Abstract
Nonspecific endonucleases hydrolyze DNA without sequence specificity but with sequence preference, however the structural basis for cleavage preference remains elusive. We show here that the nonspecific endonuclease ColE7 cleaves DNA with a preference for making nicks after (at 3′O-side) thymine bases but the periplasmic nuclease Vvn cleaves DNA more evenly with little sequence preference. The crystal structure of the ‘preferred complex’ of the nuclease domain of ColE7 bound to an 18 bp DNA with a thymine before the scissile phosphate had a more distorted DNA phosphate backbone than the backbones in the non-preferred complexes, so that the scissile phosphate was compositionally closer to the endonuclease active site resulting in more efficient DNA cleavage. On the other hand, in the crystal structure of Vvn in complex with a 16 bp DNA, the DNA phosphate backbone was similar and not distorted in comparison with that of a previously reported complex of Vvn with a different DNA sequence. Taken together these results suggest a general structural basis for the sequence-dependent DNA cleavage catalyzed by nonspecific endonucleases, indicating that nonspecific nucleases could induce DNA to deform to distinctive levels depending on the local sequence leading to different cleavage rates along the DNA chain.
INTRODUCTION
Nonspecific endonucleases are a group of enzymes capable of degrading DNA and/or RNA molecules without sequence specificity. They participate in many cellular processes, including DNA digestion in nucleotide salvage pathways (1), DNA fragmentation during cell apoptosis (2), nucleotide deletion in DNA repair (3), mRNA turnover in transcription regulation (4) and mRNA degradation in gene silencing (5,6). In contrast to site-specific endonucleases, such as restriction enzymes which cleave DNA only at specificsites, nonspecific endonucleases cleave phosphodiester linkages at many positions along the DNA/RNA chain but cleavage efficiencies vary between different sites. Although the sequence-dependent cleavage preference of nonspecific endonucleases has been noticed for decades, the molecular basis for the selectivity of preferred cleavage sites remains elusive.
Early studies showed that DNase I cleaved the P-O3′ bond of a double-stranded DNA with a preference for cleavage at the 5′-side of T residues in alternating poly(AT) sequences and it was suspected that the varied twist angles between base pairs contributed to its different cleavage rates (7,8). DNA footprint assays further identified a global component in DNase I digestion patterns that the two strands of a duplex DNA were cleaved at a correlated rate even though DNase I cleaved one strand at a time (9). DNA groove width and flexibility were thus suggested to account for the global variation that DNA with narrower grooves and stiffer conformation was a poorer substrate for DNase I. However, in addition to the global component, a local unknown variation was also suggested to be an important determinant in cleavage since DNA groove width and helix flexibility are slowly changing parameters which cannot explain the drastic difference in cleavage rates by DNase I of neighboring nucleotides. The suggestion of a global determinant for cleavage preference was latter supported by the crystal structure of DNase I in complex with a cleaved DNA which showed an exposed loop binds at the widened minor groove in the bent DNA, suggesting that a more flexible DNA with an easily widened minor groove is likely to be more accessible for cleavage (10,11).
Further crystal structure analysis of DNase I in complex with an uncleaved octamer DNA seemed to support the hypothesis that the high twist angles between the alternating pyrimidine–purine sequences may lead to the pronounced preference of DNase I for cutting 5′ to pyrimidine residues in alternating pyrimidine–purine sequences (12). More comprehensive sequence analysis of the DNase I cleavage sites showed a preferred sequence of RRATY-Y (13) and AYA-AVN (14) (-, cleavage site; A, A or T; R, purine; Y, pyrimidine; V, not-T; N, any bases). Several other nonspecific endonucleases show different sequence preferences: DNase II avoids pyrimidine–purine steps (9); Serratia nuclease and NucA prefer GC-rich regions and avoid cleavage of poly(A) and poly(T) tracts in dsDNA (15,16); the nuclease domain of ColE9 cleaves tracts of GC poorly and has a preference for cleaving at the 3′O-side of thymine of the tyrT promoter (17); the apoptotic endonuclease DFF40/CAD prefers purine/pyrimidine blocks with rotational symmetry whereas EndoG preferentially attacks DNA 5′ of G residues (2,18); human topoisomerase I prefers to cleave between AT-N and TT-N steps (19). In summary, it is generally believed that nonspecific endonucleases are sensitive to global features of DNA, such as the width of minor groove (9) and stiffness of DNA (20). In addition, local sequence-dependent interactions between nucleases and DNA may contribute a second-level determinant for cleavage preference (14,15). However it has remained unclear what kind of structural feature underlying in DNA is recognized by these enzymes.
We used two endonucleases ColE7 and Vvn to re-visit the unresolved question, of what are the local structural variations in DNA that are recognized by nonspecific endonucleases and thus to determine DNA cleavage preference. ColE7 is an Escherichia coli released toxin containing an endonuclease domain, capable of digesting DNA and RNA in sensitive bacteria cells to induce cell death (21,22). Vvn is a Vibrio vulnificus endonuclease which digests nucleic acid molecules in the periplasm to prevent the cell from uptake foreign DNA during transformation (23,24). Both nucleases cleave the P-O3′ bond nonspecifically in single-stranded and double-stranded DNA and RNA molecules (25). The endonuclease domain of ColE7 (referred to N-ColE7 hereafter) contains an H-N-H motif which has been identified in hundreds of homing endonucleases, restriction and DNA repair enzymes (26–29). Crystal structures of N-ColE7 in free form (30), in complex with its protein inhibitor Im7 (31), in complex with an 8 bp DNA without metal ions (22) and in complex with a 12 bp DNA with a metal ion (32) have been reported previously. These structures elucidate the enzyme inhibition and activation mechanisms and show that the H-N-H motif, folded in a ββα-metal finger structure (33–35), binds DNA at the minor groove. Vvn shares no sequence identity to ColE7, but it also contains an endonuclease active site folded in a similar ββα-metal finger structure. The crystal structure of Vvn in complex with an 8 bp DNA shows that the metal finger motif is bound to the minor groove of DNA and that Vvn interacts with DNA primarily at the phosphate backbone (24). Since the sequence cleavage preferences of ColE7 and Vvn were not known before this study, it was intriguing to find out whether these two enzymes with structural but no sequence homology at their respective active sites cleave DNA with a similar preference.
