Abstract
A RING finger-containing protein (AO7) that binds ubiquitin-conjugating enzymes (E2s) and is a substrate for E2-dependent ubiquitination was identified. Mutations of cation-coordinating residues within AO7’s RING finger abolished ubiquitination, as did chelation of zinc. Several otherwise-unrelated RING finger proteins, including BRCA1, Siah-1, TRC8, NF-X1, kf-1, and Praja1, were assessed for their ability to facilitate E2-dependent ubiquitination. In all cases, ubiquitination was observed. The RING fingers were implicated directly in this activity through mutations of metal-coordinating residues or chelation of zinc. These findings suggest that a large number of RING finger-containing proteins, with otherwise diverse structures and functions, may play previously unappreciated roles in modulating protein levels via ubiquitination.
Modification with chains of ubiquitin (Ub) constitutes a primary mechanism by which proteins are targeted for proteasomal degradation (1). Ubiquitination involves the sequential action of Ub-activating enzyme (E1), Ub-conjugating enzymes (UBCs or E2s), and Ub protein ligases (E3s). This process initiates with formation of a thiol ester linkage between E1 and the C terminus of Ub, followed by transfer of Ub to the active-site Cys of an E2. There are at least 17 mammalian E2s, and these are characterized by a conserved 14- to 16-kDa core, with some having N- or C-terminal extensions. For the most part, formation of isopeptide bonds between the C terminus of Ub and lysines on substrates involves an additional protein or protein complex known as an E3. E3s recognize E2 and facilitate the transfer of Ub from E2 to substrate, in some cases forming thiol esters with Ub. E3s play important roles in catalyzing the formation of chains of Ub molecules on substrates (multiubiquitination or polyubiquitination) that are crucial for recognition by proteasomes.
Despite the large number of substrates, relatively few E3s have been characterized on a molecular level (1). E3s for which the amino acid sequences are known include the N end rule E3s of yeast and mammals (2) and members of the HECT (homologous to E6-AP C terminus) family. Mammalian HECT E3s include E6-AP, which targets p53 for ubiquitination in the presence of human papilloma virus E6 (3), and Nedd4, which ubiquitinates epithelial sodium channel subunits (4, 5). Other E3s include Mdm2, which catalyzes both its own ubiquitination and that of p53 (6); the anaphase-promoting complex (APC); and other F box and cullin-containing complexes whose substrates include Sic1p, G1 cyclins, IκB, and β-catenin (7).
In an attempt to identify additional Ub protein ligases, a member of a family of highly conserved human core E2s (UbcH5 family; refs. 8 and 9) was used in a yeast two-hybrid screen. This screen resulted in the identification of a previously uncharacterized RING finger protein, AO7, that undergoes ubiquitination in the absence of eukaryotic proteins other than E1 and E2. Characterization of AO7 led us to determine that six additional, otherwise-unrelated RING proteins also have the capacity for E2-dependent ubiquitination.
MATERIALS AND METHODS
Two-Hybrid Screen.
Saccharomyces cerevisiae HF7c was transfected with pGBT9-UbcH5B followed by 300 μg of pACT mouse T cell lymphoma library cDNA (CLONTECH). Transformants were selected according to the manufacturer’s instructions. An expressed sequence tag (GenBank accession no. W81753) that extended 5′ of the yeast two-hybrid isolate of AO7 was from Genome Systems (St. Louis). To define binding partners for AO7 in a yeast two-hybrid screen, W81753 was cloned into pGBT9 from EcoRI to PstI.
Generation of Plasmids.
AO7 in pACT was inserted from EcoRI to HindIII in pBluescript SK(+) (Stratagene) and then from XbaI to HindIII in glutathione S-transferase (GST) encoding pGEX-KG to generate GST-AO7. GST-AO7T was generated by digestion of GST-AO7 with KpnI and HindIII, followed by religation. GST-AO7T2-T6 was created by using restriction sites intrinsic to AO7 and ligation into pGEX vectors (Amersham Pharmacia). Hemagglutinin (HA)-tagged AO7 was generated by PCR by using W81753 as template with a 5′ oligonucleotide that encoded an EcoRI site, followed by an HA tag and an overlap with the predicted N terminus of W81753 and an antisense oligonucleotide beginning distal to an internal BamHI site. The product was digested with EcoRI and BamHI and inserted into similarly digested W81753. The insert was excised with EcoRI and NotI and inserted into pcDNA3 (Invitrogen). Praja1 was cloned into pcDNA3.1(+) from pGEX-Praja1 (10) by using EcoRI and NotI. An expressed sequence tag (GenBank accession no. AA429297) encoding amino acids 540–686 of kf-1 was subcloned into pGEX-4T-2 from EcoRI to NotI. An expressed sequence tag (GenBank accession no. AA166821) encoding amino acids 477–665 of TRC8 was used as template for PCR, and the product was cloned into pGEX5X-1 from EcoRI to NotI. A fragment of the NF-X1 cDNA (11) encoding amino acids 207–645 was cloned into pGEX-KG. cDNA encoding amino acids 1–788 of BRCA1 was cloned into pGEX5X-1 as a BamHI fragment. Coding regions of pcDNA3-Flag-Siah-1 and Flag-RING-deleted Siah-1 (12) were cloned into pGEX4T-1 by using EcoRI and a 3′ blunt end ligation. GST-Nedd4 and GST-E6AP have been described (9). Site-specific mutagenesis was carried out with the Stratagene Quick Change kit. Oligonucleotide sequences are available on request.
