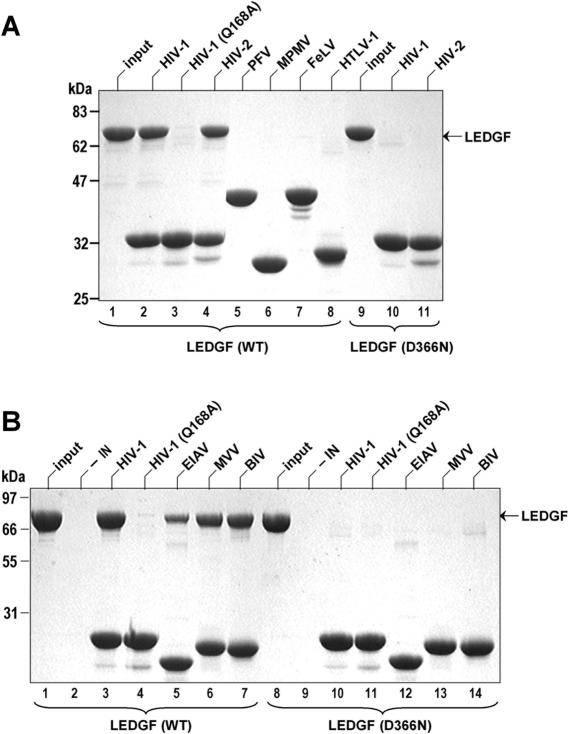
A series of IN proteins of lentiviral and non-lentiviral origins produced with identical C-terminal His6-tags were tested for interaction with recombinant human LEDGF in a pull-down assay using Ni-NTA agarose beads (25,31). In agreement with published observations (20,25,31,34), HIV-1 and HIV-2 INs readily bound LEDGF (Figure 1A, lanes 2 and 4). LEDGF was not recovered when IN was omitted (Figure 1B, lane 2), and the Q168A substitution close to the HIV-1 IN binding interface (20,32) precluded the interaction (Figure 1A, lane 3). Under identical conditions betaretroviral MPMV, gammaretroviral FeLV, deltaretroviral HTLV-1 and spumaretroviral PFV INs failed to pull-down LEDGF (Figure 1A, lanes 5–8). In contrast, lentiviral EIAV, MVV and BIV INs reproducibly retained LEDGF on the Ni-NTA beads (Figure 1B, lanes 5–7). LEDGF residue Asp366 plays a crucial role in recognition of HIV-1 IN, forming a bidentate hydrogen bond with main chain amides of IN residues Glu-170 and His-171 (32), and LEDGF mutants D366A and D366N are deficient for interaction with HIV-1 IN (31). Similar to HIV-1 IN (Figure 1A and B, lanes 10), none of the lentiviral INs studied here pulled down the LEDGF D366N mutant (Figure 1A, lane 11; Figure 1B, lanes 12–14), suggesting that divergent lentiviral INs share the structural basis for the host factor recognition.
Figure 1.
Interaction with LEDGF is limited to and conserved among lentiviral INs. (A) His6-tagged HIV-1, HIV-2, PFV, MPMV, FeLV and HTLV-1 INs (as indicated) were tested for the ability to pull-down WT (lanes 2–8) or D366N LEDGF (lanes 10 and 11). Proteins recovered on Ni-NTA agarose beads were separated in SDS–PAGE gels and detected by staining with Coomassie-R250. Lanes 1 and 9 contained input quantities of WT and D366N LEDGF, respectively. The migration positions of LEDGF and molecular weight markers (kDa) are indicated. (B) His6-tag pull-down assays with lentiviral HIV-1, EIAV, MVV and BIV INs (as indicated) and WT (lanes 3–7) or D366N LEDGF (lanes 10–14). Lanes 1 and 8 contained input quantities of WT and D366N LEDGF. IN was omitted from reactions in lanes 2 and 9.
Recent results indicate that applied to the IN-LEDGF interaction, yeast two-hybrid assays can be more sensitive than in vitro pull-down experiments. Residual interactions between LEDGF and several point mutants of HIV-1 IN (e.g. Q168A, V165A and R166A) could be revealed by yeast two-hybrid analyses, while evading detection by His6-tag pull-down (20,31,54). In agreement with the results of the pull-down assays described here, no interaction between LEDGF326–471 and non-lentiviral INs (MPMV, FeLV, HTLV-1 and PFV) could be detected in a GAL4-based yeast two-hybrid assay, under conditions that readily revealed binding of the co-factor to HIV-1 and FIV INs (Supplementary Figure S1). Furthermore, the D366N mutation in LEDGF disrupted the interactions with both HIV-1 and FIV INs (Supplementary Figure S1), corroborating a common structural mechanism of host factor recognition by divergent lentiviral INs.
