Abstract
In vitro analyses of plant GATA transcription factors have implicated some proteins in light-mediated and circadian-regulated gene expression, and, more recently, the analysis of mutants has uncovered further diverse roles for plant GATA factors. To facilitate function discovery for the 29 GATA genes in Arabidopsis (Arabidopsis thaliana), we have experimentally verified gene structures and determined expression patterns of all family members across adult tissues and suspension cell cultures, as well as in response to light and signals from the circadian clock. These analyses have identified two genes that are strongly developmentally light regulated, expressed predominantly in photosynthetic tissue, and with transcript abundance peaking before dawn. In contrast, several GATA factor genes are light down-regulated. The products of these light-regulated genes are candidates for those proteins previously implicated in light-regulated transcription. Coexpression of these genes with well-characterized light-responsive transcripts across a large microarray data set supports these predictions. Other genes show additional tissue-specific expression patterns suggesting novel and unpredicted roles. Genome-wide analysis using coexpression scatter plots for paralogous gene pairs reveals unexpected differences in cocorrelated gene expression profiles. Clustering the Arabidopsis GATA factor gene family by similarity of expression patterns reveals that genes of recent descent do not uniformly show conserved current expression profiles, yet some genes showing more distant evolutionary origins have acquired common expression patterns. In addition to defining developmental and environmental dynamics of GATA transcript abundance, these analyses offer new insights into the evolution of gene expression profiles following gene duplication events.
Plant GATA-binding proteins were first identified during studies on light-responsive promoters (Lam and Chua, 1989; Buzby et al., 1990; Gilmartin et al., 1990; Lam et al., 1990; Schindler and Cashmore, 1990; Sarokin and Chua, 1992; Borello et al., 1993) following the identification of conserved GATA motifs within promoters that were up-regulated in response to light, including RbcS and Cab (Dean et al., 1985; Grob and Stuber, 1987; Castresana et al., 1988; Giuliano et al., 1988; Gidoni et al., 1989; Gilmartin et al., 1990; Arguello-Astorga and Herrera-Estrella, 1998). Subsequent studies implicated these elements in the regulation of circadian-responsive genes, and a number of in vitro analyses using plant nuclear extracts led to the definition of several proteins with specificity for GATA elements (Carré and Kay, 1995). The presence of sequences within light-responsive promoters matching the GATAAGG motif previously defined as the binding site for the fungal GATA-binding proteins AreA (Kudla et al., 1990) and Nit2 (Fu and Marzluf, 1990) and the vertebrate GATA transcription factors (Evans et al., 1988; Evans and Felsenfeld, 1989; Tsai et al., 1989; Orkin, 1992; Merika and Orkin, 1993) created the possibility that plant GATA-binding proteins could also be type IV zinc finger proteins (Gilmartin et al., 1990; Daniel-Vedele and Caboche, 1993; Teakle and Gilmartin, 1998).
The identification of a gene, Ntl1, from tobacco (Nicotiana tabacum) encoding a plant type IV zinc finger protein following degenerate PCR (Daniel-Vedele and Caboche, 1993) provided the first evidence for this class of protein in plants. Subsequent expressed sequence tag (EST) and genome sequence data revealed the presence of a gene family of related sequences in Arabidopsis (Arabidopsis thaliana; Teakle and Gilmartin, 1998). The encoded proteins share extensive sequence similarity over the zinc finger domain to animal and fungal GATA factors but differ from typical animal GATA factors that typically contain two Cx2C-x17-Cx2C zinc finger domains by having a single zinc Cx2C-x18-Cx2C zinc finger (Teakle and Gilmartin, 1998). This configuration is also present within fungal GATA factors WC1 (Ballario et al., 1996) and WC2 (Linden and Macino, 1997) involved in blue light and circadian responses. Subsequently, an additional plant-specific zinc finger configuration, Cx2C-x20-Cx2C, was identified in Arabidopsis and rice (Oryza sativa; Nishii et al., 2000; Riechmann et al., 2000; Jeong and Shih, 2003; Reyes et al., 2004), and full genome sequence analysis confirms the absence of the animal and fungal type Cx2C-x17-Cx2C zinc finger domains in plants (Arabidopsis Genome Initiative, 2000; Riechmann et al., 2000). In vitro binding studies using recombinant proteins have demonstrated the specificity of this class of zinc finger protein for DNA sequences containing GATA motifs (Teakle et al., 2002; Jeong and Shih, 2003; Sugimoto et al., 2003).
Although plant GATA factors were initially implicated in light-mediated (Castresana et al., 1988; Giuliano et al., 1988; Buzby et al., 1990; Donald and Cashmore, 1990; Gilmartin et al., 1990; Lam et al., 1990; Schindler and Cashmore, 1990; Borello et al., 1993) and circadian-responsive gene expression (Carré and Kay, 1995; Teakle and Kay, 1995), they have also been predicted to play a role in the control of nitrogen metabolism (Daniel-Vedele and Caboche, 1993; Bi et al., 2005) based on the involvement of GATA factors in the regulation of nitrogen balance in fungi (Fu and Marzluf, 1990; Kudla et al., 1990; Scazzocchio, 2000). However, there is now a growing body of data, both from analysis of mutant phenotypes arising from disruption of GATA genes and from expression and bioinformatic analyses of members of this gene family in wild-type plants, which connects GATA factors to a wide range of different biological functions.
The Arabidopsis genome contains 29 GATA factor genes (Riechmann et al., 2000; this article). Mutations arising from disruption or overexpression of only four of these genes have so far been reported. These studies have identified effects on a range of processes; the ZIM (GATA25) overexpression phenotype shows altered cell elongation (Nishii et al., 2000; Shikata et al., 2004), mutation of HANABU TARANU (HAN; GATA18) in the han mutant shows defects in flower and shoot apical meristem development (Zhao et al., 2004), and disruption of BME3 (GATA8) shows defects in seed germination (Liu et al., 2005). Mutation of GNC (GATA21; Bi et al., 2005) reduces chlorophyll levels and produces defects in regulation of expression of a range of genes involved in sugar metabolism. Interestingly, expression of GNC is nitrate inducible (Bi et al., 2005). Although some of these effects involve light-regulated processes, none of the currently available evidence conclusively implicates any of these GATA genes as key regulators of photosynthetic gene expression.
With the availability of near full-genome coverage microarray platforms and extensive publicly available microarray data sets representing a broad spectrum of growth conditions and mutants, it is possible to identify changes in transcript abundance for those GATA factor genes represented on the arrays. In addition, Web-based tools, such as NASCArray tools, Genevestigator, and others, provide opportunities for data mining to characterize expression patterns of individual GATA factor genes (Craigon et al., 2004; Zimmermann et al., 2004; Jen et al., 2006; Manfield et al., 2006) that may provide insight into potential biological function. This approach has been used previously to predict roles for other poorly characterized genes in secondary cell wall thickening, leading to the identification of mutant phenotypes (Persson et al., 2005). Coexpression analysis tools such as the Arabidopsis Coexpression Tool (ACT; Jen et al., 2006), in conjunction with tools such as Genevestigator (Zimmermann et al., 2004), can therefore be used to identify information facilitating gene function prediction. However, only 21 of the 29 GATA genes are represented by probe sets on the Affymetrix ATH1 array that is the source of the data used by NASCArray tools, Genevestigator, and ACT. Similarly, some of the GATA factor genes are not represented in the extensive datasets generated by Massively Parallel Signature Sequencing (Meyers et al., 2004) and are therefore not amenable to bioinformatics analysis of expression patterns.
Bioinformatic analysis of the Arabidopsis GATA family (Reyes et al., 2004) has provided insight into the evolutionary relationships of the different GATA family members, but experimentally confirmed gene structures are not available for the majority of these genes. As a prerequisite for a comprehensive functional genomics analysis of the Arabidopsis GATA factors, we analyzed the entire GATA gene family to experimentally confirm predicted gene structures, including definition of 5′ and 3′ untranslated regions (UTRs). The identification of transcription start sites by 5′ RACE and discovery of introns within several 5′ UTR sequences have provided experimental confirmation of the location of upstream regulatory sequences. In addition, as part of our on-going studies to define biological functions for members of this family, we have undertaken gene-specific expression analysis using quantitative PCR (qPCR) with different tissues and growth conditions to obtain an integrated expression profile for the whole family. These data complement extensive bioinformatic analysis of GATA expression profiles for those genes represented on the Affymetrix ATH1 array and provide new insights into the biological significance of several members of this gene family. These results are used to elucidate the divergence and convergence of expression profiles following gene and genome duplications.
RESULTS
Defining Membership of the GATA Factor Family
A number of families of zinc finger transcription factors containing a C2-C2 zinc-binding domain have been defined in plants, including the CONSTANS and CONSTANS-LIKE family (Griffiths et al., 2003), the Dof family, which includes DAG1 and DAG2 (Gualberti et al., 2002), and the GATA family (Teakle and Gilmartin, 1998). These families of proteins contain members that share some common features, for example, similar spacing between the paired Cys residues, and this has sometimes resulted in the consideration of members of different families under the general term GATA factors (e.g. Putterill et al., 1995; Nemoto et al., 2003; Umemura et al., 2004). However, for this study, we used conserved features of GATA factor family members across all kingdoms (Lowry and Atchley, 2000) to identify all GATA factor gene family members within the Arabidopsis genome. The criteria for inclusion based on the zinc finger configuration C-x2-C-x18/20-C-x2-C are: (1) the presence of two pairs of Cys residues within the predicted zinc finger domain that are each separated by two amino acids; (2) a loop of 18 or 20 amino acids between the two pairs of Cys residues; (3) conservation of the amino acid sequence LCNACG around the second Cys pair; and (4) the presence of conserved TPQWR or TPMMR motifs within the X18/20 loop.