Binding and Functional Studies.
GST fusion proteins were expressed in log-phase Escherichia coli BL-21(DE3) (Novagen) induced with 0.1 mM isopropyl β-d-thiogalactoside for 1 h at 22°C. Bacterial pellets were resuspended in 50 mM Tris, pH 7.4/1 mM EDTA/1% Triton X-100/5 mM DTT/2 mM PMSF (sonication buffer), lysed by probe sonication followed by clarification at 28,000 × g, and stored at −70°C. Levels of GST fusion proteins were estimated by binding sonicates to glutathione Sepharose (GS), washing, and quantification by Coomassie Blue by using BSA as standards. Relative quantification by immunoblotting with anti-GST was in agreement with estimates based on Coomassie Blue.
For binding and functional studies with GS-bound material, fusion proteins (20 pmol, except where noted) were bound to GS followed by washing in PBS. Ubiquitination assays with GS-bound material, unless indicated, were carried out by using 20 ng each of wheat E1 (13) and UbcH5B (8) expressed in E. coli. The ubiquitination reaction buffer contained 50 mM Tris (pH 7.4), 0.2 mM ATP, 0.5 mM MgCl2, 0.1 mM DTT, 1 mM creatine phosphate, 15 units of creatine phosphokinase, and 2 μl of bacterial lysate from BL-21 transformed with pET15B. Reactions (30 μl) were done for 1.5 h at 30°C with 2 × 104 cpm of 32P-labeled Ub. GST-Ub was 32P-labeled on GS (14), followed by thrombin cleavage and depletion with benzamidine Sepharose (Amersham Pharmacia). Samples in β-mercaptoethanol-containing buffer were resolved by SDS/PAGE and analyzed with a Storm PhosphorImager (Applied Biosystems). Anti-Ub (15) immunoblotting experiments were carried out as above, except that 0.3 μg of wild-type Ub or Ub with no lysines (UbK0) (a gift from C. Pickart, Johns Hopkins University, Baltimore, MD) was used. [35S]methionine-labeled UbcH5B was generated by in vitro transcription and translation in wheat germ by using TnT kits (Promega). Binding studies were carried out by tumbling 105 cpm of UbcH5B with GS-bound proteins for 16 h at 4°C in 25 mM Tris, pH 7.4/50 mM NaCl/5 mM DTT/0.5% Nonidet P-40 followed by washing at 4°C in the same buffer.
For chelation experiments, GS-bound proteins were incubated for 18 h at 4°C with three changes of PBS plus 5 mM of EDTA, disodiumtriaminopentaacetic acid (DTPA), or tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) (Sigma). For zinc reconstitution experiments, TPEN-chelated GST-AO7T was washed with PBS and then with 50 mM Tris (pH 7.4) before addition of ZnCl2 for 3 h at 4°C, followed by sequential washing with 50 mM Tris and PBS.
Transfections of and immunoprecipitations from COS cells were performed as described (9); 12CA5 (anti-HA; Roche Biomolecular Chemicals) was used for immunoprecipitations. Immunoblotting was performed with 12CA5, anti-Ub, or rabbit polyclonal anti-UbcH5B (1388).
Praja1, Siah-1, and mutated Siah-1 were labeled with [35S]Met by in vitro translation by using TnT. Translation reactions of Praja1 (2 μl) or Siah-1 (6 μl) were evaluated for ubiquitination in 50 mM Tris⋅HCl, pH 7.4/5 mM MgCl2/2 mM ATP/2 mM DTT/100 ng of E1/100 ng of UbcH5B/10 μg of Ub (Sigma). Reactions were for 1 h at 30°C.
RESULTS
Isolation of a UbcH5B Binding Partner.
By using cDNA encoding UbcH5B in a yeast two-hybrid screen of a murine T cell library, a cDNA that encodes a protein (AO7) of 440 aa was isolated. Analysis of murine tissues identified a ubiquitously expressed ≈1.5-kilobase mRNA (not shown). A 1,490-nt GenBank expressed sequence tag was characterized that encoded the entire amino acid sequence found in the yeast two-hybrid isolate with an additional 16 aa at the N terminus, including a translation start site. A RING consensus sequence (16) is evident between amino acids 134 and 198; otherwise, AO7 bears no overall homology to known proteins. To isolate proteins with which AO7 interacts, a cDNA encoding the ORF was used in a second yeast two-hybrid screen. Five clones were obtained, all of which are murine E2s, including UBCM4 (murine UbcH5B), UBCM2, and UBCM3.
