Abstract
Parthenogenetic embryonic stem (ES) cells with two oocyte-derived genomes (uniparental) have been proposed as a source of autologous tissue for transplantation. The therapeutic applicability of any uniparental cell type is uncertain due to the consequences of genomic imprinting that in mammalian uniparental tissues causes unbalanced expression of imprinted genes. We transplanted uniparental fetal liver cells into lethally irradiated adult mice to test their capacity to replace adult hematopoietic tissue. Both maternal (gynogenetic) and paternal (androgenetic) derived cells conveyed long-term, multilineage reconstitution of hematopoiesis in recipients, with no associated pathologies. We also establish that uniparental ES cells can differentiate into transplantable hematopoietic progenitors in vitro that contribute to long-term hematopoiesis in recipients. Hematopoietic tissue in recipients maintained fidelity of parent-of-origin methylation marks at the Igf2/H19 locus; however, variability occurred in the maintenance of parental-specific methylation marks at other loci. In summary, despite genomic imprinting and its consequences on development that are particularly evident in the androgenetic phenotype, uniparental cells of both parental origins can form adult-transplantable stem cells and can repopulate an adult organ.
Keywords: Androgenetic, gynogenetic, parthenogenetic, imprinting, stem cell, hematopoietic
Normal mammalian development requires both an oocyte- and a sperm-derived genome. This is exemplified by the failure of parthenogenesis in mammals, when an unfertilized oocyte commences development, but does not form a viable fetus in the absence of a male genome. Embryos with two genomes from the same sex that are generated by the exchange of maternal and paternal pronuclei between zygotes have similarly limited developmental potential (McGrath and Solter 1984; Surani et al. 1984). Androgenetic (AG) embryos with two paternal genomes can form blastocysts and implant, but rarely develop to or past early somite stages, with abundant trophoblast but retarded development of the embryo proper (Barton et al. 1984). Diploid parthenogenetic (PG) embryos with two genomes from the same oocyte and developmentally equivalent gynogenetic (GG) embryos with two maternal genomes from different oocytes can occasionally develop to later somite stages but typically lack extraembryonic tissues (Kaufman et al. 1977; Surani and Barton 1983). The inability of PG/GG and AG embryos to form viable fetuses is attributed to the functional nonequivalency of maternal and paternal genomes, which results from epigenetic modifications that occur during germ cell development and lead to parental allele-specific expression of imprinted genes. PG/GG and AG embryos can, however, complete preimplantation development and can give rise to pluripotent embryonic stem (ES) cell lines (Robertson et al. 1983; Mann et al. 1990). When combined with normal fertilized embryos to form chimeras, cells of PG/GG and AG origin can contribute to tissues of fetal and postnatal mammals as observed in the mouse (Stevens et al. 1977; Surani et al. 1977; Paldi et al. 1989; Mann et al. 1990; Barton et al. 1991; Mann and Stewart 1991; Allen et al. 1994), including the germline (Stevens 1978; Narasimha et al. 1997); bovine (Boediono et al. 1999); and, in a single reported case, human (Strain et al. 1995). PG/GG and AG cells exhibit different capacities to participate in chimera development, with a parent-of-origin-related bias in distribution of cells to tissues (Surani et al. 1987; Fundele et al. 1989, 1990, 1995; Nagy et al. 1989; Barton et al. 1991), and the formation of specific phenotypes that correlate with abnormal expression of imprinted genes (Allen et al. 1994; Eggenschwiler et al. 1997; McLaughlin et al. 1997). Androgenetic cells cause severe defects and lethality in chimeras (Mann et al. 1990; Barton et al. 1991; McLaughlin et al. 1997) and exhibit a propensity for malignant transformation in vitro (Hernandez et al. 2003). Conversely, parthenogenetic cells often exhibit a reduced ability to proliferate (Nagy et al. 1989; Fundele et al. 1990; Jagerbauer et al. 1992; Hernandez et al. 2003).