Table I presents a comparison of selected amino acid sequences from plant, animal, and fungal GATA factors and highlights differences between the CONSTANS and Dof zinc finger configuration. By these criteria, GATA29 (At3g20750; Table I) is the most divergent gene we consider to encode a GATA factor even though the spacing between the first Cys pair is four amino acids rather than the classical two. Gene At4g16141 (Table I) has been considered by some (Riechmann et al., 2000; Bi et al., 2005) to be a GATA factor. However, this assessment would appear to be based solely on the presence of the LCNACG motif; it does not match any of the other defined criteria, and we have therefore excluded it from consideration as a GATA factor. Similarly, At3g17660, used as an outgroup in phylogenetic analyses (Reyes et al., 2004), lacks the necessary motifs for inclusion in the family (Table I). Our bioinformatics analyses resulted in the identification of 29 members of the GATA gene family. During the course of our work, similar database searches were reported (Jeong and Shih, 2003), and we have adopted the nomenclature defined by these authors for our experimental structure and expression analysis of the GATA factor family.
Table I.
Comparison of amino acid sequences of zinc finger motifs
Amino acid sequences of zinc finger motifs from selected Arabidopsis GATA factors representing the four subfamilies (Reyes et al., 2004) and related representative sequences from chicken (Gallus domestica; Evans and Felsenfeld, 1989), Neurospora crassa (Fu and Marzluf, 1990), and Aspergillus nidulans (Kudla et al., 1990) are compared. Highly conserved residues are shown in bold with conserved Cys residues also underlined. The zinc finger domains of other Arabidopsis proteins (encoded by At4g16141, At5g15850, and At3g61850) and previously considered to be GATA factors due to the presence of conserved Cys pairs (shown in bold and underlined) are also presented. Gene identifiers and database accession numbers are shown. Dashes indicate genes without names.
| Species | Gene Identifier | Gene Name | Zinc Finger Motif |
|---|---|---|---|
| Arabidopsis | At3g24050 | GATA1 | CQHCGAE-K-TPQWRAGPAGPKTLCNACG |
| Arabidopsis | At5g49300 | GATA16 | CADCGTS-K-TPLWRGGPVGPKSLCNACG |
| Arabidopsis | At4g24470 | GATA25 (ZIM) | CTHCGISSKCTPMMRRGPSGPRTLCNACG |
| Arabidopsis | At4g17570 | GATA26 | CYHCGVT-N-TPLWRNGPPEKPVLCNACG |
| Arabidopsis | At3g20750 | GATA29 | CTNMNCNALNTPMWRRGPLGPKSLCNACG |
| G. domestica | P17678 | cGATA1-N | CVNCGATA--TPLWRRDGTGHY-LCNACG |
| cGATA1-C | CSNCQTST--TTLWRRSPMGDP-VCNACG | ||
| A. nidulans | X52491 | AreA | CTNCFTQT--TPLWRRNPEGQP-LCNACG |
| N. crassa | P78714 | WC2 | CTDCGTLD---SPEWRKGPSGPKTLCNACG |
| Arabidopsis | At4g16141 | – | CLIDVIMCIHSLGMRALLLLDQSLCNACG |
| Arabidopsis | At3g17660 | – | CADCRSKAPRWASVNLGIFICMQCS |
| Arabidopsis | At5g15850 | CONSTANS-N | CDTCRSNACTVYCHADSAYLCMSCD |
| CONSTANS-C | CESCERAPAAFLCEADDASLCTACD | ||
| Arabidopsis | At3g61850 | DAG1 | CPRCNSTNTKFCYYNNYSLTQPRYFCKGCR |
Gene Structures
Bioinformatic analysis of GATA factor genes in genome sequence has been used to predict transcription units, including the location of introns (Reyes et al., 2004), but to investigate gene function, accurate and experimentally defined gene structures are needed to confirm transcription start sites, as well as to confirm intron splice junctions and delineate 5′ and 3′ UTRs to facilitate identification of regulatory sequence motifs.
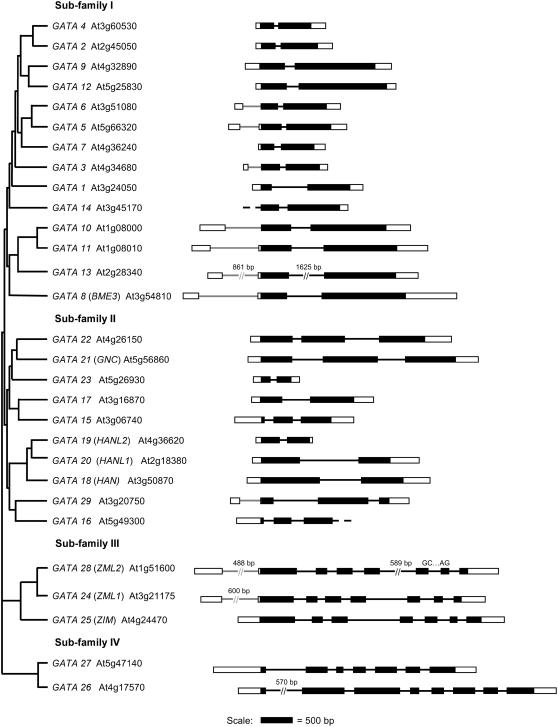
We used EST database sequence information, where available, to assemble full-length cDNA sequences for the Arabidopsis GATA genes. No cDNA sequence was available for seven of the predicted genes, and 5′ and 3′ cDNA end sequence was incomplete for eight and nine other genes, respectively. We therefore used reverse transcription (RT)-PCR with RNA from a range of tissues to confirm or identify exon-intron boundaries and performed 5′ and 3′ RACE-PCR to determine the limits of the transcription unit for those genes where full-length EST sequences were unavailable. These analyses identified the transcription start and end points, as well as intron splice junctions, for gene family members. Sequences have been deposited at GenBank under accession numbers DQ875127 to DQ875134. In the case of GATA14, we were unable to obtain any 5′ UTR data to confirm the transcription start site of this gene, and for GATA16, we were unable to obtain 3′ UTR information. However, cDNA sequences were identified for all genes, providing evidence that therefore there are no untranscribed pseudogenes in the family. Assembled gene structures for the GATA genes are presented in Figure 1. The phylogenetic relationships of the different family members, as defined previously (Reyes et al., 2004), are represented diagrammatically.
Figure 1.
GATA factor gene structure and phylogeny. Genes are numbered following published work (Jeong and Shih, 2003) with Arabidopsis Genome Initiative codes and gene names also presented. Gene structure diagrams are presented in the order and in the subfamilies reported by Reyes et al. (2004). Exons are indicated by black boxes with 5′ and 3′ UTRs of the mature mRNA represented by white boxes. Where there is no data for UTR length (GATA14 and GATA16), this is indicated by a dashed line. Introns are represented by lines in dark gray or in light gray where the intron is present in the 5′ leader. A nonconsensus splice donor and acceptor junction is indicated by the appropriate bases. Gene structure diagrams are to scale (see scale bar) except for very long regions where the length is given in base pairs above the region.
Our analyses have identified features within the genes that could not have been predicted using in silico analysis alone, including introns within the 5′ UTRs of 10 of the GATA genes, as well as the absence of a predicted short exon in GATA13 (Reyes et al., 2004; Fig. 1). We have not identified any alternative splicing of GATA transcripts, although one gene, GATA28, features a nonconsensus donor-acceptor splice junction, GC-AG. A number of GATA genes contain short upstream open reading frames (suORFs) in the 5′ UTR in addition to motifs involved in modulating RNA stability and translation (Supplemental Table S1). The average lengths of GATA 5′ leader and 3′ UTR sequences, 153 ± 98 nucleotides (nt) and 217 ± 90 nt, respectively, are similar to transcriptome averages of 125 nt and 248 nt (Kawaguchi and Bailey-Serres, 2005). Such information and knowledge of 5′ leader intron size and position are prerequisites for the delineation of promoter elements and construction of promoter-reporter constructs. These analyses will also inform searches of T-DNA insertion lines to identify disruptions within transcription units as well as support in silico predictions of transcription regulatory motifs.
Gene Expression Patterns
Comprehensive microarray data sets are available for some members of the GATA gene family, but several of the GATA factor genes (indicated by asterisks in Table II) are not represented on the Affymetrix ATH1 microarray. Gene-specific confirmation of microarray expression data and in-depth analyses on individual genes, using, for example, northern and in situ expression analysis, is available for a very limited number of genes in the GATA family. Phylogenetic analysis of GATA factor genes based on protein sequence data has identified four subfamilies (Reyes et al., 2004). However, these bioinformatic analyses do not provide any insight into gene function or divergence of expression profiles following gene duplication during genome evolution. To obtain a comprehensive overview of expression dynamics of the GATA family and evaluate the expression profiles in relation to phylogenetic relationships, we have undertaken a qRT-PCR analysis of all 29 family members.
Table II.