Sequence analysis of bovine and equine LEDGF cDNAs
An issue in this study was the use of human LEDGF in various assays with INs from non-primate retroviruses. LEDGF is known to be well conserved in mammals (24). For instance, the human and feline orthologs share 96.4% identical and 97.9% similar residues (30). To justify using human LEDGF in enzymatic assays involving BIV and EIAV INs, sequences of bovine and equine LEDGF cDNAs were analyzed. A DNA fragment spanning the entire CDS of the E.caballus LEDGF cDNA could be amplified by RT–PCR from RNA of horse NBL-6 cells, whereas the complete CDS of the Bovis taurus LEDGF cDNA could be reconstructed from available partial sequences. Alignments of the predicted protein sequences revealed only minor differences between the human, bovine and equine LEDGF orthologs (Supplementary Figure S2). Thus, bovine and equine LEDGF orthologs share ∼97% identical and ∼99% similar residues with the human protein. Of importance, all functional and structural elements so far identified in LEDGF (i.e. the PWWP domain, the IBD, the NLS and the core A/T-hook motifs) are identical in the human, mouse, feline, equine and bovine orthologs.
LEDGF augments enzymatic activity of divergent lentiviral INs
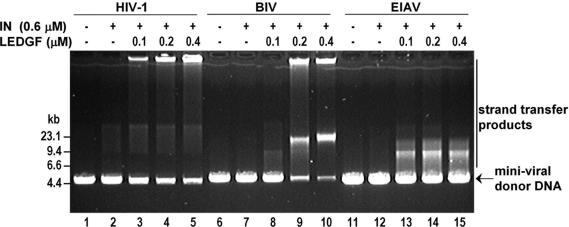
LEDGF enhances the strand transfer activity of HIV-1 IN as much as 150-fold under certain in vitro conditions (18,24,31,33). Notably, the LEDGF-dependent reactions take place in the absence of organic solvents or molecular crowding agents, otherwise required to observe efficient magnesium-dependent activity of HIV-1 IN in vitro (51,55–59). To establish whether divergent lentiviral INs respond to LEDGF in a similar way, strand transfer activities of recombinant HIV-1, BIV and EIAV INs on their respective mini-viral substrates were studied side-by-side. Mini-HIV (51), mini-BIV and mini-EAIV substrates, ∼4.5 kb linear blunt-ended DNA molecules flanked by U3 and RU5 fragments of the viral LTRs, were designed to mimic the viral cDNA. In the simplest form of the assay, mini-viral DNA serves as donor and target DNA and strand transfer products can be detected by staining with ethidium bromide following electrophoresis in agarose gels (18,51). Under close-to-physiological solution conditions [115 mM NaCl, 5 mM MgCl2, 10 mM DTT and 20 mM Hepes (pH 7.5)] HIV-1, BIV and EIAV INs were almost inactive in the absence of LEDGF, yielding barely detectable levels of strand transfer products (Figure 2, lanes 2, 7, 12). In agreement with previous observations (18), addition of LEDGF unleashed the strand transfer activity of HIV-1 IN (Figure 2, lanes 3–5). More than 50% of input mini-HIV DNA was converted into various products, including those that failed to enter the 0.8% agarose gel, in the presence of 0.4 μM LEDGF (Figure 2, lane 5). Similarly, LEDGF stimulated strand transfer activities of BIV and EIAV INs on their respective mini-viral DNA substrates (Figure 2, lanes 8–10, 13–15). Of note, the range of reaction products was IN-specific, each enzyme generating a reproducible and unique pattern.
Figure 2.
LEDGF stimulates gross strand transfer activity of HIV-1, BIV and EIAV INs. Reactions containing mini-HIV (lanes 1–5), mini-BIV (lanes 6–10) or mini-EIAV (lanes 11–15) DNAs were incubated in the presence of 0.6 μM (∼20 μg/ml) non-tagged recombinant HIV-1 (lanes 2–5), BIV (lanes 7–10) or EIAV (lanes 12–15) IN. LEDGF was added to the reactions in lanes 3–5, 8–10 and 13–15 at indicated concentrations. The proteins were omitted from the mock reactions in lanes 1, 6 and 11.
Fidelity of LEDGF-dependent strand transfer in vitro varies among INs
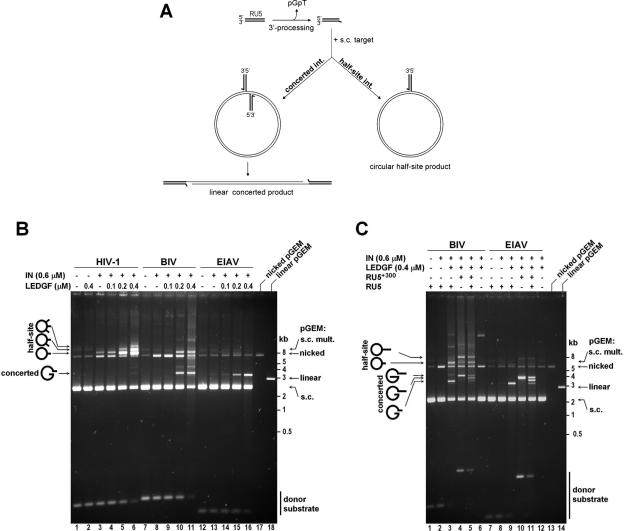
Although INs mediate concerted integration of both ends of the retroviral cDNA in vivo, their pure recombinant forms are prone to uncoupled single-LTR (termed half-site) strand transfer. IN assays utilizing long mini-viral DNA substrates proved useful to visualize gross strand transfer activity (18,51). However, the complexity of the reaction products hampers their finer analyses. The use of shorter donor substrates in the presence of excess supercoiled target DNA greatly reduce complexity of the reaction products, the major being circular half-site, resulting from integration of a single-LTR into one strand of the target plasmid, and/or linear concerted, from integration of pairs of LTRs into opposing strands of the target DNA (Figure 3A) (56,60). To compare fidelity of LEDGF-dependent strand transfer by HIV-1, BIV and EIAV INs (herein, fidelity refers to relative efficiency of concerted versus half-site strand transfer), the enzymes were incubated with blunt-ended donor DNA molecules containing respective viral RU5 sequences in the presence of supercoiled plasmid DNA (pGEM), and the reaction products were separated in 1.5% agarose gels. Although the reaction products were clearly IN-specific, LEDGF stimulated strand transfer activities of all three enzymes, in a concentration-dependent manner (Figure 3B). In the presence of the host factor, HIV-1 IN produced a distinct ladder of products that migrated above the nicked form of the target DNA (Figure 3B, compare lanes 1, 6 and 17), as expected for products of single and multiple half-site integration events.