Determining the full extent of contribution and functionality that uniparental cells could potentially attain in adult tissues is not feasible using adult developmental chimeras as a model, due to the restriction or lethality that is associated with uniparental cells during development. Ascertaining whether cells of uniparental origin can function normally in an adult, however, is highly relevant to the potential therapeutic use of PG ES cells (Cibelli et al. 2002). Therefore, to test the capacity of uniparental maternal and paternal cells to engraft into adults and form functional cells in adult tissues, we transplanted fetal liver cells from AG and GG ES cell chimeras into lethally irradiated adult mice. Normal mammalian fetal liver, at a stage often attained by uniparental chimeras, contains hematopoietic stem cells (HSC) capable of long-term, multilineage reconstitution of the hematopoietic system of adults (Morrison et al. 1995). Here we report that both androgenetic and gynogenetic fetal liver cells from mouse chimeras engraft in adult recipients, and can entirely populate hematopoiesis from the uniparental component, providing long-term, multilineage reconstitution. Uniparental ES cells also exhibit hematopoietic differentiation capacity in vitro, and when genetically modified to express the HoxB4 gene (Kyba et al. 2002), can engraft in lethally irradiated adults and contribute to long-term hematopoiesis.
Results
Fetal livers from uniparental chimeric mice repopulate hematopoiesis in adult recipients
To obtain gynogenetic and androgenetic fetal liver cells for transplantation into adult mice, we first produced GG and AG embryos, from which we derived ES cell lines. We then generated chimeras by injection of AG and GG ES cells into blastocysts, and recovered fetal liver cells for transplantation from developing (13.5–14.5 d of gestation = days post-coitum, dpc) chimeras (Fig. 1A). Both GG and AG embryos were transgenic for eGFP (Okabe et al. 1997), such that AG and GG cells could be identified by eGFP fluorescence in fetal chimeras and, after transplantation, in adult recipients. The contribution and differentiation phenotype of the uniparental ES cells in chimeras and teratomas (Supplementary Table 1; Supplementary Fig. 1) were consistent with previous observations in AG and PG ES cells and chimeras (Mann et al. 1990; Allen et al. 1994; Szabo and Mann 1994) and validated their use for this study.
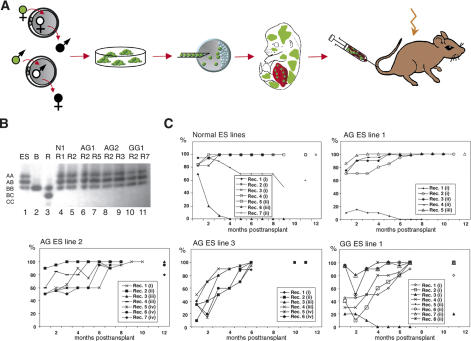
Figure 1.
Multilineage reconstitution by androgenetic and gynogenetic cells. (A) Experimental design. EGFP-expressing ES cell lines derived from androgenetic and gynogenetic embryos produced by pronuclear transfer between zygotes were injected into blastocysts. After embryo transfer, fetuses were recovered at 13.5–14.5 dpc, chimeras were identified by eGFP-fluorescence, and fetal liver from chimeras was transplanted into lethally irradiated recipient mice. (B) Analysis of GPI-1 isoenzymes to identify contribution of ES cell-derived cells to the peripheral blood of recipients. Lanes 1–3 show the GPI-1 isoenzyme dimers present in the ES cells ([ES] A and B isoforms), blastocysts ([B] B isoform only), and adult recipients ([R] B and C isoforms), respectively. (GPI-1 forms homo- and heterodimers, such that cells containing A and B isoforms contain AA, AB, and BB dimers; all dimers are indicated on the left.) Lanes 4–11 show the predominance of ES cell-derived cells (A, B isoforms) in the peripheral blood of individual recipients (R) 6–8 mo after transplantation of ES cell chimeric fetal liver (ES line indicated on top). (C) Presence of ES cell-derived cells in peripheral blood of recipients over time as determined by GPI-1 analysis. Numbers in parentheses indicate transplanted fetal liver cell pools, with identical numbers referring to the same pool.
For hematopoietic reconstitution, fetal liver cells from chimeras, consisting of both blastocyst-derived and injected ES cell-derived cells, were transplanted into lethally irradiated adult B6129Sv mice. Fetal liver transplants from normal (N; derived from fertilized embryos), AG, and GG ES cell chimeras reconstituted recipients with similar efficacy. Contribution of ES cell- and blastocyst-derived cells to the peripheral blood of recipients was monitored monthly after transplantation by analysis of mouse strain-specific glucose-6-phosphate isomerase-1 isoforms (GPI-1 isozyme gel electrophoresis) and revealed high levels of the N, AG, and GG ES cell-derived component in animals (Fig. 1B,C). Ultimately, recipients had a normal life span and no pathologies associated with uniparental cell transplants.