Comparison of GATA expression families with subfamilies based on coding sequence similarities
Sequence subfamilies (indicated by different numbers in the Sequence Subfamily column) are presented as previously defined (Reyes et al., 2004), while expression families are based on expression information from Figures 2, 4, and 5. Genes not represented by probe sets on the Affymetrix ATH1 array are indicated with an asterisk against the gene name.
| Expression Clade | Sequence Subfamily | GATA Factor | Mutant Name | Gene Identifier |
|---|---|---|---|---|
| 1 | 1 | GATA 8 | (BME3) | At3g54810 |
| 1 | GATA 14* | At3g45170 | ||
| 1 | GATA 3 | At4g34680 | ||
| 2 | GATA 16* | At5g49300 | ||
| 2 | GATA 19 | (HANL2) | At4g36620 | |
| 2 | GATA 23 | At5g26930 | ||
| 2 | 1 | GATA 9 | At4g32890 | |
| 1 | GATA 2 | At2g45050 | ||
| 1 | GATA 4 | At3g60530 | ||
| 3 | 1 | GATA 11* | At1g08010 | |
| 1 | GATA 12 | At5g25830 | ||
| 1 | GATA 10 | At1g08000 | ||
| 1 | GATA 7* | At4g36240 | ||
| 1 | GATA 5 | At5g66320 | ||
| 1 | GATA 6* | At3g51080 | ||
| 2 | GATA 18 | (HAN) | At3g50870 | |
| 2 | GATA 20 | (HANL1) | At2g18380 | |
| 2 | GATA 15 | At3g06740 | ||
| 4 | 2 | GATA 21 | (GNC) | At5g56860 |
| 2 | GATA 22 | At4g26150 | ||
| 5 | 1 | GATA 1 | At3g24050 | |
| 2 | GATA 17 | At3g16870 | ||
| 3 | GATA 25 | (ZIM) | At4g24470 | |
| 3 | GATA 24 | (ZML1) | At3g21175 | |
| 3 | GATA 28 | (ZML2) | At1g51600 | |
| 4 | GATA 26 | At4g17570 | ||
| 6 | 1 | GATA 13* | At2g28340 | |
| 4 | GATA 27* | At5g47140 | ||
| 7 | 2 | GATA 29* | At3g20750 |
We designed primers specific for each of the GATA factor genes (Supplemental Table S2) and performed RT-qPCR to generate a comprehensive expression analysis of all members of the family. This approach provides the greatest sensitivity and quantitative detection of genes expressed at low levels (Czechowski et al., 2004). We performed an analysis of cDNA from light-grown and dark-grown etiolated seedlings to define which members of the GATA gene family are developmentally light regulated. We then determined the influence of circadian regulation on the gene family. We also analyzed cDNA from a clearly defined set of adult tissues, namely roots, stems, leaves, flowers, and siliques, as well as cell culture tissue, to generate an expression dataset that together with the light-grown and dark-grown seedling expression data was used to compare expression profiles between different family members and support the grouping of genes based on this simple set of expression criteria. The expression data are presented as three sets: Figure 2 presents expression data derived from light-grown and dark-grown seedlings, Figure 3 shows the analysis of circadian regulation, and Figure 4 presents the developmental expression in roots, stems, leaves, flowers, siliques, and cell culture. The cDNAs used for analysis of light- and dark-grown seedlings and dissected organs were prepared and analyzed by qPCR in parallel. It was these data that were used in combination to produce a cladogram, grouping genes by the similarities of their expression (Fig. 5). An overview of the results is initially presented followed by a detailed analysis of the different expression clades identified by expression pattern clustering.
Figure 2.
Expression of GATA genes in light- and dark-grown seedlings Transcript abundance for each GATA gene was measured by qPCR analysis of cDNA from light-grown (photomorphogenetic) and dark-grown (skotomorphogenetic) seedlings. Results for light-grown and dark-grown samples are indicated by white and black bars, respectively. Values are the result of duplicate analysis of two biological replicates and are shown with se. The ratio of expression levels in light- and dark-grown tissues is presented.
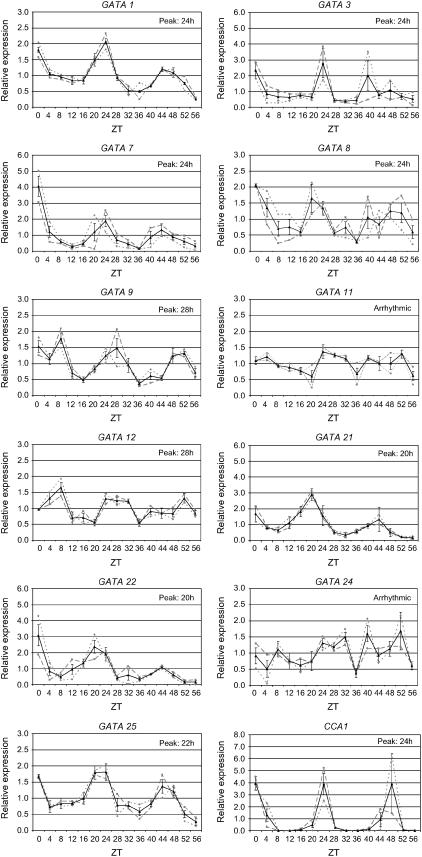
Figure 3.
Circadian expression patterns for selected GATA genes. The expression of genes with a significant level of expression in light-grown seedlings collected over a circadian time course was analyzed over a time course designed to identify genes showing evidence of clock regulation in the absence of light signaling, following standard procedures. Gene expression was measured by qPCR analysis of cDNA from samples of light-grown seedlings collected every 4 h. Results presented show two biological replicates (dashed lines) and the average (with se bars) of duplicate analysis of the two biological replicates.
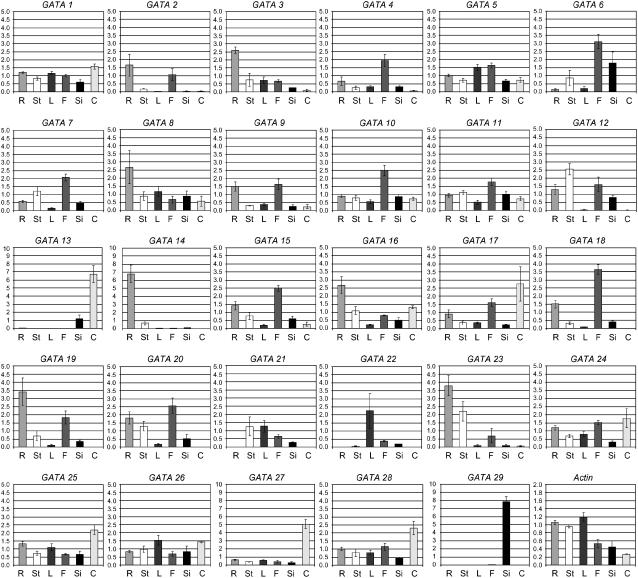
Figure 4.
GATA expression profiles in adult tissues. Transcript abundance for each GATA gene was measured by qPCR analysis of cDNA from roots, stems, leaves, flowers, siliques, and cell culture (abbreviated as R, St, L, F, Si, and C, respectively). Values are the result of duplicate analysis of two biological replicates and are shown with se.
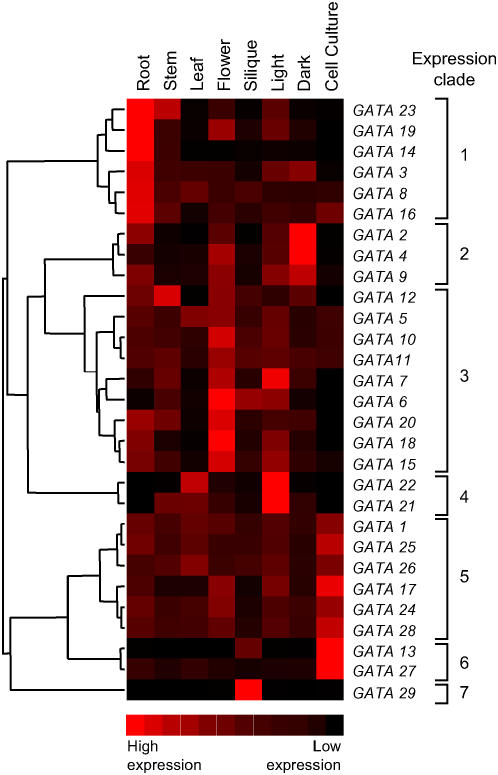
Figure 5.
Clustering of Arabidopsis GATA factors based on expression patterns. Expression profile data for all GATA genes in roots, stems, leaves, flowers, siliques, light-grown seedlings, dark-grown seedlings, and cell culture was used to construct a cladogram grouping genes according to the similarity of their expression patterns. Increasing expression level is indicated by squares from black to red.
Reports of DNA-binding activities in plant nuclear extracts recognizing GATA motifs in the promoters of light-responsive genes (Lam and Chua, 1989; Buzby et al., 1990; Gilmartin et al., 1990; Lam et al., 1990; Schindler and Cashmore, 1990; Sarokin and Chua, 1992; Borello et al., 1993) and information from published microarray data indicating light-regulated expression of some GATA genes (e.g. Harmer et al., 2000; Tepperman et al., 2001; Monte et al., 2004) underpins the perceived involvement of GATA factors in the control of light-responsive transcription. To identify those family members that are up- and down-regulated during photo- and skotomorphogenesis, we compared expression in 7-d-old light-grown seedlings with 7-d-old etiolated seedlings. The majority of genes are expressed during one or both of these growth conditions. Only GATA13, GATA14, and GATA29 show minimal expression in either sample. A number of genes show greater than 2-fold higher expression in light-grown than dark-grown seedlings (Fig. 2), with the greatest differences observed for GATA6, GATA7, GATA21 (GNC), GATA22, and GATA23, with GATA22 showing a 75-fold difference in expression level. In contrast, four genes show stronger expression in etiolated over light-grown seedlings, namely GATA2, GATA4, GATA9, and GATA12, with GATA2 showing a 5-fold difference in expression.