AO7 Undergoes E2-Dependent Ubiquitination.
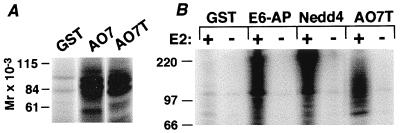
To determine whether AO7 might have a role in ubiquitination, an in vitro assay was used (8, 9). This assay employs recombinant proteins expressed in E. coli that do not express components of the Ub-conjugating system, avoiding issues related to contaminating or copurifying eukaryotic proteins such as E3s. Proteins being evaluated were expressed as GST fusions and isolated by binding to GS. 32P-labeled recombinant Ub and recombinant E1 and E2 (UbcH5B) in E. coli lysate were added. GST fusion proteins serve as potential substrates, as do added bacterial proteins. Evaluation of GST-AO7 (Fig. 1A) revealed substantial ubiquitination above the GST background. A GST fusion of a C-terminal truncation of AO7 at amino acid 363 (GST-AO7T) also was ubiquitinated (Fig. 1A, right lane). Because GST-AO7 is poorly expressed, GST-AO7T was used for subsequent analyses. As with GST fusions of HECT E3s, ubiquitination in the presence of GST-AO7T was E2-dependent (Fig. 1B). Other UbcH5 family members also functioned with AO7 (not shown). For AO7, as for the well characterized HECT E3s, when the soluble and GS fractions from ubiquitination reactions are separated, the large majority of the ubiquitinated material was GS-bound (not shown), indicative of addition of multiple Ub molecules to the fusion proteins. Thrombin cleavage of GST-AO7 showed that residues both on the GST moiety and within AO7 are ubiquitinated (not shown). Thus, the E2-dependent ubiquitination is not limited to the AO7 portion of the fusion protein.
Figure 1.
AO7 is involved in E2-dependent ubiquitination in vitro. (A) Equimolar amounts of the indicated GS-bound fusion proteins or GST alone were incubated in the presence of E1, E2 (UbcH5B), proteins from bacterial lysates, and 32P-labeled Ub for 1.5 h at 30°C. The entire reaction was resolved under reducing conditions by SDS/PAGE. (B) GST fusion proteins were incubated as described for A in the absence or presence of E2 (UbcH5B). GST fusions of AO7T, Nedd4, and E6-AP migrate at approximately 69, 120, and 115 kDa, respectively.
Defining a Region Required for E2-Dependent Ubiquitination.
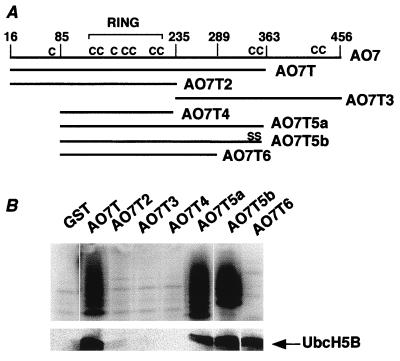
A series of N- and C-terminal truncations of AO7 were generated as GST fusion proteins (Fig. 2). The minimal region that manifested both E2-dependent ubiquitination and E2 binding spans amino acids 85–363 (AO7T5a). When AO7T5a was truncated at amino acid 289 (AO7T6), E2 binding was maintained, but no significant ubiquitination was detected (Fig. 2B, right lane). These findings indicate that sequences C-terminal to amino acid 289 are required for ubiquitination but not for E2 binding.
Figure 2.
A region required for E2-dependent ubiquitination. (A) Schematic representation of AO7 and AO7 truncations fused to GST at their N termini. The RING and positions of cysteines (C) are indicated. (B) Equimolar amounts of the indicated GST fusions were evaluated for ubiquitination by using 32P-labeled Ub (Upper) or for binding to 35S-labeled UbcH5B (Lower). Upper and Lower are from separate experiments.
AO7 Ubiquitination Depends on an Intact RING-H2 Finger.