Regardless of the initial level of ES cell-derived cells in fetal liver transplants that varied between 10% and 60%, the contribution of ES cell-derived cells in recipients typically increased with time, and at 6–9 mo post-transplantation, the peripheral blood of many recipients had high levels of ES cell-derived cells or was entirely ES cell-derived (Fig. 1C). This predominance of ES cell-derived cells did not correlate with parental origin, and in a few animals of each experimental group (N: 1/7; AG: 3/20; GG: 2/8), blastocyst-derived cells (B6C3 or B6 strain) persisted and, in a control experiment of transplanting only blastocyst-derived fetal liver, repopulated hematopoiesis (data not shown). Competitive secondary transplantation experiments did not reveal any predominance of uniparental or normal ES cell-derived (B6129S1) over B6129Sv bone marrow cells (see below). The frequent predominance of the ES cell-derived cell component from the chimeric fetal liver cells is thus possibly associated with mouse-strain-specific properties. Such strain-specific stem cell properties have been previously associated with observations in mouse chimeras between DBA/2 and B6 strains, in which hematopoiesis over time as well as long-term engraftment into secondary recipients was accomplished by B6-derived cells (Van Zant et al. 1990, 1992).
Uniparental HSCs can establish long-term, multilineage engraftment of primary and secondary hosts
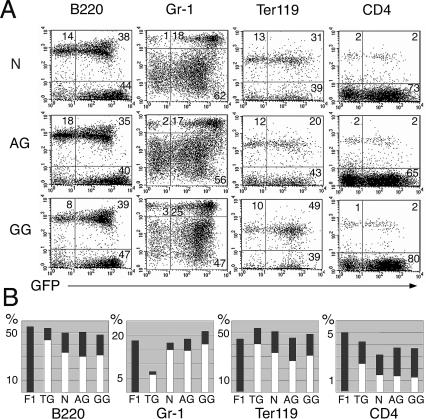
Parthenogenetic/gynogenetic and androgenetic ES cells are distinct from each other and normal ES cells in their ability to differentiate into various cell types both in vitro and in vivo (Mann et al. 1990; Allen et al. 1994), and cells of AG and GG origin may be limited in their differentiation potential into hematopoietic lineages. Using lineage-specific surface markers, we determined the contribution of N, AG, and GG eGFP-expressing cells to lymphoid (B220, CD4-positive), myeloid (Gr-1-positive), and erythroid (Ter119-positive) cell populations of the peripheral blood of recipients. The percentage of peripheral blood lymphocytes positive for each lineage marker, and the percentage of eGFP-expressing cells in each of these populations, were similar in animals with N, AG, and GG derived blood and eGFP transgenic mice (Fig. 2 A,B), indicating no limitation or bias in the hematopoietic lineage formation of uniparental cells. Steady-state hematology of peripheral blood from recipients reconstituted from N, AG, and GG ES cell-derived fetal liver cells was similar to B6129Sv or eGFP transgenic (B6Osb) mice (Table 1). High levels of ES cell-derived cells were detected in hematopoietic organs (spleen, thymus, bone marrow) (Supplementary Table 2). Animals with an entirely N, AG, or GG ES cell-derived hematopoietic system exhibited normal maturation of T- and B-lymphocytes, indicated by the presence of CD4 and CD8 double- and single-positive cells in thymus and peripheral blood, respectively, and the expression of IgM and B220 in splenic and peripheral lymphocytes (Supplementary Fig. 2). Bone marrow and spleen cells of animals reconstituted entirely from N-, AG-, or GG-derived cells exhibited normal in vitro myeloid colony-forming activity in a CFU-C assay in methylcellulose (data not shown).
Figure 2.