The identification of GATA genes showing differential regulation between light-grown and dark-grown seedlings, coupled to the implications of GATA factor involvement in circadian regulation, led us to perform RT-qPCR analysis of circadian regulation of the GATA gene family. We followed published methods (Millar et al., 1995; Harmer et al., 2000) and used primers designed to the transcript for CCA1 (Wang et al., 1997) as a control (Fig. 3). Data are presented from two independent biological replicates, each containing two technical replicates. Dotted lines indicate the biological replicates and the solid lines represent an average of the two data sets. A very clear and circadian regulation of CCA1 transcript levels was observed peaking at subjective dawn with increases in transcript level preempting subjective dawn (Fig. 3), confirming clock entrainment of seedlings.
Thirteen GATA genes were expressed at a sufficient level in the assays to evaluate their circadian regulation (Fig. 3). Of these 13, nine revealed rhythmic expression. Five genes, GATA1, GATA3, GATA7, GATA8, and GATA25, showed an expression peak coinciding with CCA1 at 24 h (subjective dawn), while GATA21 (GNC) and GATA22 produced a circadian peak at 20 h, preempting dawn. In contrast, expression of GATA9 and GATA12 peaked at 28 h, 4 h after subjective dawn. A number of genes, namely GATA1, GATA3, GATA7, GATA21, and GATA22, showed damping in the amplitude of the second peak of transcript abundance. Analysis of GATA2 revealed rhythmic behavior, but independent biological replicates showed different phases of peak transcript abundance; averaging data from these duplicate experiments therefore does not portray a single clear rhythm (data not shown). The different phases of the rhythm in these samples are surprising, as the assays were done using the same RNA samples used for the analyses shown in Figure 3. GATA11, GATA24, and GATA28 were arrhythmic. These results identify a set of clock-regulated GATA factor genes showing different phases of expression. In addition, these data reveal that not all light-modulated GATA genes are under the control of the circadian oscillator and that some of the gene family members under strong circadian control are not directly influenced by growth in the light and dark.
The analysis of GATA factor gene expression in differentiated tissues and cell culture material identified six genes that show less than 2-fold expression variation in different parts of the plant, namely GATA1, GATA5, GATA11, GATA25 (ZIM), GATA26, and GATA28 (Fig. 4). Other family members reveal different levels of differential expression in different samples, with some showing enhanced expression levels in flowers, others predominantly expressed in roots. Only GATA22 is expressed predominantly in leaves. GATA13, GATA14, and GATA29, none of which are represented on the ATH1 Affymetrix gene chip, show highly specific expression in cell culture, roots, and siliques, respectively.
Analysis of Expression Clades
To facilitate inferences of functional relationships, including potential redundancy between genes, and to investigate whether evolved expression profiles correlate with previously defined phylogenetic groupings based on amino acid sequence, we integrated expression pattern data for all 29 genes across eight different RNA samples by clustering with respect to similarities in expression pattern (Fig. 5). Strikingly, few GATA genes are expressed in predominantly one specific tissue; rather, the expression profiles show expression in most RNA samples analyzed with the different relative levels of expression revealing major expression groupings that we define as seven expression clades (Fig. 5).
All of the samples analyzed, with the exception of the suspension culture, consist of complex mixtures of cell types with some commonality of cell types between the different tissues. This situation may contribute to the broad expression profiles of some genes, but the data clearly illustrate that the clustering of family members by expression profile does not correlate precisely with sequence-derived phylogenies. These differences are summarized in Table II.
Expression clade 1 represents those GATA genes that are predominantly expressed in roots. The six genes in this clade are also expressed to varying degrees in other tissues, but GATA14 shows the strongest preferential expression in roots with only limited expression in other samples (Fig. 4). GATA14 shows extremely low levels of expression in seedlings (Fig. 2), indicating a difference in expression between adult soil-grown roots and roots from agar-grown seedlings.
GATA23 and GATA19 are members of sequence subfamily II (Fig. 1) that have not arisen by gene duplication (Reyes et al., 2004) but show very similar patterns of expression in relation to organ specificity (Fig. 4) and in response to light (Fig. 2). The key difference in expression of these two genes is reflected by different relative expression levels in stems, flowers, and roots. Analysis of GATA23 gene expression patterns, using ACT (Jen et al., 2006; Manfield et al., 2006) and Genevestigator (Zimmermann et al., 2004) tools (data not shown), also reveals this gene and the 20 genes showing strongest coexpression are most strongly expressed in the root elongation zone. Similar bioinformatic analyses for GATA19 using Genevestigator and ACT (Zimmermann et al., 2004; Jen et al., 2006) are compromised by the relatively low level of expression of this gene, as detected by microarray analysis. However, in agreement with our RT-qPCR analysis, the 20 genes most strongly coexpressed with GATA19 (data not shown) are expressed in roots and flowers, and, more specifically, stamens. The enhanced expression in flowers is evident in the heat map shown in Figure 5. This gene pair also shares similar expression profiles in light-grown and dark-grown seedlings where the differences in signal intensity possibly reflect differences in the sizes of the root system in the photomorphogenic and skotomorphogenic seedlings. GATA3, GATA8 (BME3; Liu et al., 2005), and GATA16 also present similar expression profiles within expression clade 1, although GATA3 is not expressed in suspension culture cells and shows slightly higher expression in seedlings. None of these three genes cluster through sequence alignment (Fig. 1); indeed, GATA8 and GATA16 represent members of different GATA subfamilies (Reyes et al., 2004). GATA29, which encodes a protein that aligns most closely to that encoded by GATA16 (Reyes et al., 2004), is the only GATA factor gene that shows highly specific expression in siliques and represents the sole member of expression clade 7. The GATA29 zinc finger is also the most divergent of all family members, as it contains an unusual Cx4C Cys pair within the zinc finger domain (Table I). Within this clade, only GATA19 and GATA23 show differences in expression between light-grown and etiolated seedlings, and GATA3 and GATA8 are the only two genes under circadian control.
Expression clade 2 is characterized by the enhanced expression of the three members, GATA2, GATA4, and GATA9, in dark-grown seedlings and in mature light-grown plants; the strongest expression is in roots and flowers. Phylogenetic analysis identifies GATA2 and GATA4 as having arisen from a common origin via a genome duplication event (Reyes et al., 2004). Our analyses also highlight GATA9 and GATA12 as a gene pair sharing extensive gene structure and sequence similarity (Fig. 1). These four genes cluster as members of subfamily I (Reyes et al., 2004). The exclusion of GATA12 from expression clade 2 appears to arise due to the high levels of expression of this gene in stem, a tissue where GATA2, GATA4, and GATA9 expression is limited; GATA12 is grouped in expression clade 3. However, all four of these genes show significant down-regulation in light-grown as opposed to dark-grown seedlings and share other expression characteristics (Figs. 2 and 3). Microarray analyses report the circadian cycling of transcript abundance of GATA4 (Harmer et al., 2000) and of GATA2, GATA4, GATA9, and GATA12 (Edwards et al., 2006). Our qPCR analyses reveal circadian regulation of GATA9 and GATA12 with a peak of expression 4 h after subjective dawn (Fig. 3). However, we were unable to demonstrate robust circadian control of GATA2 and GATA4. As discussed above, GATA2 expression cycles but the phase is variable, and GATA4 expression in light-grown seedlings (Fig. 2) is too low to reveal a robust rhythm.
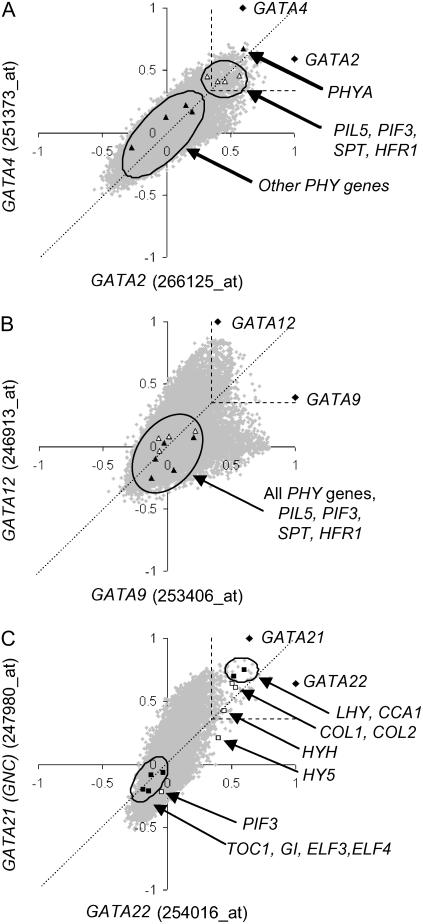
Analysis of gene expression patterns using ACT indicates that GATA2 and GATA4 show strong coexpression with each other as well as a significant number of genes with roles in cell wall assembly, including expansins, arabinogalactan proteins, and glycosyl hydrolases (data not shown). These analyses also reveal that GATA2 and GATA4 are coexpressed with PHYA but not with genes encoding other phytochromes (shown as black triangles in Fig. 6A); PHYA is the eighth-most strongly coexpressed gene with GATA4 (r value, 0.67; P value for the observed correlation occurring by chance, 2 × 10−44). In addition, a number of genes involved in photoresponse signaling, including transcription factors PIL5/PIF1, PIF3, SPT, and HFR1 that act downstream of PHYA signaling, show correlation of expression with GATA4 (P values < 1 × 10−14; these genes ranked in the best-correlated 3% of genes). A P value of 1 × 10−10 is shown on these graphs as a guideline significance threshold. Experimentation, directed by these results, will identify the biological significance of these correlations. Other genes encoding components involved in light and clock signaling, namely HY5, HYH, LHY, and CCA1, showed low correlation r values (between 0.2 and −0.2), indicating uncorrelated expression with GATA2 and GATA4 (data not shown). The similar expression of GATA2 and GATA4 (represented by black diamonds in Fig. 6A) over all array experiments in the ACT database is reflected in the alignment of all data points along the 45° bisecting dotted line.