Figure 3.
Strand transfer activities of HIV-1, BIV and EIAV INs in the presence of LEDGF. (A) Schematic of in vitro integration with circular DNA target and the expected linear concerted and circular half site strand transfer products. Coordinated insertion of a pair of substrate DNA molecules into opposing strands of target DNA results in a linear concerted integration product (left). Uncoupled insertion of a single substrate molecule into one strand of the target plasmid gives a circular half site product (right). Thick lines are substrate and thin are target-derived DNA. (B) HIV-1 (lanes 1–6), BIV (lanes 7–11), or EIAV (lanes 12–16) RU5 substrates and supercoiled pGEM target DNA were incubated with 0.6 μM (∼20 μg/ml) recombinant HIV-1, BIV, or EIAV INs and 0–0.4 μM LEDGF, as indicated. Deproteinized reaction products were separated in 1.5% agarose gels and detected by staining with ethidium bromide. Lanes 17 and 18 contained nicked circular and linearized pGEM DNA respectively. Migration positions of DNA markers (kb), pGEM DNA forms [supercoiled multimer (s.c. mult.), nicked circular, linear, and supercoiled (s.c.)], substrate RU5 DNAs, and the reaction products (concerted, half-site, and multiple half-site) are indicated. (C) BIV (lanes 1–6) and EIAV (lanes 7–12) integration reactions were carried out with the respective RU5 or/and RU5+300 substrates. Lanes 13 and 14 contained nicked circular and linearized pGEM DNA, respectively. Migration positions of the two circular half site and three linear concerted integration products derived from RU5 and RU5+300 substrates are indicated.
Reactions containing EIAV IN and the 120 bp EIAV RU5 substrate revealed a major product band migrating as a ∼3200 bp linear DNA fragment (Figure 3B, lanes 13–16). The difference in electrophoretic mobility between this product and linearized pGEM (lane 18) was consistent with concerted integration of a pair of EIAV RU5 substrate molecules into the 2912 bp target (see Figure 3A). When EIAV IN was incubated with a 300 bp longer substrate (RU5+300, Figure 3C) the resulting product migrated as a ∼3800 bp DNA fragment, consistent with concerted integration of a pair of 420 bp substrate molecules into pGEM (Figure 3C, lane 10). Furthermore, a reaction containing both RU5 and RU5+300 EIAV substrates resulted in a triplet of bands, of which two migrated as products of reactions containing only RU5 or RU5+300 plus one at an intermediate position (Figure 3C, lane 11). Such a mixed pattern is expected only when two substrate molecules are used simultaneously in a strand transfer reaction; half-site integration would result in only two different product bands. Of note, degradation of target DNA was not observed when donor DNA substrates were omitted, ruling out contamination of the protein preparations by a nuclease activity (Figure 3C, compare lanes 7 and 12). In addition, digests of the material from the ∼3800 bp band with KpnI or StuI, which restrict within the RU5+300 sequence, further confirmed that it consists of linear DNA species flanked by pairs of donor substrate molecules (data not shown). In sharp contrast to its HIV-1 counterpart, EIAV IN produced only a minor fraction of circular half-site products, which were barely detectable following staining with ethidium bromide (Figure 3B, lanes 13–16; Figure 3C, lanes 8–11).
Under similar conditions, BIV IN was proficient in both concerted and half-site integration (Figure 3B, lanes 7–11; Figure 3C, lanes 1–6). The products migrating as a distinct band at ∼3400 bp (Figure 3B, lanes 10 and 11; Figure 3C, lane 3) resulted from concerted integration of pairs of 217 bp BIV RU5 molecules into pGEM. The identity of this band was confirmed by an ∼4000 bp product obtained in a reaction with a 517 bp BIV RU5+300 substrate (Figure 3C, lane 4) and by the characteristic triplet at ∼3400/3700/4000 bp revealed when both RU5 and RU5+300 BIV substrates were used in one reaction (Figure 3C, lane 5). In addition, digests of the 3.7 kb product with KpnI or XhoI, which restrict within RU5+300 DNA, confirmed its identity (data not shown). The circular half-site products obtained in reactions with BIV RU5 substrate migrated above the nicked form of pGEM (Figure 3B, lanes 10 and 11; Figure 3C, lane 3). A slower migrating band of circular half-site products was obtained with the longer RU5+300 BIV substrate (Figure 3C, lane 4). A reaction containing both BIV RU5 and RU5+300 substrates revealed a pair of bands migrating at the positions identical to those from reactions containing either RU5 or RU5+300 (Figure 3C, lane 5), consistent with a single-LTR strand transfer.