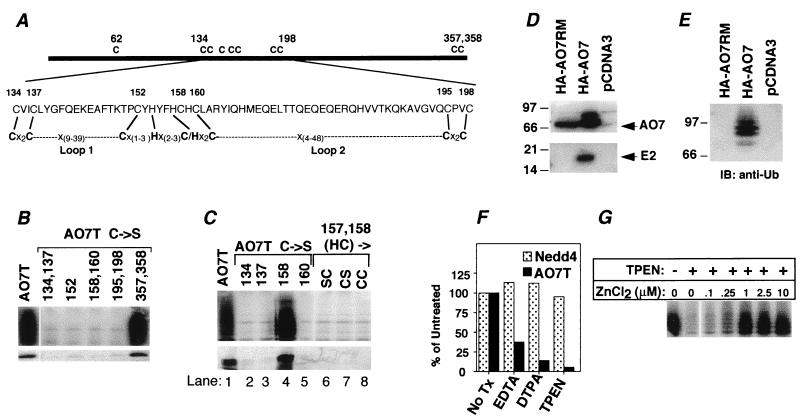
Because the HECT E3s require an active-site Cys for their activity (1) and because AO7 includes a RING consensus sequence, the requirements for Cys residues in the activity of AO7T were evaluated. AO7T has 10 cysteines; all but one (C62) are retained in AO7T5a. In addition to the seven cysteines in the RING (Fig. 2A), AO7T5a has two cysteines (C357 and C358) near its C terminus. When both of these were mutated to Ser (AO7T5b), neither E2-dependent ubiquitination nor E2 binding was affected adversely (Fig. 2B, second lane from right). Cysteines within the RING (see Fig. 3A for AO7 RING sequence) were evaluated by mutation to Ser (C → S), either individually (C152) or in pairs (C134 and 137; C158 and C160; C195 and C198). All of these resulted in loss of both ubiquitination (Fig. 3B Upper) and E2 binding (Fig. 3B Lower), establishing a requirement for at least four RING cysteines. Cysteines outside of the RING were required neither for activity nor for E2 binding (Fig. 3B and not shown). These findings suggest that the RING finger structure itself may be crucial for the E2 binding and ubiquitination seen with AO7.
Figure 3.
Ubiquitination of AO7 depends on RING-H2 and zinc. (A) Schematic of AO7 and RING-H2 domain. Cysteines are numbered. Eight residues predicted to be crucial to Zn2+ coordination are indicated by lines connecting AO7 and the RING consensus (16). Variable loops between the second and third (Loop 1) and between the sixth and seventh (Loop 2) predicted Zn2+ coordinating residues are indicated. (B and C) GST fusion proteins were assayed for ubiquitination (Upper) and binding of labeled UbcH5B (Lower). Numbers above lanes denote residues that were mutated. (C, lanes 6–8) Labeling indicates amino acids to which H157 and C158 were changed. Upper and Lower are from separate experiments. (D) COS-7 cells were transfected with plasmid encoding HA-tagged AO7 (HA-AO7), a RING mutant (HA-AO7RM), or empty vector (pcDNA3). HA-tagged proteins were immunoprecipitated and resolved by SDS/12% PAGE. The membrane was probed with anti-HA (Upper) or polyclonal anti-UbcH5B (Lower). (E) Anti-HA immunoprecipitates of cells transfected as described for D were resolved by SDS/8% PAGE. The membrane was probed with anti-Ub. (F) GS-bound GST-AO7T and GST-Nedd4 were incubated with the indicated chelators before assaying ubiquitination. Data are normalized to samples not exposed to chelators. (G) TPEN chelation of GS-bound GST-AO7T was followed by incubation with ZnCl2 for 3 h. GS was washed extensively before evaluation of ubiquitination.
RINGs, defined by eight cysteines and histidines that coordinate two zinc ions (see Fig. 3A for consensus sequence), vary substantially in length and composition. RINGs have cysteines in the first three and last three coordination sites and a His in the fourth site. Additionally, proteins bearing this motif have either a Cys [C3HC4 RING (RING-HC)] or a His [C3H2C3 RING (RING-H2)] in the fifth position. The first, second, fifth, and sixth cysteines/histidines coordinate one cation, and the third, fourth, seventh, and eighth coordinate the second (16). As determined with consensus sequences, AO7 falls into the RING-H2 category, with the amino acids indicated predicted to be coordination sites (Fig. 3A). Thus, if the RING provides the physical basis for E2 binding and ubiquitination, then mutation of either C134 or C137 should result in loss of ubiquitination. For the next closely spaced pair of cysteines (C158 and C160), C160 is predicted to be the sixth coordination site, but H157 rather than C158 is predicted to be the fifth site. Consistent with the RING-H2 being crucial for this activity, mutation of C134, C137, C160, or H157 resulted in loss of both ubiquitination and E2 binding (Fig. 3C, lanes 2, 3, 5, and 6), whereas mutation of C158 affected neither (Fig. 3C, lane 4). Notably, mutation of H157 to Cys, creating a potential RING-HC, also abrogated both ubiquitination and E2 binding, regardless of whether C158 was retained or converted to Ser (Fig. 3C, lanes 7 and 8). These data establish that AO7’s ubiquitination and E2 binding both depend on an intact RING-H2 finger.