Figure 6.
ACT scatter plot comparison of duplicate gene expression. Scatter plots of r values for correlation of expression of all genes in the ACT database against pairs of GATA genes with similar expression patterns. A, GATA2 and 4. B, GATA9 and 12. C, GATA22 and GNC. On each diagram, the query genes are represented by black diamonds. In A, phytochrome apoprotein genes and a selection of transcription factor genes (see text for details) are represented by black and white triangles. In C, genes encoding clock-related components and other transcription factor genes are represented by black and white squares, respectively. The position of the r value corresponding to a P value of 1 × 10−10 is shown as a guideline significance threshold.
Similar analyses of GATA9 and GATA12 using ACT reveals no clear overrepresentation of gene ontology terms that might have suggested functions for these genes (data not shown). GATA2, GATA4, GATA9, and GATA12 all show down-regulation in light-grown seedlings. However, scatter plot analysis for GATA9 and GATA12 (Fig. 6B) reveals that genes highlighted in relation to GATA2 and GATA4 (Fig. 6A) show no expression correlation to GATA9 and GATA12. The heart-shaped distribution of data points (Fig. 6B) reveals that there are sets of genes distributed along the x axis and above the 45° bisecting dotted line that show stronger correlation with GATA9 than with GATA12 and vice versa. This divergence of correlation data sets suggests that this conserved gene pair has partially diverged not only in their own regulation but in relation to the genes with which they are coexpressed and potentially regulate.
Expression clade 3 (Fig. 5) represents those genes that predominantly show strongest expression in flowers. GATA12 is the outlier of this group with strongest expression in stems, and this gene has already been considered in relation to GATA9 above. All other genes in this clade, with the exception of GATA11 and GATA20, show greater than 2-fold higher expression in light-grown versus dark-grown seedlings, and, with the exception of GATA5, all show low levels of expression in light-grown leaves. This observation suggests that the observed seedling expression profiles in this clade reflect developmental rather than light-responsive expression patterns. The only remaining gene in this clade under circadian control is GATA7, with expression peaking in phase with CCA1 at subjective dawn.
GATA10 and GATA11 show very similar expression profiles (Figs. 4 and 5) across the spectrum of samples analyzed and neither is under circadian control (Fig. 3). These genes arose via tandem duplication, and the similarities in expression suggest that they have not diverged significantly either in relation to their encoded proteins or their expression dynamics. Another pair of genes within clade 3 with very similar expression profiles is GATA18 (HAN; Zhao et al., 2004) and GATA15. Phylogenetic analysis (Reyes et al., 2004; Fig. 1) identifies HAN (GATA18) as one of three closely related genes, with GATA19 (HANL2) and GATA20 (HANL1) in subfamily II. GATA20 also forms part of expression clade 3 and shows similar expression dynamics to GATA18, but, as discussed above, GATA19 falls within expression clade 1, as this gene is expressed more strongly in roots than in flowers. Based on sequence analysis, GATA15 is most similar to GATA17, but these genes do not share expression profiles, with GATA17 residing in expression clade 5. The close similarity of expression profiles for GATA18 (HAN; Zhao et al., 2004) and GATA15 (Fig. 5) could provide insight into the potential site of GATA15 function.
The final pair of genes within expression clade 3 is GATA6 and GATA7. These genes, together with GATA5, represent a group of three closely related sequences within subfamily I that arose through two different segmental duplication events (Reyes et al., 2004). GATA5 and GATA6 retain greater sequence similarity to each other than either do to GATA7 (Reyes et al., 2004; Fig. 1), yet in terms of expression profiles, GATA5 is the most divergent with almost constant expression across all adult plant organs analyzed, while neither GATA6 nor GATA7 show strong expression in leaves, roots, or suspension culture cells.
Expression clade 4 contains only two members, GATA21 (GNC; Bi et al., 2005) and GATA22, and these genes are characterized by showing significantly high levels of expression in light-grown seedlings. Together with GATA23, these genes show the strongest differential expression between light-grown and etiolated seedlings, with GATA22 showing a 75-fold difference in expression levels (Fig. 2) and the highest level of expression of any GATA gene in adult leaf (Fig. 4). These three genes cluster in subfamily II with GATA21 and GATA22 arising from a duplication event between chromosome 4 and 5 (Reyes et al., 2004). GATA23 is the most divergent of the three as revealed by gene structure (Fig. 1) and also by expression profile (Fig. 5). Notwithstanding the significant differences in expression between light- and dark-grown seedlings for these three genes, GATA23 is not part of this clade, as its expression in light-grown seedlings is relatively weak, and in contrast with GATA21 and GATA22, it shows high transcript levels in root and stem (Fig. 5). GATA21 and GATA22 show no expression in roots or in nonphotosynthetic cell culture but do show strong expression in green photosynthetic tissues, and available array data suggests both circadian and diurnal changes of transcript abundance (Smith et al., 2004; Edwards et al., 2006). Our circadian analyses by RT-PCR (Fig. 3) reveal that GATA21 (GNC) and GATA22 are under circadian regulation with peaks of expression preempting subjective dawn by 4 h.
Bioinformatic analysis of GATA21 (GNC) using the Gene Ontology tool within ACT (Manfield et al., 2006) reveals coexpression with a number of light-regulated genes encoding proteins destined for the chloroplast, including those involved in carotenoid biosynthesis (data not shown). In contrast, GATA22 shows coexpression with genes showing overrepresentation of transcription factor and circadian rhythm Gene Ontology terms (data not shown). The ACT scatter plot analysis of GATA21 (GNC) and GATA22 is shown in Figure 6C. The skew of data points representing coexpressed genes away from the 45° bisecting line indicates that of those genes that are coexpressed with both GATA genes, the expression of the majority is more strongly correlated with GATA21 than with GATA22. Although GATA21 and GATA22 share a common origin and present similar conserved expression profiles (Fig. 5), they have diverged in terms of the genes with which they are coregulated and perhaps regulate. This divergence of expression correlation is less dramatic than seen for GATA9 and GATA12 (Fig. 6B) but reveals a number of genes that stand apart from the main cluster. These genes include a number of transcription factor genes involved in or subject to light and circadian regulation, including CCA1, LHY, CONSTANS-LIKE1, CONSTANS-LIKE2, HY5, and HYH. The probability of these observed expression correlations occurring by chance is extremely small (P = 8 × 10−14), and these genes all rank within the best-correlated 0.5% of genes with GATA22. Several of these genes are also coexpressed with GATA21 (GNC) but at much lower rankings (Fig. 6C). The expression correlation of these morning-phased genes with GATA22 contrasts with the anticorrelation of evening-phased genes, such as PIF3, TOC1, GI, ELF3, and ELF4, with both GATA21 and GATA22.
Those GATA genes that are ubiquitously expressed but more strongly expressed in suspension culture cells are grouped within expression clade 5. These genes also include family members that show the least differential expression across different parts of the whole plant. This aspect distinguishes them from genes in expression clade 6 that are expressed in the suspension culture cells but show only limited expression in differentiated tissues. Clade 5 contains the three members of subfamily III (Reyes et al., 2004) that contain a 20-amino acid spacer between the Cys pairs of the zinc finger, GATA25 (ZIM), GATA24 (ZML1), and GATA28 (ZML2; Shikata et al., 2004). GATA24 and GATA28 are most closely related based on gene structure and protein sequence (Fig. 1) and also show conservation of expression dynamics. GATA25 (ZIM) and GATA1 also represent a pair of genes with very similar expression patterns despite representing very different GATA factor lineages (Fig. 1). Similarly, GATA17 and GATA26 represent different subfamilies indicating divergent origins but having overlapping expression profiles. Only GATA17 shows greater than 2-fold higher expression in light- compared to dark-grown seedlings, but both GATA1 and GATA25 (ZIM) show robust circadian regulation with peaks of expression at subjective dawn and 2 h prior to subjective dawn, respectively. Expression of the two other members of subfamily III, GATA24 and GATA28, is not influenced by the circadian clock, suggesting either that GATA25 has come under circadian control since the gene duplication event or that GATA24 and GATA28, or their progenitor gene, have lost this aspect of regulation. Although ZIM, ZIML1, and ZIML2 proteins all contain a CCT motif, also present in CCA1, CONSTANS, and TOC1 proteins (Reyes et al., 2004), only GATA25 (ZIM) shows evidence of regulation by the clock (Figs. 2 and 3).
The convergence of expression patterns for genes from different sequence subfamilies discussed above is also seen for GATA13 and GATA27, comprising expression clade 6, with strongest expression in suspension culture cells and very little expression in differentiated plants. These genes also show divergent expression patterns from the genes with most similar sequence; for GATA13, the related GATA10, GATA11, and GATA8 (BME3) are expressed in clades 1 and 3, while for GATA27, the related GATA26 is expressed in clade 5.