Normal lineage differentiation of AG and GG cells in recipients. (A) FACS analysis of peripheral blood of representative recipient mice from each experimental group (N, AG, GG) 5–7 mo post-transplantation. The fluorescence intensity of eGFP (marking ES cell-derived cells) is indicated on the X-axis, the fluorescence intensity of differentiation markers (B220, CD4, Ter119, Gr-1) is indicated on the Y-axis. Gating was based on forward-scatter and side-scatter profiles typical for lymphocytes/granulocytes. Numbers indicate the percent of gated cells. (B) Summary of lineage analysis. Columns represent average values for groups of four to eight mice. (F1) B6129, not transgenic; (TG) B6Osb (eGFP transgenic); both are controls to demonstrate the similarity of lineage- and eGFP-positive percentages between ES reconstituted and normal mice. (Dark-gray bars) The percent of gated cells positive for the lineage marker; (white bars) the percent of eGFP-positive = ES cell-derived cells within the lineage-positive population. No difference was detected between AG, GG, and N ES cell reconstituted recipients. ANOVA was performed with α = 0.050, and normality tests were passed (P > 0.050). The P values were as follows: B220 total, P = 0.087; B220GFP, P = 0.126; Gr-1 total, P = 0.228; Gr-1GFP, P = 0.635; Ter119 total, P = 0.304; Gr-1/GFP, P = 0.165; CD4GFP, P = 0.077. For CD4 total, Kruskal-Wallis ANOVA on ranks was applied, P = 0.803.
Table 1.
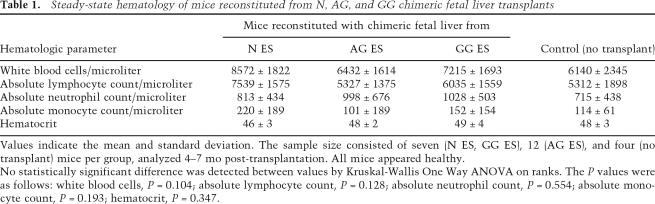
Steady-state hematology of mice reconstituted from N, AG, and GG chimeric fetal liver transplants
Values indicate the mean and standard deviation. The sample size consisted of seven (N ES, GG ES), 12 (AG ES), and four (no transplant) mice per group, analyzed 4–7 mo post-transplantation. All mice appeared healthy.
No statistically significant difference was detected between values by Kruskal-Wallis One Way ANOVA on ranks. The P values were as follows: white blood cells, P = 0.104; absolute lymphocyte count, P = 0.128; absolute neutrophil count, P = 0.554; absolute monocyte count, P = 0.193; hematocrit, P = 0.347.
To establish the presence of long-term repopulating HSC, bone marrow from recipients with entirely ES cell-derived hematopoietic systems was transplanted into lethally irradiated secondary recipients. All secondary recipients (seven recipients receiving bone marrow from donors with N-derived blood; 19 recipients receiving bone marrow from donors with AG-derived blood; 10 recipients receiving bone marrow from donors with GG-derived blood) survived for >12 mo after transplantation, and exhibited N, AG, or GG ES cell-derived peripheral blood. In competitive transplantation assays of bone marrow from primary recipients with entirely N-, AG-, or GG-derived blood mixed with bone marrow cells from B6129F1 animals, cells of all parental types exhibited stable levels of contribution for at least 11 mo subsequent to transplantation, suggesting neither a competitive disadvantage nor advantage compared with normal cells (data not shown).
Uniparental ES cells differentiate into hematopoietic progenitors in vitro
To determine if uniparental hematopoiesis is limited to the codevelopment of uniparental with normal cells within a chimera, or if uniparental cells have the intrinsic capacity to undergo hematopoietic commitment, we ascertained hematopoietic in vitro differentiation of uniparental ES cells using established protocols for murine ES cells (Kennedy and Keller 2003). Specifically, we analyzed the formation of committed hematopoietic progenitors at day 6 of ES cell differentiation by plating ES cell derivatives at this stage in methylcellulose media containing a mix of hematopoietic cytokines that enabled us to evaluate the formation of primitive erythroid, definitive erythroid, megakaryocyte, macrophage, and multilineage colonies. Both uniparental maternal (GG line 1; three PG ES cell lines, PG lines 1–3) and a paternal (AG line 3 [MM9], previously characterized for imprinting-related phenotypes) (McLaughlin et al. 1997) ES cell lines showed commitment to the same hematopoietic progenitor types in vitro as the N ES cells (N ES line 2) (Hooper et al. 1987), which was consistent with previous observations (Kennedy and Keller 2003). Similar numbers of hematopoietic colonies were obtained for N, AG, GG, and PG ES cell derivatives (N: 10; AG: 13; GG: 14; PG 1–3: 31; eight and 11 colonies/100,000 cells at day 6 of differentiation).