RING Finger-Dependent E2 Association and Ubiquitination of AO7 in Cells.
To determine whether AO7 binds E2s in a RING-dependent manner in cells, COS-7 cells were transfected either with plasmids encoding full-length AO7 fused to a HA N-terminal tag (HA-AO7) or with a plasmid in which C134 and C137 were changed to Ser (HA-AO7RM). Consistent with in vitro data, wild-type HA-AO7, but not the RING mutated form, coimmunoprecipitated endogenous E2s recognized by polyclonal anti-UbcH5B (Fig. 3D Lower). When the same membrane was immunoblotted with anti-HA, bands corresponding to HA-AO7 and HA-AO7RM were evident (Fig. 3D Upper). Additional species above the predominant band were apparent with only HA-AO7. These were determined to represent ubiquitinated AO7 by resolution on a lower percentage gel and anti-Ub immunoblotting (Fig. 3E). Reprobing of blots with anti-HA confirmed that these represented ubiquitinated HA-AO7 (not shown). Thus, AO7 undergoes RING-dependent ubiquitination in cells.
A Role for Zinc in AO7 Ubiquitination.
A requirement for divalent cations in the ubiquitination seen with AO7 was established by the loss of activity that accompanied chelation of divalent cations, with the more potent chelators, DTPA and TPEN, having greater effects than EDTA (Fig. 3F). Notably, cation chelation did not inhibit Nedd4, a HECT E3. To determine whether Zn2+ reconstitutes activity, GS-bound AO7 was treated with TPEN, followed by incubation with ZnCl2. Restoration of ubiquitination was observed, establishing a role for Zn2+ in AO7’s ubiquitination (Fig. 3G).
Unrelated RING-H2 Finger Proteins Mediate E2-Dependent Ubiquitination.
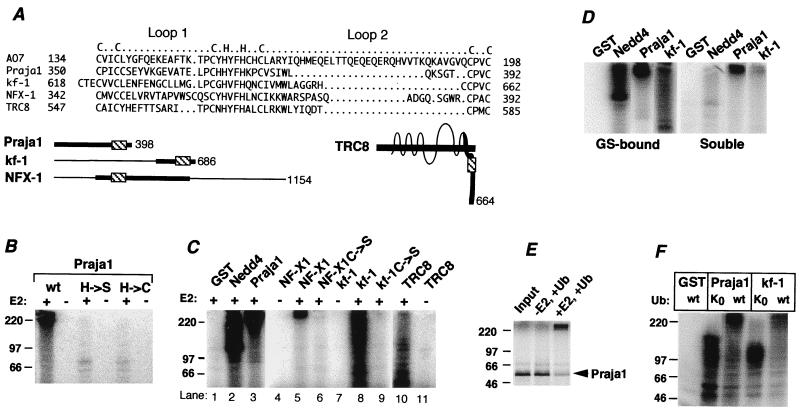
The results obtained with AO7 led us to address the possibility that RING-H2-containing proteins in general have the intrinsic capacity for ubiquitination. To evaluate this possibility, four RING-H2 proteins that are otherwise unrelated to AO7 and for which there was no a priori reason to suspect roles in ubiquitination were selected for evaluation. Praja1 is a 398-aa protein of unknown function (Fig. 4A; ref. 10). The eighth coordination site of the Praja1 RING-H2 is within 7 aa of its C terminus. This location is of note given the dependence of AO7’s activity on sequences distal to the RING. Strikingly, full-length GST-Praja1 manifests substantial E2-dependent ubiquitination (Fig. 4B, right two lanes). Mutation of the His in the fifth coordination position to Ser (H → S) resulted in loss of activity. As with AO7, mutation of this His to Cys (H → C), creating a potential RING-HC, still resulted in loss of ubiquitination (Fig. 4B).
Figure 4.
Evaluation of four RING-H2 proteins. (A) Alignment of RING-H2 regions of proteins. Loops and predicted coordination sites are indicated above. kf-1 has two possible alignments beginning with either C618 or C621; the alignment shown begins with C621. Full-length forms of proteins are schematized below. Heavy lines indicate regions fused to GST (Praja1 is full length), and RING finger regions are indicated (▧). TRC8 is a multispanning membrane protein. (B) GST-Praja1 and mutants of the fifth putative coordination site to either Ser (H → S) or Cys (H → C) were evaluated for ubiquitination. (C) GST fusions of the indicated proteins were evaluated as described for B. The kf-1, NF-X1, and TRC8 fusions included amino acids 538–686, 207–645, and 477–664, respectively. The C → S mutation of kf-1 was at amino acid 621; NF-X1 C → S was a double mutation at C342 and C345. Because of difficulty expressing GST-NF-X1, only 5 pmol was used, and 20 pmol of NF-X1 C → S was used. (D) Soluble material was removed from ubiquitination reactions carried out as described for C, and GS beads were washed extensively. Both fractions were resolved by SDS/PAGE. (E) 35S-labeled Praja1 was resolved directly on SDS/PAGE (Input) or subjected to a ubiquitination reaction in E1-containing reaction buffer with or without UbcH5B. (F) GS-bound proteins were evaluated for ubiquitination with either wild-type Ub or UbK0. After Western transfer, membranes were immunoblotted with anti-Ub. GST-Praja1 and GST-kf-1 migrate at approximately 85 kDa and 50 kDa, respectively.