DISCUSSION
Gene and Protein Structures
Our analyses have focused on 29 members of the GATA family in Arabidopsis. We defined membership by conservation of specific sequence elements within the zinc finger domain across the GATA families of all kingdoms. By these criteria, At4g16141 (Table I), which has been considered by others to represent a 30th member of the family (Riechmann et al., 2000; Bi et al., 2005), is not included in our analyses. We, however, included At3g20750 (GATA29) as the most divergent and potential family member. GATA29 differs from the consensus Cx2C-x18/20-Cx2C configuration with Cx4C in place of the first Cys pair but contains many other signature amino acids within the zinc finger. Neither GATA29 nor the possible 30th member show conservation of sequence domains C terminal to the zinc finger identified by Reyes et al. (2004). Sequences C terminal to the zinc finger have been shown to be required for DNA binding in chicken GATA-1 (Omichinski et al., 1993) and in AreA (Manfield et al., 2000). The corresponding regions in plant GATA factors are highly conserved with different sequences represented in the different subfamilies, but these are distinct from the fungal and animal proteins. This region has been proposed, following deletion and site-directed mutagenesis, to be required for both DNA binding and transactivation, leading to the suggestion that plant GATA factors fold to create a DNA-binding domain more similar to the yeast (Saccharomyces cerevisiae) GAL4 Cys4His2 zinc binuclear cluster motif (Sugimoto et al., 2003). This model indicates a 2:1 zinc:protein molar ratio rather than the expected 1:1 and this could be tested using colorimetric zinc-binding assays (Manfield et al., 2000). However, we note that the critical His residues are conserved only within proteins belonging to subfamily I (Reyes et al., 2004). This assay would also determine whether GATA29 and the more divergent At4g16141 encode proteins that can bind zinc.
Previous analyses of the Arabidopsis GATA factor genes (Jeong and Shih, 2003; Reyes et al., 2004) focused on bioinformatic approaches to classify the members of the family. Jeong and Shih (2003) defined 25 available members of the family into three classes based upon the alignment of the zinc finger and C-terminal tail regions, and Reyes et al. (2004) classified 29 full-length proteins into four subfamilies. These analyses provide important information on the sequence relationships and origins of the family members but do not directly address aspects of GATA factor function. As a first step toward a systematic analysis of GATA factor function, we have undertaken a comprehensive analysis to experimentally define the gene structures and expression profiles of the 29 GATA family members.
Bioinformatic gene structure predictions provide an important framework for gene organization and expression analyses but cannot accurately predict many important gene features such as transcription initiation and termination sites, introns within 5′ UTRs, alternative splice sites, and splice variants. Although available full-length cDNA clones contribute significantly to the accurate mapping of a transcription unit, these were not available for many of the GATA factor genes; therefore, accurate identification of regulatory sequences and promoter elements has not been possible. Experimental confirmation of predicted gene structures is therefore an important aspect of functional genomics. Accurate gene structure data in conjunction with gene expression analysis can also provide valuable information on the regulatory mechanisms influencing transcript abundance.
Our analyses have provided experimentally verified gene structure models for the GATA gene family using both 5′ and 3′ RACE to confirm available existing full-length cDNA sequences as well as generating data for genes where only partial cDNA sequences were available. However, we could not obtain complete cDNA sequences for GATA14 and GATA16. Although we have confirmed that both these genes are expressed (Fig. 4), we were unable to amplify the 5′ end of GATA14 and the 3′ end of GATA16 by RACE. There is only partial EST sequence data available for these genes, and we conclude that their transcripts must contain sequences that make them resistant to cDNA synthesis. Our analyses have defined and confirmed the presence of introns within the 5′ UTRs of 10 GATA genes, corrected the misprediction of an exon in GATA13, and confirmed a nonconsensus splice donor site in GATA28. This information will not only permit the accurate prediction of upstream regulatory sequences for promoter and gene expression analysis but has identified the 5′ UTR sequences.
Analysis of gene expression by steady-state transcript analysis reflects a balance between transcription rate and RNA stability. Many of the elements regulating transcript stability are located within the 5′ and 3′ UTRs. Accurate prediction of these regions is therefore an essential component of gene expression analysis. Examination of sequences across the GATA family has revealed the presence of several motifs with defined roles in the regulation of RNA stability. Ten leader sequences contain a CAUU element defined as a dark-destabilizing motif (Dickey et al., 1997; Hansen et al., 2001), and seven contain the functionally similar ferredoxin-A ACAAAA motif (Dickey et al., 1997; Hansen et al., 2001). These genes include, but are not exclusively, those with higher transcript levels in light- versus dark-grown seedlings. Four of the 3′ UTRs contain AU-rich repeats previously defined as instability motifs (Newman et al., 1993), and 11 genes have leader sequences containing C/T-rich motifs that have a role in controlling tissue-specific gene expression (Bonaventure and Ohlrogge, 2002).
Eight transcripts contain suORFs in their 5′ UTRs. Upstream AUGs have been reported as overrepresented in genes with key regulatory roles (Morris and Geballe, 2000). In plants, the role of suORFs has been well documented in relation to Suc control (SC), with SC-suORFs encoding a peptide in the leader sequences of a subset of bZIP transcription factors (Wiese et al., 2004). Other than the suORFs of related GATA genes that encode similar peptides, none of the GATA suORFs show any similarity to each other, to the SC-suORF, or to any other sequence of the genome predicted to be found in a transcript leader sequence. It is possible that any functional role for the GATA suORFs is mediated not through the specific sequence of the suORF but rather by having an effect on the efficiency of translation reinitiation at the downstream authentic AUG (Kozak, 2000). The presence of sequences involved in RNA stability and translational control within some GATA transcripts suggests potential posttranscriptional regulation. These analyses are summarized in Supplemental Table S1.
Light Regulatory Roles for GATA Factors and Expression Divergence following Gene Duplication
The consequences of divergence on the expression of closely related gene pairs are best illustrated by consideration of three gene pairs: GATA2 and GATA4, GATA9 and GATA12, and GATA21 (GNC) and GATA22. All six of these genes provide multiple lines of evidence to implicate them in aspects of light regulation. The first and last of these gene pairs arose following large chromosomal duplications between 53 and 97 million years ago (Reyes et al., 2004), whereas GATA9 and GATA12 possibly arose from a small duplication event not previously identified. Based on sequence, gene structure, and elements of their expression profiles, GATA2, GATA4, GATA9, and GATA12 could share a common ancestry. While the best measures of divergence of duplicated gene function have come from analysis of mutants (e.g. Causier et al., 2005), the availability of databases of microarray data has allowed the comparison of paralogs by expression patterns (Casneuf et al., 2006). Similarly, our scatter plot analyses reveal the correlations of expression patterns of two query genes with 21,890 other genes over 322 arrays comprising 52 experiments using the ATH1 Affymetrix array.
GATA2, GATA4, GATA9, and GATA12 all show higher expression levels in dark-grown over light-grown seedlings (Fig. 2), and this is supported by analysis of publicly available microarray data from a systematic analysis of a range of light treatments (Schmid et al., 2005). Down-regulation of GATA2 and GATA4 expression in light-grown seedlings was observed following a 4-h treatment with far-red, red, blue, and white light, but not UV-A. However, for all wavelengths of light, a 45-min treatment did not produce any changes in transcript levels. GATA9 showed similar but less marked responses to far-red and blue light but not red or UV-A, whereas GATA12 shows no such transcript responses to these various wavelengths of light. These responses to light suggest regulation by phyA (and possibly cryptochrome) action. Published array analyses identified circadian regulation of all four of these genes (Harmer et al., 2000; Edwards et al., 2006), although the amplitude for GATA2 and GATA4 is weaker than for GATA9 and GATA12. Our qPCR analyses support the strong rhythmic behavior of GATA9 and GATA12 with peaks of expression 4 h after subjective dawn. GATA2 showed rhythms with different phases between replicates, whereas for all other genes tested using these same cDNA samples, the replicates were superimposed. The low level of GATA4 expression did not permit circadian analysis of this gene. These four genes show considerable similarities in their developmental expression profiles (Fig. 4); however, GATA12 does not group within expression clade 2 due to the high levels of expression seen in stems (Fig. 5). This difference suggests that one aspect of the divergence of GATA9 and GATA12 is reflected by a gain of expression in stems by GATA12.
Scatter plot analysis using ACT reveals that GATA2 and GATA4 are coexpressed with each other, as indicated both by the close proximity of the data points representing these two genes (Fig. 6A) and by the close alignment of data points for coexpressed genes along the 45° diagonal. This observation indicates that following the gene duplication, GATA2 and GATA4 have maintained similar expression relationships with coexpressed genes, including potential target genes, suggesting some conservation of function and potential functional redundancy. We have also shown previously (Teakle et al., 2002) that recombinant GATA2 and GATA4 interact with the same DNA sequence motifs. GATA2 and GATA4 are also tightly coexpressed with PHYA, and a range of genes encoding bHLH transcription factors, including PIL5/PIF1, SPT, PIF3, and HFR1, which have defined roles in light-responsive signaling. We note that none of the other PHY genes are coexpressed with GATA2 and GATA4. The light down-regulation of GATA2 and GATA4 suggests that these GATA genes may have a role in repression of photomorphogenesis. Analysis of the promoters of genes coexpressed with GATA2 and GATA4 shows overrepresentation of G-box and abscisic acid response element-like motifs, the elements recognized by bHLH and bZIP transcription factors (data not shown) supporting common regulation of these coexpressed genes. PIL5/PIF1 and SPT have been shown to have roles integrating light, hormonal, and environmental signals during seed germination and etiolation (Oh et al., 2004; Penfield et al., 2005; Shen et al., 2005). We speculate that GATA2 and GATA4 may play a role in seed germination and seedling establishment, either within the light signaling pathway (Penfield et al., 2005) or through mobilization of lipid reserves by enzymes such as isocitrate lyase and malate synthase (Eastmond et al., 2000; Penfield et al., 2005).