In vitro hematopoietic derivatives of uniparental ES cells engraft in adult recipients and contribute to hematopoiesis in primary and secondary recipients
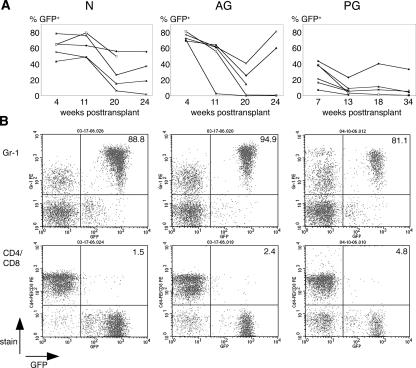
Ectopic expression of the homeodomain protein HoxB4 in differentiating ES cells has been successfully used to promote formation of progenitor cells with a definitive hematopoietic phenotype that exhibits engraftment in adult recipients (Kyba et al. 2002; Rideout et al. 2002). We applied this approach to test the capacity of uniparental ES cells to form transplantable hematopoietic progenitor cells in vitro. Using retroviral transduction with the MSCVHoxB4iGFP vector (Kyba et al. 2002), we introduced the HoxB4 and GFP genes into differentiating normal (N line 2) and uniparental (AG line 3, PG line 1) ES cells, and transplanted in vitro generated hematopoietic progenitors into lethally irradiated adult B6129Sv recipient mice. ES cell-derived progenitors constitutively expressing HoxB4 can provide extensive donor chimerism in the hematopoietic system of recipients, with predominantly myeloid engraftment (Kyba et al. 2002; Rideout et al. 2002). We observed contribution of N, AG, and PG derived cells in primary recipients for >4 mo (N: 5/5 recipients; AG: 4/5; PG: 4/6) (Fig. 3A). FACS analysis of peripheral blood of recipients demonstrated mainly monocytic lineage formation (Gr-1-positive cells) (Fig. 3B), and low levels of lymphocyte formation—CD4/CD8 (Fig. 3B) and B220 (data not shown)—of ES derivatives of N, AG, and PG origin, as previously observed for N ES cells (Kyba et al. 2002, 2003; Rideout et al. 2002). Upon transplantation of bone marrow from primary into irradiated secondary recipients, N, AG, and PG ES cell derivatives engrafted and contributed to the hematopoietic system of recipients.
Figure 3.
Engraftment and differentiation of in vitro derived hematopoietic progenitors from N, AG, and PG ES cells in irradiated recipients. (A) The percent GFP-positive, ES cell-derived cells in the peripheral blood of recipients at indicated time points post-transplantation. (N) N line 2 (E14) (Hooper et al. 1987); (AG) AG line 3 (MM9) (McLaughlin et al. 1997); (PG) B6129F1 PG ES cell line 1. The range of contribution of N-, AG-, and PG-derived cells is similar to previous observations (Kyba et al. 2002). In a different experiment, PG line 1-derived cells were also transplanted into Rag2−/−, γC−/− recipient mice (Mazurier et al. 1999), and exhibited contribution levels of up to 95% of the peripheral blood of recipients (data not shown). (B) FACS analysis of peripheral blood of representative recipient mice at 20 wk (N; AG) and 13 wk (PG) post-transplantation. The fluorescence intensity of GFP (marking ES cell-derived cells) is indicated on the X-axis, the fluorescence intensity of differentiation markers (myeloid: Gr-1; T-cell: CD4/CD8) is indicated on the Y-axis. Numbers represent the percentage of GFP-positive donor cells that express the respective differentiation antigen.