Further differences between activities of the lentiviral INs were apparent. Thus, HIV-1 and BIV INs effected substantial nicking of pGEM DNA in LEDGF- and donor DNA-independent manners (Figure 3B, compare lanes 3, 8 and 13; Figure 3C, lanes 6 and 12). HIV-1 IN is known to possess a low level of magnesium-dependent endonuclease activity (55). Also, since EIAV IN prepared in a similar way did not relax supercoiled pGEM (Figure 3B, lane 13; Figure 3C, lane 12), endonuclease activity observed in HIV-1 and BIV IN preparations was unlikely due to a contaminant but an intrinsic activity of these enzymes. A high-molecular weight band reproducibly observed in reaction with BIV IN when substrate DNA was omitted (Figure 3C, lane 6 and data not shown) suggested that BIV IN can utilize randomly nicked DNA to initiate strand transfer, though this product was not studied in detail. Multiple integration events (both half-site and concerted) presumably explain additional product bands and smears observed on the ethidium bromide-stained gels (Figure 3B and C).
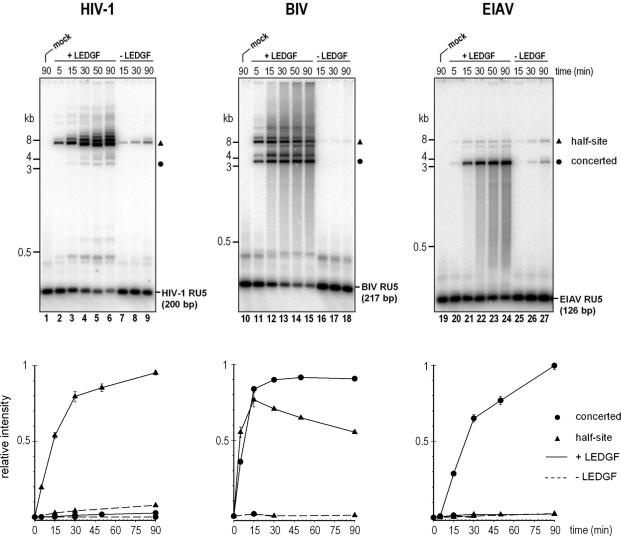
To selectively detect and quantify strand-transfer products, integration reactions were set-up with radiolabeled RU5 substrates (Figure 4). During the linear phase of the reaction, LEDGF stimulated half-site activity of HIV-1 IN ∼19.6 (±0.3) -fold. A light band migrating at ∼3.5 kb, apparently the product of concerted integration, was observed in the reactions (Figure 4, lanes 4–6, also visible in Figure 3B, lane 6). Since this activity could not be detected in the absence of the host factor, stimulation of HIV-1 concerted integration by LEDGF could not be reliably estimated. In contrast to HIV-1 IN, half-site activity of EIAV IN was not changed in the presence of LEDGF, while the concerted integration activity increased at least 60-fold (Figure 4, lanes 19–27). Both half-site and concerted integration activities of BIV IN were equally stimulated by the host factor (Figure 4, lanes 10–18). Although the circular half-site and concerted products of the LEDGF-dependent BIV reaction reached a maximum level at ∼15 min (Figure 4, lane 12), the reaction products forming secondary product bands and smears appeared to accumulate at the expense of the residual free substrate (Figure 4, lanes 13–15).
Figure 4.
Time course of LEDGF-dependent HIV-1, BIV and EIAV integration reactions with radiolabeled donor DNA substrates. HIV-1 (lanes 1–9), BIV (lanes 10–18) and EIAV (lanes 19–27) RU5 substrates were incubated with their respective INs (0.6 μM) for 5–90 min (as indicated), in the presence (lanes 2–6, 11–15 and 20–24) or absence (lanes 7–9, 16–18 and 25–27) of 0.4 μM LEDGF. Deproteinized reaction products separated in 1.5% agarose gels were detected and quantified by phosphor autoradiography. Lanes 1, 10 and 19 contained mock samples where IN and LEDGF were omitted. Migration positions of DNA standards (kb) are indicated. The plot below each gel shows accumulation of the concerted (filled circles) or half-site (triangles) integration products in the presence (discontinuous line) or absence (dashed line) of LEDGF in terms of relative band intensity. Error bars represent SDs calculated from duplicate measurements.