Here we report not only the sequence cleavage preferences of N-ColE7 and Vvn, but also the crystal structures of the mutated N-ColE7 (H545Q) and Vvn (H80A) in complex with an 18 bp and a 16 bp DNA, respectively. Based on the comparison of the cleavage preferences and enzyme–DNA complex structures, a structural basis is suggested to explain the sequence-dependent cleavage preference of nonspecific endonucleases. For the first time, a distorted DNA backbone, but not base pair stacking parameters, has been identified to be the general local determinant recognized by nonspecific endonucleases for the more efficient phosphodiester bond cleavage.
MATERIALS AND METHODS
Protein expression and purification
The nuclease-ColE7 mutant H545Q and Im7 proteins were expressed using the vector pQE70 (Qiagen) in E.coli M15 as described earlier (32). The protein complex was first purified by Ni-NTA affinity column (Qiagen, Germany) since the constructed Im7 contained a C-terminal six-histidine affinity tag. The complex was then separated by lowering the pH from 8 to 3 and the N-ColE7 was further purified by chromatographic methods using a Sepharose-SP (Pharmacia) and a heparin column (HiTrap SP; Pharmacia). The free N-ColE7 H545Q mutant containing residues 444–576 was then dialyzed three times in solutions containing 10% glycerol, 1–2 mM ZnCl2 and 20–50 mM Tris–HCl (pH 7.5). The molecular mass as measured by mass spectroscopy for mutant H545Q was 15 724 Da (calculated mass: 15 721.0).
DNA footprinting
The plasmid pQE30 was isolated from a culture of E.coli and used as a template for the PCR amplification of the 260 bp DNA fragments. Two pairs of 5′-32P-labeled forward and reverse primers were used to amplify the DNA fragments from the multiple cloning site region in pQE30. The PCR products were then purified using 0.1% agarose gel and a Gel-M™ Gel Extraction Kit (Viogene). The cleavage reaction was carried out by mixing 0.7 μM 260 bp DNA with 4 μM N-ColE7 (or 600 nM Vvn) in a buffer containing 20 mM Tris–HCl (pH 8) and 2 mM Mg2+ at 37°C for 5 min. The reactions were terminated by adding 10 mM EDTA, followed by addition of loading dye (10 mM NaOH, 95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol). The digested DNA fragments were heated to 70°C for 5 min and separated in 8% denaturing polyacrylamide gels with the sequencing reactions carried out alongside. The autoradiographs were visualized and the band intensities were measured on a phosphorimager (AlphaImage 2200).
N-ColE7-DNA crystallization, data collection and structural determination
The protein–DNA complex was prepared by mixing the zinc-containing N-ColE7 mutant H545Q (∼8 mg/ml) with an equal molar of double-stranded DNA having a sequence of 5′-GGAATTCGATCGAATTCC-3′. Complex crystals suitable for X-ray analysis were obtained by the hanging drop vapor diffusion method by mixing 1 μl complex solution and 1 μl reservoir solution consisting of 40% MPD, 0.4 M ammonium formate and 0.1 M acetate buffer (pH4.8) at room temperature. The N-ColE7–DNA complex crystallized in a tetragonal P41212 unit cell with one protein–DNA complex per asymmetric unit. X-ray diffraction data were collected at the NW12 beamline of the Photon Factory, KEK (Tsukuba, Japan) using a Quantum 210 CCD detector (ADSC, CA, USA) at −160°C.
The structure of the N-ColE7–DNA(18 bp) complex was solved by molecular replacement using the structure of N-ColE7 in complex with 8 bp DNA (PDB entry: 1PT3) as the searching model by CNS (36). The final complex model contained one N-ColE7 molecule, one duplex DNA and 95 water molecules. All the diffraction and refinement statistics are listed in Table 1. Atomic coordinates have been deposited in the PDB with accession codes of 2IVH for the N-ColE7–DNA(18 bp) complex.
Table 1.
Diffraction and refinement statistics for N-ColE7-DNA(18 bp) and Vvn–DNA(16 bp) complexes
| N-ColE7–DNA (18 bp) | Vvn–DNA(16 bp) | |
|---|---|---|
| Data collection and processing | ||
| Space group | P41212 | P1 |
| Cell dimensions (Å) | a = 106.8, c = 60.2 | a = 64.6, b = 64.7, c = 79.2 |
| Cell dimensions (deg) | α = 74.5, β = 73.6, γ = 75.6 | |
| Resolution (Å) | 2.8 | 2.9 |
| Observed reflections | 97 181 | 95 233 |
| Unique reflections | 9047 | 25 598 |
| Completeness—all data (%) | 99.6 (50.0–2.8 Å) | 98.5 (40.0–2.9 Å) |
| Completeness—last shell (%) | 99.9 (2.9–2.8 Å) | 97.2 (3.0–2.9 Å) |
| Rsym a—all data (%) | 7.9 | 7.5 |
| Rsym a—last shell (%) | 35.6 | 29.9 |
| I/σ(I), all data | 31.7 (50.0–2.8 Å) | 17.5 (40.0–2.9 Å) |
| I/σ(I), last shell | 4.0 (2.9–2.8 Å) | 3.3 (3.0–2.9 Å) |
| Refinement | ||
| Resolution range (Å) | 50.0–2.8 | 50.0–2.9 |
| Reflections (work/test) | 7835/919 | 22 675/1922 |
| R-factor/R-free (%)b | 19.4/26.3 | 23.6/28.6 |
| Non-hydrogen atoms | ||
| Protein | 1006 | 6902 |
| DNA | 731 | 1868 |
| Ion | 1 | 0 |
| Solvent molecules | 95 | 113 |
| Model quality | ||
| r.m.s.d. values in | ||
| bond lengths (Å) | 0.0064 | 0.0091 |
| bond angles (deg) | 1.04 | 1.49 |
| Average B-factor (Å2) | 46.1 | 58.8 |
| Protein atoms | 35.9 | 54.2 |
| DNA atoms | 55.5 | 73.7 |
| Solvent atoms | 39.4 | 37.2 |
| Zn | 18.5 | |
aRsym = ΣhklΣi|Ii(hkl) − <I(hkl)>|/Ii(hkl).
bR-factor = Σhkl||Fo(hkl)| − |Fc(hkl)||/Σhkl Fo(h).