In contrast, the ACT scatter plot analysis for GATA9 and GATA12 reveals significant divergence of regulation for these two genes. The heart-shaped pattern seen for GATA9 and GATA12 and the distance between the data points for these two genes suggest that these genes have diverged sufficiently in function that they are now regulated with, and potentially regulate, different sets of genes. We are currently investigating if there is a feature of the regulation of the genes best correlated with GATA 9 that distinguishes them from the genes best correlated with GATA 12 (Fig. 6B). Furthermore, although GATA9 and GATA12 share many expression characteristics with GATA2 and GATA4, including down-regulation in light-grown seedlings, they do not show coexpression with any of the genes involved in light signaling highlighted for GATA2 and GATA4 (Fig. 6B). These observations suggest that GATA9 and GATA12 have not only diverged from GATA2 and GATA4, but they are also diverging in expression from each other and would not be predicted to show functional redundancy.
GATA21 (GNC) and GATA22 represent a gene pair with similar expression profiles with strong up-regulation in light-grown seedlings (Fig. 2) and circadian regulation. ACT scatter plot analysis reveals that the majority of genes represented by the data points are located above the 45° bisecting line. This observation suggests that more genes are more closely correlated with GATA21 (GNC) than with GATA22. This observation suggests a greater divergence in expression patterns following gene duplication than between GATA2 and GATA4, but not as great as observed for GATA9 and GATA12. Two genes showing greatest correlation of expression with GATA21 (GNC) and GATA22 are the key circadian transcriptional regulators LHY and CCA1. Our analyses and those of others (Edwards et al., 2006) reveal circadian regulation of these two GATA genes, but their expression is approximately 4 h out of phase with CCA1 (Fig. 3). Mutation of GATA21 (GNC) results in a 20% reduction in chlorophyll biosynthesis and reduces expression of a number of genes involved in carbon metabolism. No mutant phenotype was observed for the two GATA22 T-DNA insertion lines characterized (Bi et al., 2005); either the positions of insertions, which do not abolish GATA22 transcripts, do not disrupt gene function, or GATA22 functions form a subset of those undertaken by GATA21 (GNC) creating partial functional redundancy (Figs. 1 and 5). GATA22 is the most highly up-regulated gene in light-grown over dark-grown seedlings, and along with GATA21 (GNC) shows a circadian peak preempting dawn. The stronger correlation of expression of genes defined in light responses, including HYH and HY5, with GATA22 than with GATA21 (GNC; Fig. 6C) leads us to speculate that GATA22 may also play a role in photoregulation along with GATA21 (GNC; Bi et al., 2005).
Additional microarray data reveals that cytokinin induces expression of both GATA22 and GATA21 (GNC; Kiba et al., 2005) and that expression of both genes is induced by red light in a PIF3-dependent manner (Monte et al., 2004). We also note that promoters of genes coexpressed with GATA22 and GATA21 (GNC) show overrepresentation of G boxes (data not shown), and we speculate that these might be among the direct targets of PIF3 or a related bHLH transcription factor. A group of genes, including LHY, CCA1, COL1, and COL2, HY5, and TOC1 have been identified as red light responsive and independent of or slightly dependent on PIF3 for this induction (Monte et al., 2004). Our bioinformatic analysis of microarray expression data using ACT allows the dissection of this group of genes reporting that LHY (Schaffer et al., 1998), CCA1 (Wang et al., 1997), COL1 (Ledger et al., 2001), and COL2 (Ledger et al., 2001) are all coexpressed with both GATA21 (GNC) and GATA22 but more highly ranked with GATA22. Calculation of correlation values using different data sets and using different statistical algorithms further supports these correlations (data not shown). In addition to PIF3, roles have been identified for TOC1, ELF3, and ELF4 in the regulation of LHY and CCA1 (Kikis et al., 2005). Analysis of the expression of GATA22 and GATA21 (GNC) in mutants for these regulatory proteins might reveal whether these are upstream regulators of these GATA genes. Furthermore, the integration of hormonal signaling with light signals (Chen et al., 2004; Cluis et al., 2004; Kiba et al., 2005) suggests that hormones may also play roles in regulation of these GATA genes.
CONCLUSION
Functional genomic approaches to define GATA factor function using T-DNA insertion lines (Bi et al., 2005; I.W. Manfield and P.M. Gilmartin, unpublished data) have shown that many T-DNA insertion lines do not lead to disruption of gene function, perhaps due to insertion in 5′ and 3′ UTRs, introns, or sequences flanking the transcription unit. The availability of experimentally confirmed gene structures will assist future identification of insertion lines that are likely to lead to perturbation of gene function. Many of the available T-DNA lines do not present obvious mutant phenotypes, possibly because the insertion does not lead to loss of the associated GATA transcript (Bi et al., 2005; I.W. Manfield and P.M. Gilmartin, unpublished data), possibly because of functional redundancy, or possibly suggesting that mutations in some of GATA genes lead to subtle mutant phenotypes. However, it is also likely that many phenotypes will be apparent only under specific environmental conditions, and the results we report here will help define conditions where GATA function defects may be expected. For example, our comprehensive expression analyses identified genes with a range of light-regulated responses in etiolated versus light-grown seedlings, some of which had not been predicted, namely, light down-regulation. This suggests that defects in skotomorphogenesis may be possible for GATA mutants. Similarly, the information about the tissue-specific expression of GATA genes, especially for genes not represented by probe sets on Affymetrix arrays, may be valuable in analyzing appropriate GATA mutants.
Our RT-PCR expression and bioinformatics coexpression analyses identified pairs of genes with very similar behavior and other groups of genes, for example HAN and HAN-LIKE genes, which may show overlapping roles but each with some unshared functions. The use of ACT and associated scatter plots represents a useful approach for comparing the expression and regulation of highly similar genes and offers a new route for charting the acquisition or differentiation of functions for duplicated genes via changes in their coexpression patterns.
In parallel to our bioinformatic analyses and GATA gene expression studies, we have analyzed a range of T-DNA insertion lines, promoter: β-glucuronidase fusions, and undertaken microarray analysis of knockout and overexpression lines for several GATA factor genes. These studies that provide further insight into the role of plant GATA factors will be reported separately.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) Columbia (Lehle seeds) plants were grown in compost:sand:perlite (3:3:1 [v/v]; Sinclair Horticulture) containing the insecticide Intercept (0.28 g/L) in a glasshouse at 22°C without supplementary light. Cell cultures were grown as described previously (Hadden et al., 2006). Seedlings for light-grown and dark-grown analysis were grown on one-half Murashige and Skoog media (Duchefa) containing 1% (w/v) Suc and 0.9% agar. Plates were placed at 22°C and 16:8 h light:dark for 7 d with plates for etiolated seedlings wrapped in two layers of aluminum foil. Seedlings were harvested under dim green light into liquid nitrogen 4 h after dawn. Seedlings for analysis of circadian patterns of gene expression were grown according to Harmer et al. (2000) in a growth chamber at 24°C with a 12:12 h light:dark cycle for 7 d. After dawn on the 7th d, lights were switched to constant light, and from subjective dawn on the 8th d, seedlings were harvested into liquid nitrogen at 4-h intervals.
RNA Purification and cDNA Synthesis
Total RNA was purified using an SDS-based extraction buffer followed by phenol/chloroform extraction, LiCl precipitation, and purification on Qiagen RNeasy spin columns with a DNase I digestion to remove contaminating genomic DNA (Hadden et al., 2006). RNA concentrations were determined by UV absorbance spectrophotometry and RNA integrity confirmed by agarose gel electrophoresis. RNA (2 μg) was converted to cDNA using an oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Superscript II, Invitrogen) in a 40-μL reaction volume but otherwise following manufacturer's instructions. For qPCR, reaction products were diluted 8-fold before analysis. Due to variation in reference gene expression across these diverse samples, to control for differences in efficiency of cDNA synthesis, the yield of cDNA product was measured directly. Concentration of nucleic acids and nucleotide removal were performed using centrifugal concentration devices (Microcon PCR, Millipore) followed by quantitation of nucleic acids by spectrophotometry (ND 1000 spectrophotometer, Nanodrop Technologies). For the results reported here, the sd was <20% of the average yield for each set of samples.
PCR Amplification of cDNA Products
To confirm exon positions, primers were designed around the predicted start and termination codons and used in a PCR under standard conditions with a pool of cDNAs from a range of tissues as the template. PCR products were ligated to pGEM T-easy cloning vector (Promega) and ligation products used to transform Escherichia coli DH5α to ampicillin resistance. The sequence of inserts was determined using dye-labeled M13 forward and reverse primers in a LI-COR sequencer.
5′ and 3′ RACE PCR
For 5′ RACE, first-strand cDNA was synthesized with an oligo(dT) primer and an adapter-tagged primer for second-strand synthesis (RLM-RACE, Ambion). For 3′ RACE, first-strand cDNA synthesis was primed with an adapter-tagged oligo(dT) primer. Gene-specific primary and nested primers were designed around 200 bp from the predicted end and used in a PCR with the adapter primer. Reaction products were cloned and sequenced, as described above.
qPCR and Primer Design
Primers (with optimal length of 20 nt and predicted melting temperature of 60°C) for qPCR were designed using the Primer 3 software at http://frodo.wi.mit.edu./cgi-bin/primer3/primer3.cgi (Rozen and Skaletsky, 2000) to give amplicons of around 100 bp. Regions of each GATA gene with similarity to other regions of the genome were identified by BLAST analysis with an E value of 1.0, and these were excluded from the regions used for primer selection. The sequence of each expected amplicon was then used in a BLAST analysis against the Arabidopsis genome, with an E value of 10, to identify any regions of weak similarity to the primers. Any amplicons with sequence similarity to untargeted regions at the primer or immediately 3′ bases were discarded. These bases were in turn excluded for selection of additional primers, with BLAST analysis again until wholly specific amplicons were predicted.