Sequencing of the BIV and EIAV concerted integration products
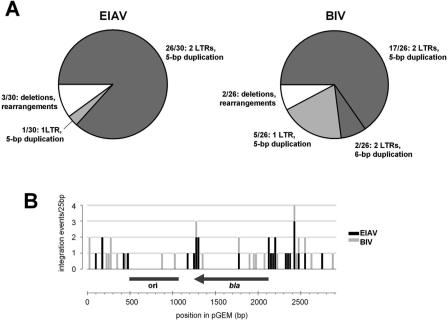
The provirus is flanked by a short duplication of chromosomal target DNA sequence (ranging from 4 to 6 bp for different retroviruses), which results from repair of short single-stranded gaps created upon integration of the retroviral cDNA termini into opposing strands (and across the major groove) of chromosomal DNA. Hence, correct duplication of target DNA sequences is a hallmark of legitimate retroviral integration. Specifically, HIV-1 and EIAV proviruses are flanked by 5 bp duplications of target DNA sequences (13,61). The 5 bp duplication can be conceivably expected for other members of Lentivirus. To further characterize the reaction products and verify target site duplications, products of concerted integration of BIV and EIAV RU5 substrates into the pGEM target isolated from agarose gels were treated with strand-displacing phi29 DNA polymerase, prior to ligation with a kanamycin-resistance cassette and transformation into E.coli cells. Plasmids isolated from randomly-picked kanamycin-resistant colonies were sequenced using primers annealing to the flanks of the cassette. Thirty clones derived from the 3.2 kb EIAV and 26 from the 3.4 kb BIV concerted linear integration products were analyzed. In all sequenced integration products, the reactive termini of BIV and EIAV RU5 substrate molecules underwent correct 3′-processing prior to strand transfer. Twenty-six (86.7%) EIAV and seventeen (65.4%) BIV product-derived plasmids had the expected structure, containing pairs of RU5 fragments inserted with 5 bp duplications of the target sequences; two (7.7%) BIV clones had 6 bp duplications (Figure 5A). Curiously, one EIAV- and five BIV- derived clones contained solo RU5 fragments while the second donor fragment was replaced by an A/T base pair, ligated to the cassette. Because these clones had 5 bp duplications of the target sequences, they were most likely derived from legitimate concerted integration. Nicking 5′ of the reactive deoxyadenosine within one of the integrated substrate molecules prior to the treatment with phi29 DNA polymerase could explain their occurrence. The remaining clones had deletions or rearrangements, possibly due to the cloning or selection procedures (Figure 5A). In total, 90% (27/30) of EIAV and 92% (24/26) of BIV integration product clones appeared to arise from legitimate concerted integration.
Figure 5.
Sequence analysis of cloned EIAV and BIV integration products. (A) Pie charts summarizing the categories of clones obtained by cloning of the gel-purified concerted linear integration products. In total, 30 EIAV- and 26 BIV- derived clones were analyzed. See the Results section for more details. (B) Distribution of the cloned concerted integration events along the pGEM target. The histogram bars represent numbers of EIAV (black) and BIV (gray) integration events within each 25 bp window along the target DNA sequence. The ruler corresponds to nucleotide coordinates within pGEM-9Zf(−) (GenBank accession no. X65312). Positions of the plasmid's replication origin (ori) and the ampicillin resistance gene (bla) are indicated.
Distribution of the cloned EIAV and BIV integration products along the target DNA did not appear entirely random; both hot and cold regions were observed (Figure 5B). Integration events into or close to the replication origin of pGEM were most likely lost due to the cloning procedure. The second avoided region, within the bla gene, was less obvious, as ampicillin was not used to select the integration product libraries. Overexpression of N-terminal fragments of β-lactamase can be toxic to E.coli cells (unpublished observations). Therefore, disruption of bla on a high-copy plasmid could lead to selective disadvantage. Alignments of local target sequences from cloned concerted integration products revealed a weak consensus for EIAV IN target sequence GTWACNNW (International Union of Biochemistry codes) (Supplementary Figure S3), which is remarkably similar to that elaborated for large datasets of HIV-1 and EIAV integration sites in vivo (6,13). BIV IN displayed less bias towards target DNA sequence. Both EIAV and BIV INs seemed to disfavor integration immediately next to a deoxythimidine; a similar observation can be made for lentiviral integration in vivo (6,13).
DISCUSSION
Earlier reports suggested that the interaction with LEDGF is a conserved and unique feature of lentiviral INs (18–20,34,37). However, Betaretrovirus and Spumavirus INs were not included in the previous studies. In addition, HIV-1 IN remained a single lentiviral IN whose enzymatic activity was examined in the presence of the co-factor (18,24,33). The present work was designed to complement and extend the prior observations, and included novel INs from Betaretrovirus (MPMV), Gammaretrovirus (FeLV), Deltaretrovirus (HTLV-1) and Spumavirus (PFV) alongside with several Lentivirus INs. The complementary approaches of in vitro His6-tag pull-down and yeast two-hybrid assays failed to detect interaction between LEDGF and any of the non-lentiviral INs from the current study. In contrast, every lentiviral IN tested here displayed affinity for the host factor, including novel EIAV, BIV and MVV INs, as well as previously studied HIV-1, HIV-2 and FIV INs (Figure 1 and Supplementary Figure S1) (18–20,34,37). These results fully confirm that the interaction with LEDGF is limited to and conserved within Lentivirus.