Vvn–DNA crystallization, data collection and structural determination
The Vvn mutant H80A was expressed and purified as described earlier (24). A protein–DNA solution (8.5 mg/ml) was prepared by mixing the 16 bp palindromic DNA (5′-GAATTCGATCGAATTC-3′) with Vvn–H80A in a one-to-one molar ratio and the complex crystals were grown by the hanging drop vapor diffusion method. The protein–DNA solution (1 μl) was mixed with reservoir solution (1 μl) containing 20% PEG3350 and 0.2 M di-sodium tartrate dehydrate and the crystals of Vvn-H80A–DNA were grown to a suitable size after one month at room temperature. Crystals were transferred into cryo-protecting buffer (reservoir supplemented with 10% PEG8000 and 20% glycerol) and flash-cooled for data collection.
The Vvn/DNA complex crystallized in a triclinic P1 unit cell with four Vvn and three DNA per cell. Diffraction data were collected at the NW12 beamline of the Photon Factory, KEK (Tsukuba, Japan) and processed by HKL2000. The structure was solved by molecular replacement using the Vvn molecule (PDB entry: 1OUP) as the searching model by the MolRep program in CCP4 suite. The final structural model contained four Vvn, three duplex DNA and 113 water molecules. The diffraction and final refinement statistics are listed in Table 1. Atomic coordinates for the Vvn–DNA(16 bp) complex have an accession code of 2IVK in the PDB.
RESULTS
Sequence-dependent DNA cleavage by ColE7 and Vvn
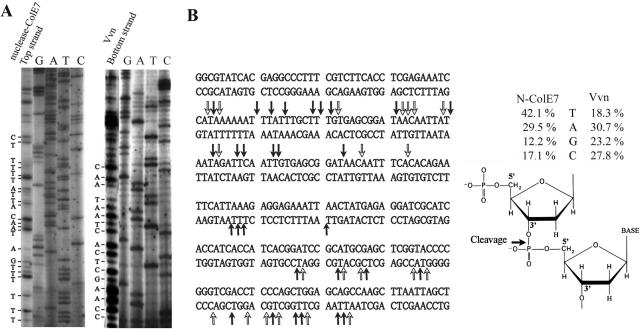
To find out the sequence cleavage preference of ColE7, 5′-32P-labeled 260 bp DNA fragments (0.7 μM) were PCR-amplified from the multiple cloning site region of pQE30 plasmids. The Zn2+-bound nuclease domain of ColE7 and Vvn were incubated, respectively, with the 260 bp DNA in the presence of 2 mM Mg2+, pH 8, and the resultant digested DNA fragments were resolved by gel electrophoresis (Figure 1). N-ColE7 cleaved double-stranded DNA with a preference at the 3′O-side (after) of T and A residues with an order of T (41.2%) > A (29.5%) > C (17.1%) > G (12.2%). The cleavage percentages were calculated by first integrating each cleavage band in the autoradiograph by a phosphorimager and then the band intensities were normalized by calculating the summation of Ibase/Nbase divided by the summation of Iall, in which Ibase is the band intensity cleaved at a specific base (A, T, C or G), Iall is the band intensity cleaved at all bases and Nbase is the number of a specific base in the DNA. A preference for cleavage after thymine for ColE7 is a similar feature to that of the homologous ColE9 which cleaves tyrT promoter sequence with >50% sites after T (17).
Figure 1.
Sequence-dependent DNA cleavage by ColE7 and Vvn. (A) The 5′-32P-labeled PCR-amplified 260 bp DNA fragments (0.7 μM) were incubated respectively with 4 μM Zn2+-bound N-ColE7 and 30 nM Vvn in the presence of 2 mM Mg2+ and 20 mM Tris–HCl buffer (pH 8) at 37°C for 5 min. The reaction was terminated by adding 10 mM EDTA. The cleaved products were separated by gel electrophoresis in 8% denaturing polyacrylamide gel with the sequencing reactions carried out alongside. Only one representative DNA digestion patterns by N-ColE7 (top strand) and Vvn (bottom strand) are shown here. (B) The sequences of top strand and bottom strand DNA used for footprint assays are listed. The DNA cleavage sites by N-ColE7 are indicated by arrows. The solid arrows show the cleavage of the 3′O-P bond with a thymidine at the 3′O-side of the scissile phosphate. A schematic diagram shows DNA cleavage by N-ColE7 with the preference for the base at the 3′O-side of the scissile phosphate: T (41.2%) > A (29.5%) > C (17.1%) > G (12.2%). Vvn is less specific in DNA cleavage with an order of A (30.7%) > G (23.2%) > C (27.8%) > T (18.3%). The cleavage percentage was estimated by first integrating each cleavage band in the autoradiography and normalized by calculating the summation of Ibase/Nbase divided by the summation of Iall, in which Ibase is the band intensity cleaved at a specific base (A, T, C or G), Iall is the band intensity cleaved at all bases and Nbase is the number of a specific base in the DNA.
In contrast to ColE7 which cleaved at about one-third of the sites, Vvn cleaved DNA at almost every site. Therefore Vvn had little sequence preference for DNA cleavage with an order of A (30.7%) > C (27.8%) > G (23.2%) > T (18.3%). Different metal ion cofactors (Mn2+ and Ca2+) were used with Vvn in similar cleavage experiments (data not shown), but the cleavage patterns were almost identical to those obtained from Mg2+, indicating that metal ion cofactors did not play a determinant role in sequence-dependent cleavage by Vvn.