Reactions were prepared using 100 pmol of each primer and cDNA products equivalent to 50 ng of template RNA in a 20-μL reaction with PCR master mix (Eurogentec or Bio-Rad). A standard curve was prepared from a pool of cDNA samples diluted through five 5-fold steps and analyzed in duplicate. Samples were analyzed using a Bio-Rad iCycler with an annealing temperature of 60°C over 40 cycles. Following the PCR, melt curve analysis was performed to distinguish the expected amplicon from primer dimers. The amount of template in unknown samples was calculated from the threshold value by the iCycler software using the standard curve results. Amplification efficiencies were typically >75%. Measured transcript levels were normalized to the average level for each gene across all eight samples tested. Data presented is from analysis in duplicate of two independent biological replicates.
Bioinformatics
DNA sequence comparisons were performed using BLAST programs available at the National Center for Biotechnology Information Web site (www.ncbi.nlm.nih.gov/BLAST). Sets of genes showing the strongest coexpression with each GATA factor were obtained using ACT (www.arabidopsis.leeds.ac.uk/ACT). Probe set identifications for GATA factors and genes coexpressed with each GATA factor were pasted into the Genevestigator MetaAnalyzer tool (Zimmermann et al., 2004) to identify tissues where each gene is expressed. Expression data were clustered using Java Treeview (http://sourceforge.net/projects/jtreeview) to produce Figure 5.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ875127 to DQ875134.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Gene structure information for Arabidopsis GATA genes.
Supplemental Table S2. Sequences of primers used for quantitative PCR analysis of GATA gene expression patterns.
Supplementary Material
Acknowledgments
We thank Denise Ashworth for DNA sequencing and Susan Marcus for growing and providing the Arabidopsis cell culture samples. We thank Tobias Joyce for assistance with BLAST searches to define the family and Ioannis Michalopoulos for plotting data producing Figure 5. We are grateful to the numerous researchers who contributed the array data upon which tools such as ACT and Genevestigator are dependent.
This work was supported by the European Union Framework V Regulatory Gene Initiative in Arabidopsis consortium and by the Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Philip M. Gilmartin (p.m.gilmartin@leeds.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49 525–555 [DOI] [PubMed] [Google Scholar]
- Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G (1996) White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J 15 1650–1657 [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44 680–692 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Ohlrogge JB (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128 223–235 [PMC free article] [PubMed] [Google Scholar]
- Borello U, Ceccarelli E, Giuliano G (1993) Constitutive, light-responsive and circadian clock-responsive factors compete for the different L-box elements in plant light-regulated promoters. Plant J 4 611–619 [DOI] [PubMed] [Google Scholar]
- Buzby JS, Yamada T, Tobin EM (1990) A light-regulated DNA-binding activity interacts with a conserved region of a Lemna gibba-rbcS promoter. Plant Cell 2 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré IA, Kay SA (1995) Multiple DNA-protein complexes at a circadian-regulated promoter element. Plant Cell 7 2039–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y (2006) Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol 7 R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana C, Garcia-Luque I, Alonso E, Malik VS, Cashmore AR (1988) Both positive and negative regulatory elements mediate expression of a photoregulated Cab gene from Nicotiana plumbaginifolia. EMBO J 7 1929–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou JL, Ingram R, Xue YB, Schwarz-Sommer Z, Davies B (2005) Evolution in action: following function in duplicated floral homeotic genes. Curr Biol 15 1508–1512 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38 332–347 [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F, Caboche M (1993) A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol Gen Genet 240 365–373 [DOI] [PubMed] [Google Scholar]
- Dean C, van den Elzen P, Tamaki S, Dunsmuir P, Bedbrook J (1985) Differential expression of the 8 genes of the Petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J 4 3055–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Sowinski DA, Hansen ER, Nguyen TT, Allen GC, Thompson WF (1997) Light effects on ferredoxin mRNA stability are mediated by translation. Plant Physiol 114 1213–1227 [Google Scholar]
- Donald RGK, Cashmore AR (1990) Mutation of either G-Box or I-Box sequences profoundly affects expression from the Arabidopsis Rbcs-1a promoter. EMBO J 9 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JCW, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Felsenfeld G (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58 877–885 [DOI] [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G (1988) An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci USA 85 5976–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Marzluf GA (1990) Site-directed mutagenesis of the zinc finger DNA-binding domain of the nitrogen-regulatory protein Nit2 of Neurospora. Mol Microbiol 4 1847–1852 [DOI] [PubMed] [Google Scholar]
- Gidoni D, Brosio P, Bond-Nutter D, Bedbrook J, Dunsmuir P (1989) Novel cis-acting elements in Petunia Cab gene promoters. Mol Gen Genet 215 337–344 [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein-binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford R, Coupland G, Laurie D (2003) The evolution of CONSTANS-like gene families in barley, rice and Arabidopsis. Plant Physiol 131 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob U, Stuber K (1987) Discrimination of phytochrome dependent light inducible from non-light inducible plant genes: prediction of a common light-responsive element (Lre) in phytochrome dependent light inducible plant genes. Nucleic Acids Res 15 9957–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci L, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden DA, Phillipson BA, Johnston KA, Brown L-A, Manfield IW, El-Shami M, Sparkes IA, Baker A (2006) Arabidopsis PEX19 is a dimeric protein that binds the peroxin PEX10. Mol Membr Biol 23 325–336 [DOI] [PubMed] [Google Scholar]
- Hansen ER, Petracek ME, Dickey LF, Thompson WF (2001) The 5′ end of the pea ferredoxin-1 mRNA mediates rapid and reversible light-directed changes in translation in tobacco. Plant Physiol 125 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Jen CH, Manfield IW, Michalopoulos I, Pinney JW, Willats WGT, Gilmartin PM, Westhead DR (2006) The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J 46 336–348 [DOI] [PubMed] [Google Scholar]
- Jeong MJ, Shih MC (2003) Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem Biophys Res Commun 300 555–562 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His -> Asp phosphorelay circuitry. Plant Cell Physiol 46 339–355 [DOI] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44 300–313 [DOI] [PubMed] [Google Scholar]
- Kozak M (2000) Do the 5′ untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics 70 396–406 [DOI] [PubMed] [Google Scholar]
- Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, Davies RW, Arst HN (1990) The regulatory gene AreA mediating nitrogen metabolite repression in Aspergillus nidulans—mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J 9 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus-35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Kano-Murakami Y, Gilmartin P, Niner B, Chua NH (1990) A metal-dependent DNA-binding protein interacts with a constitutive element of a light-responsive promoter. Plant Cell 2 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26 14–22 [DOI] [PubMed] [Google Scholar]
- Linden H, Macino G (1997) White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J 16 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Koizuka N, Martin RC, Nonogaki H (2005) The BME3 (blue micropylar end 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J 44 960–971 [DOI] [PubMed] [Google Scholar]
- Lowry JA, Atchley WR (2000) Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J Mol Evol 50 103–115 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Jen C-H, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR (2006) Arabidopsis co-expression tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res 34 W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Reynolds LA, Gittins J, Kneale GG (2000) The DNA-binding domain of the gene regulatory protein AreA extends beyond the minimal zinc-finger region conserved between GATA proteins. Biochim Biophys Acta 1493 325–332 [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH (1993) DNA-binding specificity of GATA family transcription factors. Mol Cell Biol 13 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning JC, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22 1006–1011 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163 [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang YL, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J 36 82–93 [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64 1402–1409 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM (1993) NMR structure of a specific DNA complex of Zn-containing DNA-binding domain of GATA-1. Science 261 438–446 [DOI] [PubMed] [Google Scholar]
- Orkin SH (1992) GATA-binding transcription factors in hematopoietic cells. Blood 80 575–581 [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15 1998–2006 [DOI] [PubMed] [Google Scholar]
- Persson S, Wei HR, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80 847–857 [DOI] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Sarokin LP, Chua NH (1992) Binding-sites for 2 novel phosphoproteins, 3AF5 and 3AF3, are required for Rbcs-3a expression. Plant Cell 4 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C (2000) The fungal GATA factors. Curr Opin Microbiol 3 126–131 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schindler U, Cashmore AR (1990) Photoregulated gene-expression may involve ubiquitous DNA-binding proteins. EMBO J 9 3415–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shikata M, Matsuda Y, Ando K, Nishii A, Takemura M, Yokota A, Kohchi T (2004) Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J Exp Bot 55 631–639 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and post-transcriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H (2003) Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J 36 550–564 [DOI] [PubMed] [Google Scholar]
- Teakle GR, Gilmartin PM (1998) Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem Sci 23 100–102 [DOI] [PubMed] [Google Scholar]
- Teakle GR, Kay SA (1995) The GATA-binding protein CGF-1 is closely related to GT-1. Plant Mol Biol 29 1253–1266 [DOI] [PubMed] [Google Scholar]
- Teakle GR, Manfield IW, Graham JF, Gilmartin PM (2002) Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol 50 43–57 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Martin DI, Zon LI, D'Andrea AD, Wong GG, Orkin SH (1989) Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339 446–451 [DOI] [PubMed] [Google Scholar]
- Umemura Y, Ishiduka T, Yamamoto R, Esaka M (2004) The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J 37 741–749 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YX, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM (2004) HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.