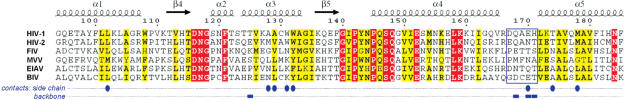
In the crystal structure of the HIV-1 IN CCD–IBD complex, the side chain of LEDGF residue Asp-366 is involved in a bidentate hydrogen bond with the main chain amides of Glu-170 and His-171 of IN (32). These contacts are essential, since D366A and D366N mutations in LEDGF ablated the interaction with HIV-1 IN (31). In agreement with previous observations (31,54), D366N LEDGF mutant failed to interact with HIV-1 IN in both pull-down and yeast two-hybrid assays. Moreover, the point mutation also ablated the interaction with all other lentiviral INs studied here (Figure 1A and B, Supplementary Figure S1), strongly arguing that the divergent INs share the mechanism of the host factor recognition. Intriguingly, while LEDGF proteins from the relevant host species are highly conserved [Supplementary Figure S2, see also (24,30)], lentiviral INs display significant sequence variations. Thus, INs from non-primate lentiviruses share only 29–37% identical residues with the HIV-1 protein; MVV IN included in this study appears to be most divergent. As revealed by the crystal structure of the HIV-1 IN CCD–IBD complex, the host factor forms several key contacts with main chain atoms of IN, and the interaction is highly sensitive to the polypeptide backbone conformation at the IN CCD dimer interface (32). Conceivably, structural conservation of lentiviral INs served to maintain the interaction within the genus. Yet, it appears that each lentivirus has evolved a distinct network of additional contacts. Strikingly, the majority of the HIV-1 IN residues that contribute side chains to the interaction are not conserved among lentiviral INs (Figure 6). The most notable differences affect positions corresponding to the HIV-1 residues Ala-128, Ala-129, Trp-131, Trp-132 and Gln-168 (Figure 6). Methyl groups of HIV-1 IN Ala-128 and Ala-129 participate in a hydrophobic pocket that buries the side chain of the LEDGF hotspot residue Ile-365 (31,32). Substitution of Ala-128 or Ala-129 for a bulkier Gln residue significantly affected apparent affinity for LEDGF (54). Nevertheless, lentiviral INs are able to accommodate residues with larger side chains at the equivalent positions, maintaining the interaction with LEDGF (Figure 6). The side chain of HIV-1 IN Trp-131 is involved in hydrophobic contacts with Phe-406 and Val-408 in LEDGF. The bulky hydrophobic Trp residue, critical to the high-affinity interaction (54), is substituted with small polar Asn in FIV IN (Figure 6). The residues 167–171 comprising a connector between α4 and α5 helices in HIV-1 IN CCD are directly involved in the interaction with LEDGF. The hydrogen bond between the side chains of Gln-168 and Trp-132 in HIV-1 IN is thought to stabilize the conformation of the connector. Although, the interaction is highly sensitive to mutations at the positions 132 and 168 in HIV-1 IN (20,54), neither residue is conserved in non-primate lentiviral INs (Figure 6). A structure of a non-primate lentiviral IN in complex with LEDGF IBD would elucidate the adaptive changes evolved to preserve the interaction. Maintenance of the interaction despite the significant differences between the INs at the sequence level, suggests that engaging LEDGF provided a strong selective advantage and played a vital role in the evolution of Lentivirus.
Figure 6.
Partial sequence alignment of lentiviral INs. Secondary structure elements from 2B4J (32) are indicated above the alignment. Residues comprising the α4/5 connector (167–171 in HIV-1 IN) (32) are boxed. Residues invariant among all six proteins are white on red background, and those with conserved biochemical properties are bold on yellow background. Blue circles and boxes under the alignment indicate residues that in HIV-1 CCD-IBD structure directly participate in the interaction with LEDGF through side chain or backbone atoms, respectively (32). The numbering corresponds to the HIV-1 sequence. The figure was generated using ESPript-2.2 (64), (http://espript.ibcp.fr).
LEDGF robustly stimulated in vitro strand transfer activity of the divergent HIV-1, BIV and EIAV INs. Importantly, in the presence of the co-factor the strand transfer occurred at the physiological salt concentration and did not require molecular crowding agents or dimethyl sulfoxide typically added to observe magnesium-dependent activity of retroviral INs (55–59). Of note, the co-solvent can alleviate dependence of other nucleotidyl transferases from protein co-factors. For instance, Mu phage transposition critically depends on a network of protein–protein and protein–DNA interactions (62). However, all elements of its intricate regulatory circuit, including the host cell factor HU, become dispensable in the presence of dimethyl sulfoxide (63). Stimulation of the divergent lentiviral INs strongly suggests that the host factor is intimately involved in regulation of lentiviral DNA integration.