Crystal structure of H545Q N-ColE7 bound with an 18 bp DNA
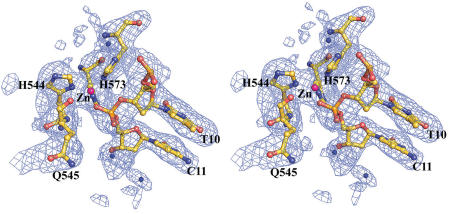
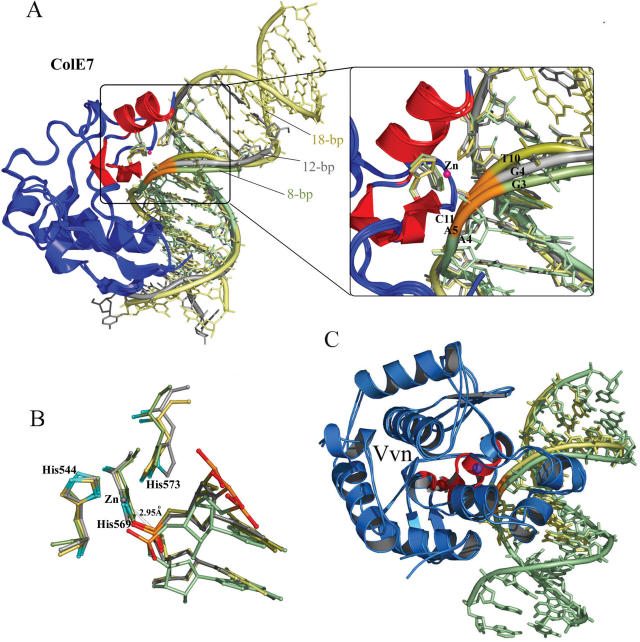
Previous structural studies had revealed two N-ColE7–DNA complex structures, both of which contained a guanine immediately before the cleavage site (GCG-ATCGC and CGGG-ATATCCCG, hyphen indicates the cleavage site) (22,32). Since these sites were the least preferred cleavage sites for N-ColE7, it was therefore intriguing to find out what would happen if N-ColE7 bound to its preferred sequences, with an A or T before the cleavage site. Hence, we carried out co-crystallization experiments mixing inactive N-ColE7 mutants, H545Q or H545E, with different lengths and sequences of DNA in the presence of metal ion cofactors. His545 functions as the general base, activating a water molecule to attack the scissile phosphate in DNA hydrolysis (32). H545Q contained a little residual endonuclease activity whereas H545E, which had been co-crystallized previously with a 12 bp DNA in the presence of Zn2+ (32), did not possess any detectable endonuclease activity. The zinc-containing N-ColE7 mutant H545Q was then successfully co-crystallized with an 18 bp DNA in a tetragonal P41212 unit cell, with one protein and one duplex DNA per asymmetric unit. The structure was solved by molecular replacement, using the N-ColE7–DNA(8 bp) structure (PDB entry: 1PT3) as the searching model. The final model contained one protein molecule (residues 449–550 and 555–576), a duplex DNA and 95 water molecules with R-factor/R-free of 19.4/26.3% for 7835/919 reflections at a resolution of 2.8 Å. The omit difference maps at the enzyme active site revealed that the scissile phosphate to be directly coordinated to Zn2+ (Figure 2).
Figure 2.
Stereo view of the electron density map of N-ColE7–DNA (18 bp) complex at active site. The omit map (Fo − Fc) contoured at 2.5 σ was calculated after omitting the active site atoms in the refinement model. The electron density of the mutated residue H545→Q and zinc ion can be clearly visualized. The scissile phosphate between T10 and C11 is coordinated to the zinc ion.
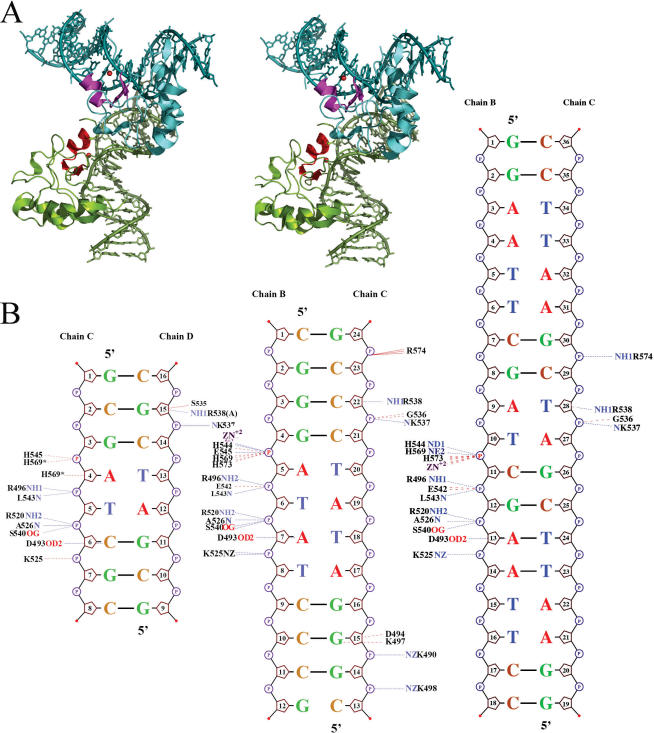
The DNA sequence in the N-ColE7–DNA(18 bp) complex structure was 5′-GGAATTCGAT-CGAATTCC-3′ (hyphen indicates the cleavage site) with the scissile phosphate located between T10 and C11, so this was a preferred cleavage site for N-ColE7 (referred to ‘preferred complex’ hereafter). The overall structure of the preferred complex is shown in Figure 3. The 18 bp DNA was bound with one protein molecule and it did not form a continuous duplex in the crystal. The H-N-H metal finger motif was bound to the minor groove of DNA, inducing DNA to bend ∼54° [calculated by Curve (37)] away from the enzyme. Smaller angles of bend have been observed in the non-preferred complexes, with an average of 9° in N-ColE7–DNA(8 bp) (22) and 19° in N-ColE7–DNA(12 bp) (32) complexes. However, the larger bending angle observed here for the 18 bp DNA was likely due to the longer DNA being used for the bending angle calculation. The 18 bp DNA in the complex bent in a similar way to the DNA in the 8 and 12 bp complexes (Discussion). The detailed interactions between N-ColE7 and DNA in the preferred and non-preferred complexes are schematically shown in Figure 3B. Most of the interactions are between protein side-chains and DNA phosphate backbones. The protein–DNA interaction patterns in the preferred complex are almost identical to those of the non-preferred complexes. The buried solvent accessible surfaces between protein and DNA are 1590 Å2 in the preferred complex (18 bp), 1933 Å2 (12 bp) and 1460 Å2 (8 bp) in the non-preferred complexes. Therefore, the preferred complex of N-ColE7–DNA(18 bp) does not contain a larger protein–DNA interface than those of non-preferred complexes.