Imprinted gene expression and methylation in uniparental hematopoietic tissues
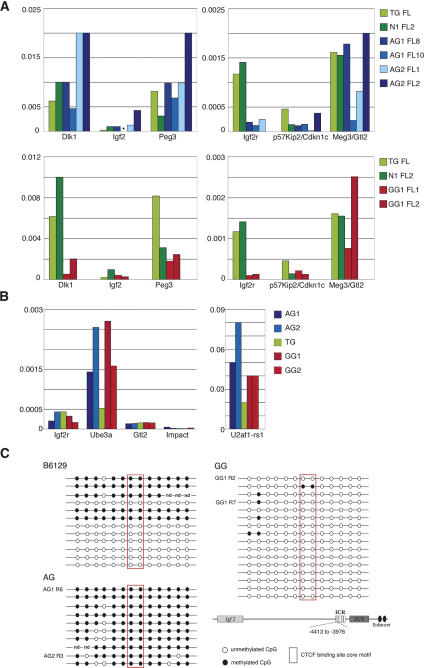
HSCs of both AG and GG origin conveyed long-term, multilineage reconstitution of primary and secondary recipients, implying that both paternal and maternal uniparental cells are functionally comparable to cells of normal origin. To ascertain how imprinting relates to this observation, we analyzed expression and methylation of regulatory regions of imprinted genes in uniparental-derived tissue prior to and post-engraftment in adult recipients. Androgenetic cells isolated from the fetal livers of individual ES cell chimeras (13.5 dpc; at the stage used for transplantation) typically had higher transcript levels of genes preferentially expressed from the paternal allele (Dlk-1, Igf2, and Peg3) (Fig. 4A, top left) compared with controls and GG-derived fetal liver cells (Fig. 4A, bottom left), and lower levels of the maternally expressed Igf2r gene compared with controls (Fig. 4A, top right), consistent with their paternal origin. Gynogenetic cells exhibited the expected allele-specific bias for genes normally expressed from the paternal allele (Dlk-1, Igf2, Peg3) Fig. (4A, bottom left), but not for the maternally expressed Igf2r, p57Kip2/Cdkn1c, and Meg3/Gtl2 genes. Expression levels of the latter were several-fold lower than the imprinted, paternal-allele-expressed genes. Uniparental cells isolated from chimeric fetal liver therefore maintain a mostly parent-of-origin-specific expression pattern of imprinted genes, with some exceptions for genes with preferential expression from the maternal allele.
Figure 4.
Expression and methylation of imprinted genes in donor cells. (A) Relative expression of imprinted genes in FACS sorted, eGFP-positive ES cell-derived fetal liver cells from individual AG, N, and GG ES cell chimeras and from an eGFP transgenic normal fetus (TG). The expression levels indicated are relative to β-actin. Each color-coded bar represents gene expression in FACS-sorted eGFP-positive cells isolated from the fetal liver (FL) of individual fetuses. AG1, AG2, and GG1 indicate the ES cell line used for chimera generation. (Left panels) Genes with bias for expression from the paternal allele. (Right panels) Genes with preferential expression from the maternal allele. (*) No data. (B) Imprinted gene expression in AG and GG ES cell-derived CD3/eGFP-positive splenocytes isolated from four adult recipients by FACS sorting. The expression levels shown are relative to β-actin. AG1, AG2, and GG1 indicate the ES cell origin of splenocytes. The genes analyzed included the maternally expressed Igf2r, Ube3, and Meg3/Gtl2 (left side of panel), and the paternally expressed Impact and U2af2-rs1 (right side of panel). (C) Conserved methylation status of the H19 differentially methylated region in bone marrow cells of recipients with entirely AG- or GG-derived hematopoietic systems. Bisulfite sequencing of the 5′ upstream region of the H19 gene (pos. −4413 to −3976; see schematic representation bottom right). This region is part of the imprinting control region that regulates reciprocal allele-specific expression of the H19 and Igf2 genes. In normal tissues, the paternal allele is methylated, and the maternal allele is unmethylated. Bone marrow from two recipients (R) with entirely AG-derived hematopoietic systems (AG1 R6, AG1 recipient 6; and AG2 R3, AG2 recipient 3), and from two recipients with GG-derived hematopoietic systems (GG 1, recipients 2 and 7) was analyzed. Each line represents a single clone. Clones derived from AG tissue exhibit a high degree of methylation, whereas clones from GG tissue are unmethylated, indicating conservation of parent-of-origin-specific methylation marks.