Surprisingly, the fidelity of the LEDGF-dependent strand transfer varied broadly: the host factor selectively stimulated concerted integration activity of EIAV IN, triggered almost exclusive half-site strand transfer by HIV-1 IN, and unleashed both activities in the BIV enzyme. Presumably, both viral cDNA termini in vivo and a pair of donor molecules in vitro must be brought together prior to concerted strand transfer. Protein–protein interactions involving IN subunits and possibly co-factors associated with the cDNA ends would mediate such synapsis. The fact that the in vitro LEDGF-dependent activities of HIV-1 and BIV INs fall short of reproducing legitimate integration might indicate a requirement for an additional host cell- or virus- derived co-factor. Conceivably, the minimal in vitro requirements for proper assembly of the lentiviral synaptic complex depend on stability of individual interfaces between the elements of the integration machinery. Thus, a tighter interaction between substrate-bound EIAV IN protomers could make the hypothetic co-factor dispensable for concerted integration in vitro. Further analyses will be necessary to explain the differences between in vitro activities of the lentiviral IVs and the mechanism of stimulation of lentiviral INs by the host factor. Intriguingly, LEDGF can both enhance and ablate concerted integration activity of HIV-1 IN in vitro, depending on the solution conditions and IN/LEDGF input ratio (K. Pandey and D. Grandgenett, personal communication). The available data collectively suggest that LEDGF can be a key regulator of lentiviral integration.
The current generations of retroviral vectors rely on the wild type viral integration machinery that evolved for maximal viral fitness. The safety of gene therapy could be drastically improved if an option of directed integration could be added to these vectors. The strong bias towards integration into transcription units appears to be a unique feature of Lentivirus (8–17), mirroring the genus-specific interaction with LEDGF. HIV-1 integration into transcription units was diminished in cells partially depleted for LEDGF, indicating that the host factor at least contributed to the integration target site selection (40). Alongside with recent work from several laboratories (42–45), the data presented here indicate that LEDGF is hardwired into the mechanism of lentiviral DNA integration. These developments provide a stronger foundation for further research into exploitation of the host factor for directed retroviral gene delivery and as a potential target for antiretroviral therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
I am grateful to Dr. Ó. Andrésson (University of Iceland) for a generous gift of KV1772 DNA (46), Dr C. Bangham (Imperial College London) for HTLV-1 DNA, Drs M. McClure and G. Patton (Imperial College London) for pcHSRV2 DNA and MPMV, Dr G. Towers (University College London) for NBL-6 cells and S. Dustan (Imperial College London) for help with DNA sequence analysis. Molecular clones pBIV127 (47), pSPeiav19 (48) and pEECC-FeLV (49) were obtained through NIH AIDS Research and Reference Reagent Program (Germantown, MD). I thank Drs A. Engelman, N. Vandegraaff, D. Grandgenett, and M. Pizzato for helpful discussions and comments on the manuscript. This study was funded by the Faculty of Medicine of Imperial College London and the Medical Research Council (MRC). Funding to pay the Open Access publication charges for this article was provided by the MRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Buchen-Osmond C. The universal virus database ICTVDB. Comput. Sci. Eng. 2003;5:16–25. [Google Scholar]
- 2.LaFemina R.L., Schneider C.L., Robbins H.L., Callahan P.L., LeGrow K., Roth E., Schleif W.A., Emini E.A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grinsztejn B., Nguyen B.Y., Katlama C., Gatell J., Lazzarin A., Vittecoq D., Gonzalez C., Chen J., Isaacs R. The_Protocol_005_Study_Team. The 13th Conference on Retroviruses and Opportunistic Infections; February 5–8, 2006; Denver, CO. 2006. [Google Scholar]
- 4.Hazuda D.J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J.A., Espeseth A., Gabryelski L., Schleif W., Blau C., et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 5.Craigie R. Retroviral DNA integration. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 613–630. [Google Scholar]
- 6.Holman A.G., Coffin J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl Acad. Sci. USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nature Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 8.Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 9.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 10.Kang Y., Moressi C.J., Scheetz T.E., Xie L., Tran D.T., Casavant T.L., Ak P., Benham C.J., Davidson B.L., McCray P.B., Jr Integration site choice of a feline immunodeficiency virus vector. J. Virol. 2006;80:8820–8823. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crise B., Li Y., Yuan C., Morcock D.R., Whitby D., Munroe D.J., Arthur L.O., Wu X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacNeil A., Sankale J.L., Meloni S.T., Sarr A.D., Mboup S., Kanki P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 2006;80:7316–7321. doi: 10.1128/JVI.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacker C.V., Vink C.A., Wardell T.W., Lee S., Treasure P., Kingsman S.M., Mitrophanous K.A., Miskin J.E. The integration profile of EIAV-based vectors. Mol. Ther. 2006;14:536–545. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Barr S.D., Leipzig J., Shinn P., Ecker J.R., Bushman F.D. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narezkina A., Taganov K.D., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trobridge G.D., Miller D.G., Jacobs M.A., Allen J.M., Kiem H.P., Kaul R., Russell D.W. Foamy virus vector integration sites in normal human cells. Proc. Natl Acad. Sci. USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowrouzi A., Dittrich M., Klanke C., Heinkelein M., Rammling M., Dandekar T., von Kalle C., Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J. Gen. Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- 18.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 19.Turlure F., Devroe E., Silver P.A., Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 20.Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 21.Van Maele B., Busschots K., Vandekerckhove L., Christ F., Debyser Z. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Ciuffi A., Bushman F.D. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Ge H., Si Y., Roeder R.G. Isolation of cDNAs encoding novel transcription co-activators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherepanov P., Devroe E., Silver P.A., Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 25.Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa Y., Usukura J., Singh D.P., Chylack L.T., Jr, Shinohara T. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (LEDGF/p75) and p52, in the nucleus. Cell Tissue Res. 2001;305:107–114. doi: 10.1007/s004410100398. [DOI] [PubMed] [Google Scholar]