Figure 3.
Overall structure of N-ColE7–DNA (18 bp) complex. (A) Two N-ColE7 molecules and two 18 bp DNA are packed in a perpendicular orientation in the crystal. The metal finger motifs, displayed in red, are bound to the minor groove of DNA. (B) The schematic diagrams for the contacts between N-ColE7 and DNA in the crystal structures of non-preferred [8 bp: 1PT3 (22) and 12 bp: 1ZNS (32)] and preferred complexes (18 bp: 2IVH, this study). Hydrogen bonds and van der Waals contacts within 3.35 Å between N-ColE7 and DNA are indicated by dotted blue and red lines, respectively.
Crystal structure of Vvn bound with a 16 bp DNA
Vvn mutant H80A was co-crystallized previously with an 8 bp DNA with a sequence of GCGATC-GC (hyphen indicates cleavage site) (24). Here we further co-crystallized Vvn with a 16 bp DNA with a sequence of GAATTCGATCG-AATTC. This Vvn–DNA complex was crystallized in a triclinic P1 unit cell, containing four Vvn and three 16 bp duplex DNA molecules per asymmetric unit. The crystal structure was solved by molecular replacement using Vvn (PDB entry: 1OUP) as the searching model. The final structural model had R-factor/R-free of 23.6/28.6 for 22 675/1922 reflections at a resolution of 2.9 Å.
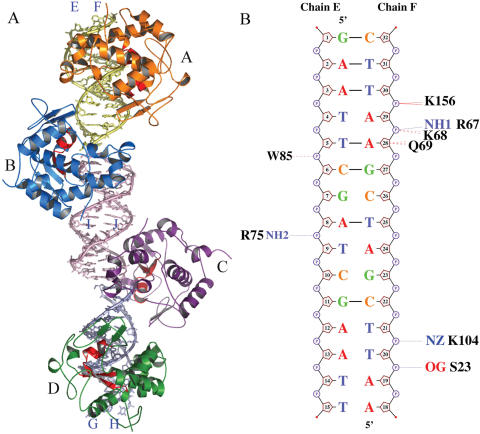
The overall structure of Vvn–DNA(16 bp) complexes of one asymmetric unit is shown in Figure 4A. Three DNA molecules were packed linearly with two Vvn (molecules A and D) bound to a single DNA molecule, respectively, and the other two Vvn (molecules B and C) bound between the junctions of two DNA molecules. The two Vvn molecules, A and D, were related to each other by a non-crystallographic 2-fold symmetry; therefore, their binding modes to DNA were similar. For clarity, only one of the Vvn–DNA complexes (molecule A) was used for further structural analysis and comparison.
Figure 4.
Overall structure of Vvn–DNA (16 bp) complex. (A) Four Vvn (A to D) and three double-stranded 16 bp DNA molecules (E-F, I-J and G-H) in one asymmetric unit in the crystal are displayed in different colors. Two Vvn (A and D) are bound to a single DNA, respectively, and the other two Vvn molecules (B and C) are bound in the junctions of two DNA duplexes. (B) The schematic diagrams for the contacts between Vvn (molecule A) and DNA (E-F). Hydrogen bonds and van der Waals contacts within 3.35 Å are indicated by dotted blue and red lines, respectively.
In the Vvn–DNA(16 bp) complex, the scissile phosphate was located between the G-A step, and therefore different from the C-G step in the 8mer complex. Vvn was bound to the minor groove of the 16 bp DNA, in a way similar to the binding mode found previously in the Vvn–DNA(8 bp) complex (24). The minor groove was also slightly widened and the DNA was bent ∼40.3° away from the enzyme. The detailed interactions between Vvn and 16 bp DNA are shown in Figure 4B, with most contacts on the DNA phosphate backbones, in a way similar to what was found previously in the 8mer complex (24). The buried protein–DNA interfaces are 1478 Å2 in Vvn–DNA(16 bp) complex and 1017 Å2 in Vvn–DNA(8 bp) complex. In summary, although the sequences of 16 and 8 bp DNA are different, the protein–DNA interaction modes in the two complexes are similar.
DISCUSSION
Structural basis for cleavage preference
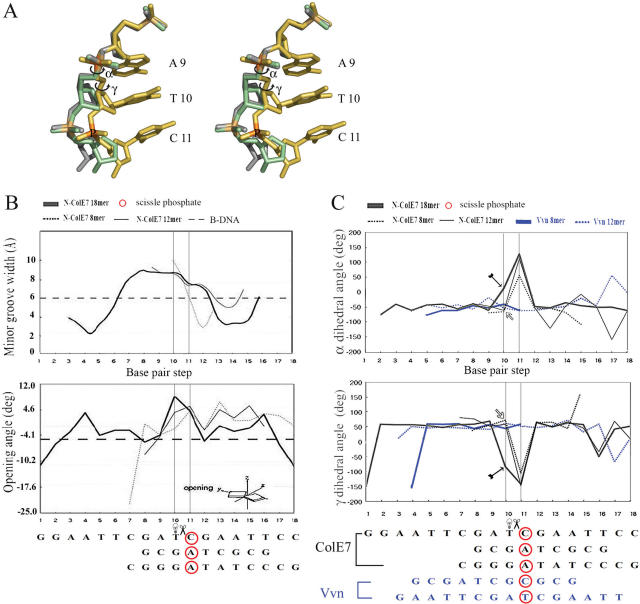
The overall structural comparison between the three ColE7–DNA complexes shows that ColE7 interacts with either preferred or non-preferred sites in a similar way and no obvious difference can be identified to explain its sequence-dependent cleavage preference. Moreover, the two Vvn/DNA complexes also look alike, with similar binding modes and interaction patterns between Vvn and DNA. Are there any other unnoticed structural features that are local determinants for the enzyme to cleave DNA at distinct rates? To find any local structural differences, the three available N-ColE7/DNA complexes, two non-preferred and one preferred complexes were superimposed by fitting only the N-ColE7 molecules in the complexes (Figure 5). The three DNA duplexes, 8, 12 and 18 bp, were lined up after the fitting and revealed similar overall bending structures. The minor grooves were all open to a similar width, from 6 Å (in a straight B-form DNA) to ∼9 Å (Figure 6B). However, a closer examination revealed that the phosphate backbones near the metal finger active site were shifted to different positions in the three complexes (Figure 5A). The phosphate backbone in the preferred complex with a thymine before the scissile phosphate, shifted the most toward the enzyme active site. The DNA backbones are shifted the most at the phosphate one step before the scissile phosphate, with a shifted distance of 3.0 Å between the two phosphorous atoms in 8mer (P4) and 18mer (P11) complexes, and 1.72 Å between the ones in 12mer (P5) and 18mer (P11) complexes. The backbones in the non-preferred complexes with a guanine residue before the scissile phosphate were located further away from the active site. The difference in the phosphate backbone distortion led to a closer Zn-P distance of 2.95 Å between the zinc ion and the scissile phosphate in the preferred complex, and longer distances of 3.18 Å (12mer) and 3.72 Å (8mer) in the non-preferred complexes. This result suggests that a DNA backbone that can be deformed more easily by the endonuclease is cleaved more efficiently.