For analysis of imprinted gene expression of uniparental-derived cells in adult recipients, we chose eGFP/CD3 double-positive splenocytes as a defined cell type, and first identified imprinted genes that are normally expressed in this cell type by microarray analysis of normal eGFP/CD3 double-positive splenocytes. Analysis criteria indicated that only a few imprinted genes examined were expressed at a significant level (Supplementary Table 3). Using quantitative real-time RT–PCR, we measured expression of these imprinted genes in eGFP/CD3 double-positive splenocytes from recipients with peripheral blood of entirely AG or GG ES cell origin. No expression bias—for example, no difference between AG- and GG-derived cells—was detected for the maternally expressed Ube3a, Igf2r, Meg3/Gtl2 and the paternally expressed impact and U2af1-rs1 genes (Fig. 4B); however, relative expression levels for all genes except U2af1-rs1 were very low. It is difficult to define whether the presence of transcripts of imprinted genes that would be expected to be silenced in AG or GG cells reflects normal basal low-level expression or tissue-specific modulation, or relaxation of the allele specificity of expression. Allele-specific expression of imprinted genes typically does not occur in an all or none pattern, with detectable, albeit lower, expression from the silenced allele (Szabo and Mann 1994, 1995a, b; Matsuoka et al. 1996; Burns et al. 2001).
Analysis of methylation of the imprinting regulatory region located 5′ of the H19 gene showed maintenance of parent-of-origin-specific marks in bone marrow cells of recipients with peripheral blood of entirely AG or GG ES cell origin (Fig. 4C). Similarly, we detected an absence of methylation at CpG islands of the differentially methylated region (DMR) 1 of the Snrpn gene in the AG ES cell lines, and in AG-derived cells in the peripheral blood of recipients, which is consistent with maintenance of male germline specific imprints (Supplementary Fig. 4A). For the DMR2 of the Igf2r gene, we observed differences between AG ES cell lines: In two of three lines used (AG1, AG3), this region was not methylated, as expected, but some clones derived from AG line 2 exhibited a substantial degree of methylation (Supplementary Fig. 4B). Peripheral blood of recipients with hematopoietic systems derived from cells of the unmethylated AG line 1 had several clones (4/14) that were methylated, indicating a change in methylation between ES cells and their engrafted derivatives. Both the Snrpn DMR1 and the Igf2r DMR2 were hypomethylated in the GG ES cell line 1 and in its derivatives in recipients, and thus not consistent with the maintenance of maternal-specific methylation marks, but also not associated with changes between ES cells and their derivatives (Supplementary Fig. 4A,B). In summary, it appears that one locus that is typically prone to methylation changes (H19/Igf2) maintains fidelity of parent-of-origin-specific marks in recipients. For two other loci (Snrpn; Igf2r), we observe heterogeneity in methylation, with mostly conserved patterns in AG ES cells, some unexpected methylation in AG-derived hematopoietic cells, but a complete absence of maternal-specific methylation in GG cells.
Discussion
We provide evidence that both gynogenetic and androgenetic cells form functional HSCs by demonstration of long-term, multilineage reconstitution of primary and secondary recipients with uniparental-derived fetal liver cell transplants. The capacity of uniparental ES cells to differentiate into adult repopulating hematopoietic progenitors in vitro, when genetically modified to express HoxB4, reveals that the ability of uniparental ES cells to commit to hematopoiesis is intrinsic and does not require codevelopment in vivo with normal cells such as in a developmental chimera. Formation of several cell types in vitro has been shown for nonhuman primate parthenogenetic ES cells (Cibelli et al. 2002), and parthenogenetic ES cell-derived dopaminergic neurons can survive in a transplantation model (Sanchez-Pernaute et al. 2005), although whether these cells can functionally integrate is uncertain. Our findings provide evidence that transplanted uniparental cells can proliferate in and even replace an adult organ, substantiating the concept of using derivatives of uniparental ES cells for transplantation. The success in repopulating adult tissue with androgenetic tissues contrasts with their developmental limitations and establishes that they are a potential source of patient-derived transplantation tissue.