- 27.Ge H., Si Y., Wolffe A.P. A novel transcriptional co-activator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell. 1998;2:751–759. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- 28.Fatma N., Singh D.P., Shinohara T., Chylack L.T., Jr Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J. Biol. Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 29.Maertens G.N., Cherepanov P., Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 30.Vanegas M., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 31.Cherepanov P., Sun Z.Y., Rahman S., Maertens G., Wagner G., Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nature Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 32.Cherepanov P., Ambrosio A.L., Rahman S., Ellenberger T., Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turlure F., Maertens G., Rahman S., Cherepanov P., Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1663–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busschots K., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 35.Cherepanov P., Pluymers W., Claeys A., Proost P., De Clercq E., Debyser Z. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000;14:1389–1399. doi: 10.1096/fj.14.10.1389. [DOI] [PubMed] [Google Scholar]
- 36.Devroe E., Engelman A., Silver P.A. Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 2003;116:4401–4408. doi: 10.1242/jcs.00747. [DOI] [PubMed] [Google Scholar]
- 37.Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E.M. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 39.Vandegraaff N., Devroe E., Turlure F., Silver P.A., Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 41.Ciuffi A., Diamond T.L., Hwang Y., Marshall H.M., Bushman F.D. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 42.Vandekerckhove L., Christ F., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z. Transient and stable knock-down of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zielske S.P., Stevenson M. Modest but reproducible inhibition of human immunodeficiency virus type 1 infection in macrophages following LEDGF/p75 silencing. J. Virol. 2006;80:7275–7280. doi: 10.1128/JVI.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelman A., Vandegraaff N., Daigle J., Cherepanov P. The American Society for Cell Biology 2006 Summer Meeting on. The Cell Biology of HIV-1 and Other Retroviruses; July 20–23, 2006; Atlanta, GA: Emory University; 2006. [Google Scholar]
- 45.Llano M., Saenz D.T., Meehan A., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M. An essential role for LEDGF/p75 in HIV Integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 46.Andresson O.S., Elser J.E., Tobin G.J., Greenwood J.D., Gonda M.A., Georgsson G., Andresdottir V., Benediktsdottir E., Carlsdottir H.M., Mantyla E.O. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology. 1993;193:89–105. doi: 10.1006/viro.1993.1106. [DOI] [PubMed] [Google Scholar]
- 47.Braun M.J., Lahn S., Boyd A.L., Kost T.A., Nagashima K., Gonda M.A. Molecular cloning of biologically active proviruses of bovine immunodeficiency-like virus. Virology. 1988;167:515–523. [PubMed] [Google Scholar]
- 48.Cunningham T.P., Montelaro R.C., Rushlow K.E. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene. 1993;124:93–98. doi: 10.1016/0378-1119(93)90766-v. [DOI] [PubMed] [Google Scholar]
- 49.Overbaugh J., Donahue P.R., Quackenbush S.L., Hoover E.A., Mullins J.I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 50.Moebes A., Enssle J., Bieniasz P.D., Heinkelein M., Lindemann D., Bock M., McClure M.O., Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cherepanov P., Surratt D., Toelen J., Pluymers W., Griffith J., De Clercq E., Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 53.Shibata T., Cunningham R.P., Radding C.M. Homologous pairing in genetic recombination. Purification and characterization of Escherichia coli recA protein. J. Biol. Chem. 1981;256:7557–7564. [PubMed] [Google Scholar]
- 54.Rahman S., Lu R., Vandegraaff N., Cherepanov P., Engelman A. Structure-based mutagenesis of the integrase-LEDGF/p75 interface uncouples a strict correlation between in vitro protein binding and HIV-1 fitness. Virology. doi: 10.1016/j.virol.2006.08.011. doi:10.1016/j.virol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Engelman A., Craigie R. Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. J. Virol. 1995;69:5908–5911. doi: 10.1128/jvi.69.9.5908-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S., Grandgenett D.P. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M., Mizuuchi M., Burke T.R., Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hindmarsh P., Ridky T., Reeves R., Andrake M., Skalka A.M., Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgan A.L., Katzman M. Subterminal viral DNA nucleotides as specific recognition signals for human immunodeficiency virus type 1 and visna virus integrases under magnesium-dependent conditions. J. Gen. Virol. 2000;81:839–849. doi: 10.1099/0022-1317-81-3-839. [DOI] [PubMed] [Google Scholar]
- 60.Li M., Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellison V., Abrams H., Roe T., Lifson J., Brown P. Human immunodeficiency virus integration in a cell-free system. J. Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu. Rev. Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 63.Mizuuchi M., Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell. 1989;58:399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- 64.Gouet P., Robert X., Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.