Figure 5.
Superposition of three N-ColE7–DNA complexes and two Vvn–DNA complexes. (A) The crystal structures of the three N-ColE7–DNA complexes, the preferred complex (18 bp: 2IVH) and the non-preferred complexes (8 bp: 1PT3 and 12 bp: 1ZNS), were superimposed by least-square-fitting of the Cα-atoms of N-ColE7 molecules. The three duplex DNA were aligned after fitting with the metal finger motif (displayed in red) bound at the minor groove, inducing DNA to bend slightly away from the enzyme. The DNA backbones are displayed in yellow for the 18 bp DNA, gray for 12 bp DNA and green for 8 bp DNA, but the backbone ribbons of the scissile phosphates are all displayed in orange. A detailed view of the phosphate backbones is shown in the right panel. The DNA backbone is distorted the most in the preferred complex with a thymine base (T10) located at the 3′-O side of the scissile phosphate as compared to the backbones in the non-preferred complexes, which contain a guanine base before the scissile phosphates (G4 in the 12 bp complex and G3 in the 8 bp complex). (B) Superposition of the active sites in the three N-ColE7–DNA complexes shows that the scissile phosphate is located closer to the endonuclease active site in the preferred complex (18 bp) with a distance of 2.95 Å between the zinc ion and phosphorus atom. (C) The crystal structures of Vvn–DNA complexes, 8 bp (PDB: 1OUP) and 16 bp (this study, PDB: 2IVK) complexes, were superimposed by a least-square-fit of the Cα atoms in Vvn. The DNA backbones fitted well and were not distorted, consistent with Vvn's low sequence-dependent endonuclease activity. Vvn are displayed in blue with metal finger motifs in red. The 16 bp DNA is displayed in green and 8 bp DNA is displayed in yellow.
Figure 6.
The transitions in α and γ dihedral angles before and after the cleavage site result in a larger DNA backbone conformational change in the preferred complex of N-ColE7–DNA (18 bp) upon enzyme binding to DNA. (A) Stereo view of the superimposed DNA in the three N-ColE7–DNA complexes shows that a larger phosphate backbone conformational change observed only in the preferred complex is resulted from the transitions in α and γ dihedral angles upon the enzyme binding to the preferred DNA site. The phosphate backbones are shown in green for the 8 bp DNA, gray for the 12 bp DNA and yellow for the 18 bp DNA; only the bases of A9, T10 and C11 in the 18 bp DNA are shown. The transitions in α and γ dihedral angles before the cleavage site in the preferred 18 bp complex are marked by circled arrows. (B) The minor groove widths in the three complexes (8 bp: 1PT3, 12 bp: 1ZNS and 18 bp: 2IVH) are all widened to ∼9 Å as compared to a canonical B-form DNA. The base pair opening angles are slightly higher in the widened minor groove regions in the preferred complex (18 bp) than those of the non-preferred complexes (8 and 12 bp). (C) The α and γ dihedral angles immediately before and after the cleavage site have non-canonical values only in the preferred 18 bp complex (marked by arrows in the figure). The α/γ before the cleavage site are 39.4°/−89.4° for 18 bp complex, −61.9/59.1° for 12 bp complex and −56.6/46.0° for 8 bp complex. The transitions in α and γ upon protein binding to the preferred site lead to the DNA backbone conformational change observed in the preferred complex.
However, why can some specific sequences of DNA be distorted by ColE7 more easily than others? We compared all the base pair stacking parameters and DNA phosphate backbone dihedral angles in N-ColE7–DNA complexes and found that it was the α and γ dihedral angles that varied most significantly between the preferred and non-preferred ColE7–DNA complexes (Figure 6). Compared to a canonical B-form DNA with α/γ of −60/49° (38), the α/γ dihedral angles were all shifted to non-canonical values of ∼100/−100° after the cleavage site in the three N-ColE7–DNA complexes. However, the α/γ before the cleavage sites were within canonical values of −61.9°/59.1° (12mer) and −56.6/46.0° (8mer) in the non-preferred complexes but shifted to non-canonical values of 39.4°/−89.4° only in the preferred complex (18mer). The transitions in α and γ before and after the cleavage site resulted in a larger DNA backbone conformational change in the preferred complex upon enzyme binding to DNA. These unusual α/γ states have been observed only in B-DNA complexed to proteins and thus has been suggested to play a role in the fine structural adjustment during protein–DNA complexation (39). We found that the preferred complex had slightly larger base pair opening angles between the T–A base pair right before the cleavage site, but the non-preferred complexes hadlower opening angle since they contained more hydrogen-bonded G–C base pairs, thus were presumably less prone to base pair opening and backbone conformational change (Figure 6B). Therefore, although the overall interactions between protein and DNA between the non-preferred and preferred complexes are similar (as listed in Figure 3), the DNA backbone is more distorted in the preferred complex. This structural difference could be the local structural determinant for preferential enzyme DNA cleavage.