GST fusions of RING-H2 regions from three other proteins were evaluated (see Fig. 4A for RING alignment and schematic of proteins evaluated). NF-X1 is a 1,154-aa protein that represses MHC class II gene expression (11). kf-1 is a 686-aa protein postulated to be involved in membranous protein sorting (17). TRC8 is a 664-aa multispanning membrane protein with similarity to Patched (18). All of these GST fusions showed E2-dependent ubiquitination (Fig. 4C). Mutation of RING cysteines was carried out for NF-X1 (lane 6), kf-1 (lane 9), and TRC8 (not shown). In all cases, E2-dependent ubiquitination was lost. As shown for kf-1 and Praja1 as well as Nedd4, most of the ubiquitinated material was GS associated, indicative of ubiquitination that is occurring predominantly on the fusion proteins (Fig. 4D). Similar results were obtained with TRC8 and NF-X1 (not shown). E2-dependent ubiquitination of the GST fusions was also observed when reactions were carried out without GS immobilization (data not shown). To assess whether ubiquitination occurs in the absence of the GST moiety, 35S-labeled full-length Praja1 was generated by in vitro translation in reticulocyte lysate and assessed for ubiquitination. When E2 was added, a substantial majority (≈80%) of Praja1 underwent a striking change in migration to the top of the gel, indicative of E2-dependent ubiquitination (Fig. 4E).
Recognition of ubiquitinated proteins by proteasomes requires multi-Ub chains (1). To determine whether RING fingers promote multi-Ub chain formation, reactions with GST-Praja1 and GST-kf-1 were carried out by using either wild-type Ub or a form of Ub that contains no lysines (UbK0) and therefore does not allow for multiubiquitination. As determined by anti-Ub immunoblotting, ubiquitination was seen with both Ub and UbK0. However, significant levels of species extending to the top of the gel were seen for both GST-Praja1 and GST-kf-1 with only wild-type Ub (Fig. 4F). These findings show that the interaction of E2s with otherwise-unrelated RING-H2 proteins can result in multi-Ub chains.
RING-HC Finger Proteins Mediate Ubiquitination.
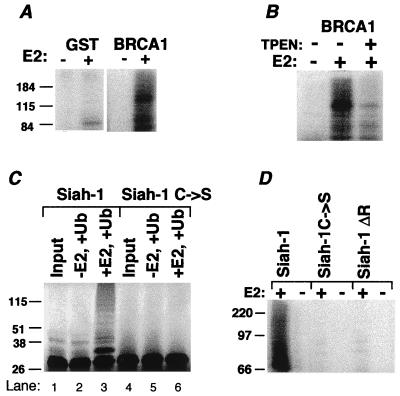
Two RING-HC finger proteins were evaluated for their capacity to facilitate E2-dependent ubiquitination. BRCA1 (19) manifests a RING-HC finger near its N terminus. When the first 788 aa of BRCA1 was expressed as a GST fusion (Fig. 5A), E2-dependent ubiquitination was observed. Consistent with RING-dependence, TPEN markedly diminished ubiquitination (Fig. 5B). Siah-1 is a 324-aa protein that targets both itself and the deleted in colorectal cancer (DCC) gene product for proteasomal degradation, which depends on an intact Siah RING (12, 20). To test whether Siah-1 undergoes ubiquitination, Siah-1 and a mutant in the second putative Zn2+ coordination site (Siah-1C → S) were evaluated after in vitro translation (Fig. 5C). Bands indicative of ubiquitinated Siah-1 were seen only in the presence of E2 and only for wild-type Siah-1 (Fig. 5C, lane 3). To confirm that this ubiquitination does not require other eukaryotic proteins present in the translation mixture, GST fusions were assessed. E2-dependent ubiquitination was detected for wild-type Siah-1 but not for the C → S mutant or for a RING-deleted form (Siah-1ΔR) (Fig. 5D). These findings suggest that Siah-1 has the capacity for RING and E2-dependent ubiquitination.
Figure 5.