Vvn has little sequence preference as regards DNA cleavage compared to ColE7. If conformational change in DNA backbone is an important factor for cleavage preference, a backbone distortion difference should not be identified between the two Vvn–DNA complexes with Vvn binding at C–G and G–A with similar cleavage rates of 27.8 and 23.2%, respectively. Indeed the superposition of the two Vvn–DNA complexes did not reveal any local phosphate backbone conformational difference between Vvn–DNA (8 bp) and Vvn–DNA(16 bp) (Figure 5C). Further analysis of backbone dihedral angles and base pair stacking parameters revealed no significant difference between the two complexes either (Figure 6C). The α and γ angles in the two Vvn–DNA complexes were within typical values of a B-form DNA, around −50° for α and 50° for γ, before and after the scissile phosphate. This result confirms our hypothesis that local DNA backbone conformational change is the structural determinant for sequence cleavage preference. Without an enzyme-induced conformational difference, little sequence preference for cleavage can be generated.
Comparison to site-specific DNA-binding proteins and restriction enzymes
Most site-specific DNA-binding proteins interact not only at specific sites with high affinity but also nonspecific sites with appreciable affinity. It is believed that the nonspecific interactions play an important role in site-specific DNA-binding proteins searching for their target sites (40). What are the differences in the DNA recognition process between site-specific DNA-binding proteins and nonspecific endonucleases? Structure studies in solution of Lac repressor in complex with specific and nonspecific DNA sites have shown that several residues in Lac repressor which interact with the DNA backbone in the nonspecific complex, switch roles to interact with the DNA bases in the specific complex (41). As a result, the DNA in the nonspecific complex adopts a canonical straight B-DNA conformation, whereas in the specific complex it is bent by ∼36°. Moreover, the specific complex has 1400 Å2 excess of buried interface as compared to the nonspecific complex. However, for the nonspecific endonuclease ColE7, protein residues interact with DNA backbones in the preferred as well as non-preferred complexes and thus the overall protein and DNA conformations in the two complexes remain almost identical. This difference in protein–DNA interactions differentiates a site-specific DNA-binding protein from a nonspecific one. In other words, only the site-specific DNA-binding protein can interact with DNA bases when it is bound to its specific site and can induce a larger DNA conformational change, leading to better protein–DNA interactions.
As compared to the site-specific restriction enzymes, nonspecific endonucleases share dissimilar but also similar features in DNA recognition and cleavage. Restriction enzymes bind to nonspecific sites mostly by interacting with DNA backbones. When they bind to the specific sites, similar to site-specific DNA-binding proteins, restriction enzymes interact with both backbones and bases (42,43). Crystal structure analyses of EcoRV and BamHI in complex with their specific and nonspecific DNA sequences have shown that the specific complexes contain larger protein–DNA interfaces (2173 Å versus 1370 Å in EcoRV; 4350 Å versus 1489 Å in BamHI), a more compact complex structure, and more direct interactions with bases (44–46). However, these structural differences are not observed for ColE7 in that neither protein conformational change nor excess buried interfaces were identified in the preferred complexes in this study as compared to the non-preferred complexes. Once again, the key difference that differentiates a nonspecific endonuclease from a site-specific one is that nonspecific endonucleases lack the ability to interact with DNA bases.
However, as with the restriction enzymes, nonspecific endonucleases appear to use a similar strategy in sequence-dependent cleavage. The extensive direct and indirect interactions between a restriction enzyme and its specific DNA trigger large conformational changes both in the enzyme and the DNA, with the result that the DNA usually becomes more kinked and the scissile phosphate moves closer to the enzyme active site (42). This is similar to what has been observed here in ColE7, that the scissile phosphate shifts closer to the active site only in the preferred complex. The distortion in the DNA backbone in the N-ColE7–DNA complex seen in this study is not as drastic as observed in restriction enzyme–DNA complexes; however, the small difference suffices for the enzyme to differentiate between preferred and non-preferred sites. Therefore, even though site-specific and nonspecific enzymes bind and interact with DNA in different ways, they discriminate between preferred and non-preferred sites for cleavage by a similar strategy of inducing DNA backbone conformational change.
In summary, nonspecific endonucleases cleave DNA with a preferred sequence context; we suggest here that a more distorted DNA backbone in the preferred enzyme–DNA complex leads to a closer distance between scissile phosphate and enzyme active site, resulting in preferential DNA cleavage. In the case of ColE7 which prefers to cleave after T or A, this is likely because the T–A base pair is more easily opened than G–C and the phosphate backbone can be deformed more easily by the rotation in α and γ dihedral angles in the phosphate ribose backbone. We conclude that nonspecific endonucleases prefer to cleave at sites where the DNA backbone can be deformed more easily by the enzyme.
Acknowledgments
This work was supported by research grants from the Academia Sinica and National Science Council (NSC94-2311-B001-012) of the Republic of China to H.S.Y. Funding to pay the Open Access publication charges for this article was provided by the Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Srinivasan E., Shankar V. Sugar nonspecific endonucleases. FEMS Microbiol. Rev. 2001;25:583–613. doi: 10.1111/j.1574-6976.2001.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 2.Widlak P., Garrard W.T. Discovery, regulation and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J. Cell. Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 3.Marti T.M., Fleck O. DNA repair nucleases. Cell. Mol. Life Sci. 2004;61:336–354. doi: 10.1007/s00018-003-3223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker R., Song H. The enzymes and control fo eukaryotic mRNA turnover. Nature Struct. Mol. Biol. 2004;11:121–126. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 5.Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nature Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 6.Lingel A., Sattler M. Novel modes of protein–RNA recognition in the RNAi pathway. Curr. Opin. Struct. Biol. 2005;15:107–115. doi: 10.1016/j.sbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Scheffler I.E., Elson E.L., Baldwin R.L. Helix formation by dAT oligomers. I. Hairpin and straight-chain helices. J. Mol. Biol. 1968;36:291–304. doi: 10.1016/0022-2836(68)90156-3. [DOI] [PubMed] [Google Scholar]