RING-HC proteins are also involved in ubiquitination. (A) A GST fusion of amino acids 1–788 of mouse BRCA1 was evaluated for ubiquitination. (B) GS-bound GST-BRCA1 was incubated with (+) or without (−) TPEN, followed by washing of GS before a ubiquitination reaction. (C) Flag-tagged Siah-1 (Siah-1) or a mutant in the third coordination site (Siah-1 C → S) was translated with [35S]Met in wheat germ lysate. Material was resolved directly on SDS/PAGE (Input) or evaluated for ubiquitination with or without UbcH5B. (D) GST fusions of Siah-1, the C → S Siah-1 mutant, or RING-deleted (Siah-1ΔR; ref. 12) were evaluated for ubiquitination.
DISCUSSION
The results presented in this study provide strong evidence that the RING finger, in the appropriate molecular context, is a module that interacts with E2s and facilitates ubiquitination. The assays employed here use the test proteins themselves, either RING finger proteins or HECT E3s, as the predominant ubiquitination substrates. Whether any of the RING finger proteins evaluated in this study also function as E3s or components of E3s for specific heterologous proteins remains to be determined. However, there are now several examples where RINGs are included in proteins or protein complexes whose primary known function is to serve as E3s. RING-containing E3s include the N end rule E3s of yeast (Ubr1p), mammals (E3α; ref. 2), and Apc11p (21). For none of these has the significance of the RING been established. More recently, Rbx1/ROC1/Hrt1, which is 35% identical to Apc11p, has been shown to be a component of cullin-containing E3s for cyclins, Sic1p, and IκB (22–26). In addition, we now know that the E3 activity of Mdm2 depends on zinc and multiple metal-coordinating RING residues (S.F., J.P.J., K. H. Vousden, and A.M.W., unpublished work). We suggest that the finding of RINGs as components of several E3s is neither a coincidence nor simply a reflection of overall similarity between proteins such as Apc11 and Rbx1. Rather, our observation that multiple, otherwise-unrelated, RING-containing proteins all mediate E2-dependent ubiquitination makes it likely that a substantial number of other RING-containing proteins will eventually be found to be E3 components.
There are several hundred cDNAs encoding RING finger proteins in the GenBank database. For some of these, such as the TRAFs (27), IAPs (28), Cbl (29–31), Hrd1p/Der3p (32, 33), and Herpes Simplex Virus Vmw110 (34, 35), there are data to suggest roles associated with ubiquitination or regulated degradation. For these, the possible role of RING-mediated ubiquitination now awaits determination. Most RING finger proteins have either unknown functions or functions not obviously related to ubiquitination, as is the case for the majority of the proteins evaluated in this study. For example, TRC8 is a member of the sterol-sensing Patched family (18), and both NF-X1 and BRCA1 have RING-independent roles in transcription (11, 36). Although a role for the RING finger of RAG1 in ubiquitination is yet to be assessed, RAG1’s RING finger is not required for its recombinase activity (37). For RING finger proteins with functions not apparently related to ubiquitination, an attractive possibility is that RING-mediated ubiquitination targets the RING-containing protein or associated proteins for degradation in a regulated manner. One can envisage that, in vivo, changes in conformation, alterations induced by phosphorylation, and changes in intermolecular associations such as dimerization may influence the capacity of RING finger proteins to function with E2s in ubiquitination. In this way, the compact RING module could confer an additional regulatory role on a wide array of eukaryotic proteins.
Of potential significance regarding BRCA1 is the finding of a deubiquitinating enzyme associated with its N terminus, which has led to speculation that BRCA1 may be ubiquitinated in vivo (38). As RING mutations of BRCA1 are associated with familial carcinomas (39), it now becomes important to determine the physiological significance of ubiquitination mediated through BRCA1’s RING.
A question that arises now is how RING finger proteins function with E2s to catalyze ubiquitination. One possibility is that, analogous to HECT E3s, RING-containing proteins both bind E2 and form catalytic thiol ester intermediates with Ub. However, for AO7, only those cysteines predicted to coordinate zinc are required for function, making this possibility less likely. Also against this mechanism is the partial resistance of AO7 and the total resistance of Praja1 and Hrt1 to concentrations of an alkylating agent that fully inactivates HECT E3s (J.P.J. and A.M.W., unpublished data, and ref. 26).
Alternatively, the RING may provide a site of interaction with E2 that allows for the direct transfer of Ub from E2 to target lysines. However, for a RING-containing AO7 truncation (AO7T6), E2 binding is not sufficient to support ubiquitination. Furthermore, some of the RING finger proteins for which the highest levels of ubiquitination are seen show little detectable E2 binding under conditions in which E2 binding by AO7 is readily detectable (K.L.L., J.P.J., S.F., and A.M.W., unpublished data). Thus, the degree of E2 binding does not seem to be a direct predictor of activity. Therefore, we favor a model in which the RING and surrounding regions not only associate with E2-Ub but also provide a favorable environment for the transfer of Ub from E2 to an available lysine. Such a mechanism is analogous to the mechanism in models for the function of N end rule E3s. (1).
Despite the large number of proteins known to be ubiquitinated, the identification of Ub protein ligases has proven difficult (1). Our findings suggest that the RING finger, a discrete module intrinsic to a large number of proteins, confers on proteins a capacity for E2-dependent ubiquitination. Some of these RING finger proteins likely function primarily as E3s. However, in other instances, RING finger-dependent ubiquitination likely provides proteins that have other important cellular roles with a previously unappreciated self-regulatory function.
Acknowledgments
We thank E. Fearon, P. Howley, G. Hu, J. Huibregtse, B. Mishra, L. Mishra, H. Ono, S. Ono, C. Pickart, S. Sharan, and J. You for reagents and our colleagues in the National Institutes of Health Intramural program for discussions and critical reviews of this manuscript. S.H. was supported in part by the Japanese Society for the Promotion of Science. A.M.O. was supported by the Howard Hughes Medical Institute National Institutes of Health Scholars Program.
ABBREVIATIONS
- Ub
ubiquitin
- HECT
homologous to E6-AP C terminus
- APC
anaphase-promoting complex
- GS
glutathione Sepharose
- GST
glutathione S-transferase
- HA
hemagglutinin
- DTPA
disodiumtriaminopentaacetic acid
- TPEN
tetrakis(2-pyridylmethyl)ethylenediamine
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF171060).
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Kwon Y T, Reiss Y, Fried V A, Hershko A, Yoon J K, Gonda D K, Sangan P, Copeland N G, Jenkins N A, Varshavsky A. Proc Natl Acad Sci USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 4.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 5.Dinudom A, Harvey K F, Komwatana P, Young J A, Kumar S, Cook D I. Proc Natl Acad Sci USA. 1998;95:7169–7173. doi: 10.1073/pnas.95.12.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koepp D M, Harper J W, Elledge S J. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 8.Jensen J P, Bates P W, Yang M, Vierstra R D, Weissman A M. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 9.Hatakeyama S, Jensen J P, Weissman A M. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 10.Mishra L, Tully R E, Monga S P, Yu P, Cai T, Makalowski W, Mezey E, Pavan W J, Mishra B. Oncogene. 1997;15:2361–2368. doi: 10.1038/sj.onc.1201405. [DOI] [PubMed] [Google Scholar]
- 11.Song Z, Krishna S, Thanos D, Strominger J L, Ono S J. J Exp Med. 1994;180:1763–1774. doi: 10.1084/jem.180.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield P M, Callis J, Vierstra R D. J Biol Chem. 1990;265:15813–15817. [PubMed] [Google Scholar]
- 14.Scheffner M, Huibregtse J M, Howley P M. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cenciarelli C, Wilhelm K J, Jr, Guo A, Weissman A M. J Biol Chem. 1996;271:8709–8713. doi: 10.1074/jbc.271.15.8709. [DOI] [PubMed] [Google Scholar]
- 16.Saurin A J, Borden K L, Boddy M N, Freemont P S. Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 17.Yasojima K, Tsujimura A, Mizuno T, Shigeyoshi Y, Inazawa J, Kikuno R, Kuma K, Ohkubo K, Hosokawa Y, Ibata Y, et al. Biochem Biophys Res Commun. 1997;231:481–487. doi: 10.1006/bbrc.1996.6033. [DOI] [PubMed] [Google Scholar]
- 18.Gemmill R M, West J D, Boldog F, Tanaka N, Robinson L J, Smith D I, Li F, Drabkin H A. Proc Natl Acad Sci USA. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 20.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J, Mann M, Nasmyth K. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 22.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G J, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 23.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 24.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z Q. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 25.Ohta T, Michel J J, Schottelius A J, Xiong Y. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 26.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duckett C S, Thompson C B. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu Z L, McKinsey T A, Liu L, Gentry J J, Malim M H, Ballard D W. Proc Natl Acad Sci USA. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Yeung Y G, Langdon W Y, Stanley E R. J Biol Chem. 1996;271:17–20. doi: 10.1074/jbc.271.1.17. [DOI] [PubMed] [Google Scholar]
- 30.Miyake S, Lupher M L J, Druker B, Band H. Proc Natl Acad Sci USA. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bordallo J, Plemper R K, Finger A, Wolf D H. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampton R Y, Gardner R G, Rine J. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson J, Lees-Miller S P, Everett R D. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everett R D, Earnshaw W C, Findlay J, Lomonte P. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 37.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen D E, Proctor M, Marquis S T, Gardner H P, Ha S I, Chodosh L A, Ishov A M, Tommerup N, Vissing H, Sekido Y, et al. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 39.Brzovic P S, Meza J, King M C, Klevit R E. J Biol Chem. 1998;273:7795–7799. doi: 10.1074/jbc.273.14.7795. [DOI] [PubMed] [Google Scholar]