- 8.Lomonossoff G.P., Butler P.J.G., Klug A. Sequence-dependent variation in the conformation of DNA. J. Mol. Biol. 1981;149:745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- 9.Drew H.R., Travers A.A. DNA structure variations in the E.coli tyrT promoter. Cell. 1984;37:491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- 10.Suck D., Lahm A., Oefner C. Structure refined to 2 Angstrom of a nicked DNA octanucleotide complex with DNase I. Nature. 1988;332:464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- 11.Lahm A., Suck D. DNase I-induced DNA conformation. 2 Angstrom structure of a DNase I–octamer complex. J. Mol. Biol. 1991;221:645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- 12.Weston S.A., Lahm A., Suck D. X-ray structure of the DNase I–d(GGTATACG)2 complex at 2.3 Angstrom resolution. J. Mol. Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 13.Fox K.R. Probing the conformation of eight cloned DNA dodecamers: CGCGAATTCGCG, CGCGTTAACGCG, CGCGTTATACGCG, CGCGATATCGCG, TCGCAAATTTGCG, GCGTTTAAAGCG, CGCGGATCCGCG and CGCGGTACCGCG. Nucleic Acid Res. 1992;20:6487–6493. doi: 10.1093/nar/20.24.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera J.E., Jonathan B.C. Characterization of preferred deoxyribonuclease I cleavage sites. J. Mol. Biol. 1994;236:405–411. doi: 10.1006/jmbi.1994.1152. [DOI] [PubMed] [Google Scholar]
- 15.Meiss G., Friedhoff P., Hahn M., Gimadutdinow O., Pingoud A. Sequence preferences in cleavage of dsDNA and ssDNA by the extracellular Serratia marcescens endonuclease. Biochemistry. 1995;34:11979–11988. doi: 10.1021/bi00037a040. [DOI] [PubMed] [Google Scholar]
- 16.Meiss G., Franke I., Gimadutdinow O., Urbanke C., Pingoud A. Biochemical characterization of Anabaena sp. strain PCC 7120 nonspecific nuclease NucA and its inhibitor NuiA. Eur. J. Biochem. 1998;251:924–934. doi: 10.1046/j.1432-1327.1998.2510924.x. [DOI] [PubMed] [Google Scholar]
- 17.Pommer A.J., Cal S., Keeble A.H., Walker D., Evans S.J., Kuhlmann U.C., Cooper A., Connolly B.A., Hemmings A.M., Moore G.R., et al. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 2001;314:735–749. doi: 10.1006/jmbi.2001.5189. [DOI] [PubMed] [Google Scholar]
- 18.Widlak P., Li P., Wang X., Garrard W.T. Cleavage preferences of the apoptotic endonuclease DFF40 (Caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Stable C., Shen C.C., Shen C.-K.J. Enrichment and depletion of Hela topoisomerase I recognition sites among specific types of DNA elements. Nucleic Acid Res. 1988;16:7975–7993. doi: 10.1093/nar/16.16.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan M.E., Austin R.H. Importance of DNA stiffness in protein–DNA binding specificity. Nature. 1987;329:263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- 21.Chak K.-F., Kuo W.-S., Lu F.-M., James R. Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 1991;137:91–100. doi: 10.1099/00221287-137-1-91. [DOI] [PubMed] [Google Scholar]
- 22.Hsia K.-C., Chak K.-F., Liang P.-H., Cheng Y.-S., Ku W.-Y., Yuan H.S. DNA binding and degradation by the HNH endonuclease ColE7. Structure. 2004;12:205–214. doi: 10.1016/j.str.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Wu S.-I., Lo S.-K., Shao C.-P., Tsai H.-W., Hor L.-I. Cloning and characterization of a periplasmic nuclease of Vibrio vulnificus and its role in preventing uptake of foreign DNA. Appl. Environ. Microbiol. 2001;67:82–88. doi: 10.1128/AEM.67.1.82-88.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C.-L., Hor L.-I., Chang Z.-F., Tsai L.-C., Yang W.-Z., Yuan H.S. DNA binding and cleavage by the periplasmic nuclease Vvn: A novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsia K.-C., Li C.-L., Yuan H.S. Structural and functional insight into the sugar-nonspecific nucleases in host defense. Curr. Opin. Struct. Biol. 2005;15:126–134. doi: 10.1016/j.sbi.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya A.E. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shub D.A., Goodrich-Blair H., Eddy S.R. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 28.Chevalier B.S., Stoddard B.L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acid Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui M.-J., Tsai L.-C., Hsia K.-C., Doudeva L.-G., Chak K.-F., Yuan H.S. Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions. Protein Sci. 2002;11:2947–2957. doi: 10.1110/ps.0220602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Y.-S., Hsia K.-C., Doudeva L.G., Chak K.-F., Yuan H.S. The crystal structure of the nuclease domain of ColE7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J. Mol. Biol. 2002;324:227–236. doi: 10.1016/s0022-2836(02)01092-6. [DOI] [PubMed] [Google Scholar]
- 31.Ko T.-P., Liao C.-C., Ku W.-Y., Chak K.-F., Yuan H.S. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 32.Doudeva L.G., Huang H., Hsia K.-C., Shi Z., Li C.-L., Shen Y., Yuan H.S. Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+ Protein Sci. 2006;15:269–280. doi: 10.1110/ps.051903406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedhoff P., Franke I., Meiss G., Wende W., Krause K.L., Pingoud A. A similar active site for nonspecific and specific endonucleases. Nature Struct. Biol. 1999;6:112–113. doi: 10.1038/5796. [DOI] [PubMed] [Google Scholar]
- 34.Miller M.D., Cai J., Krause K.L. The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol. 1999;288:975–987. doi: 10.1006/jmbi.1999.2729. [DOI] [PubMed] [Google Scholar]
- 35.Grishin N.V. Treble clef finger—a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger A.T., Adams P.D., Clore G.M., Delano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges N., Pannu N.S., et al. Crystallography and NMR System (CNS): a new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dynam. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 38.Djuranovic D., Hartmann B. Conformational characteristics and correlations in crystal structures of nucleic acid oligonucleotides: evidence for sub-states. J. Biomol. Struct. Dyn. 2003;20:771–788. doi: 10.1080/07391102.2003.10506894. [DOI] [PubMed] [Google Scholar]
- 39.Várnai P., Djuranovic D., Lavery R., Hartmann B. α/γ transitions in the B-DNA backbone. Nucleic Acid Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acid Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalodimos C.G., Biris N., Bonvin A.M.J.J., Levandoski M.M., Guennuegues M., Boelens R., Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein–DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 42.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 44.Winkler F.K., Banner D.W., Oefner C., Tsernoglou D., Brown R.S., Heathman S.P., Bryan R.K., Martin P.D., Petratos K., Wilson K.S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 46.Viadiu H., Aggarwal A.K. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]