Studies on postnatal uniparental chimeras reveal that uniparental cells are limited in their contribution to certain adult tissues and organs, both quantitatively and functionally (Mann et al. 1990; Jagerbauer et al. 1992; Allen et al. 1995; Narasimha et al. 1997). The analysis of parthenogenetic ES cell chimeras has indicated some capacity of parthenogenetic cells for normal hematopoietic maturation in adult chimeras, however, at low contribution levels to the hematopoietic system (<10%) (Sturm et al. 1997). Androgenetic cell contribution to postnatal chimeras is severely restricted and frequently associated with lethality (Mann et al. 1990; Mann and Stewart 1991; Narasimha et al. 1997). Here, we did not observe a limit in the contribution of either uniparental cell type to the hematopoietic system and cell types thereof, with replacement of the whole organ in some recipients. An explanation could be that transplantation of fetal-stage uniparental cells into the adult bypasses developmental stages when abnormal expression of imprinted genes becomes particularly consequential, such as those involved in regulation of fetal and perinatal growth (Eggenschwiler et al. 1997; McLaughlin et al. 1997; Smith et al. 2006). This would be consistent with the observation that postnatal uniparental chimeras that survive weaning stabilize with respect to the manifestation of imprinting-related defects (Mann et al. 1990; Narasimha et al. 1997; Sturm et al. 1997). It is also conceivable that in some tissues or organs, a very large contribution of uniparental cells may be compatible with their normal development and function. In androgenetic chimeras, this would not be observed due to prior lethal effects in tissues that are susceptible to imprinting-related defects. In adult parthenogenetic mouse chimeras, despite higher levels of contribution, an entire contribution of parthenogenetic cells to specific tissues or organs has not been described (Fundele et al. 1989, 1990; Jagerbauer et al. 1992; Sturm et al. 1997). The question as to whether uniparental cells can form other adult organs could be broadly tested by tissue replacement of various adult tissues using uniparental cells.
Unbalanced expression of imprinted genes such as occurs in uniparental embryos and cells and those with uniparental duplication of imprinted chromosome regions is associated with fetal, perinatal, and postnatal defects and lethality (Solter 1988; Cattanach et al. 2006). While imprinting persists in the adult, understanding of its role in normal adult tissues and organs is minimal (Wood and Oakey 2006). Allele-specific expression of various imprinted genes is often regulated in a developmental- and tissue-specific manner (Latham 1995). In the adult, allele-specific expression of imprinted genes can be restricted to cell types or regions within tissues, such as seen for Ube3a, for which maternal-specific expression occurs in certain neurons and subregions of the brain (Albrecht et al. 1997), while other regions of the CNS and several somatic tissues express the gene in a biallelic manner. Imprinting in the mammalian hematopoietic system has almost exclusively been studied in the human, particularly in respect to the imprinted insulin-like growth factor II (IGF2) gene and the loss of imprinting associated with tumorigenesis. In human blood mononuclear cells, IGF2 is usually expressed monoallelically from the paternal allele. A study on patients with malignant lymphoma revealed that abnormal biallelic expression of IGF2 was not only observed in patients with tumors, but also in up to 20% of healthy controls (Vorwerk et al. 2003), suggesting that both mono- and biallelic expression is compatible with normal hematopoiesis. Biallelic expression of IGF2 occurs in bone marrow and activated lymphocytes, putatively associated with developmental stages or proliferation (Morison et al. 2000; Hofmann et al. 2002). It therefore appears that considerable variation in the expression and parental allele-specificity of expression of imprinted genes can be associated with normal hematopoiesis. The unique report of a human patient with blood leukocytes entirely of uniparental maternal origin (Strain et al. 1995) and our observations on hematopoietic engraftment and differentiation of murine gynogenetic and androgenetic cells suggest that the hematopoietic system is either extremely flexible with respect to the consequences of the presence of two maternal or two paternal genomes, or that imprinting in uniparental hematopoietic cells becomes relaxed to a degree that permits stem cell formation and subsequent differentiation into progenitors and differentiated cell types.
We observe that expression and parent-of-origin-specific methylation marks of imprinted genes are mostly conserved in androgenetic ES cells and their fetal derivatives, and that methylation patterns remain consistent with androgenetic origin at two of three loci analyzed in androgenetic cells in adult recipients. In contrast, gynogenetic ES cells exhibit loss of the maternal-specific methylation mark at two of three imprinted loci, and this hypomethylation remains in gynogenetic cells in adult recipients. Considerable epigenetic variability of ES cells and their fetal and postnatal derivatives has been described, but the functional consequences of this variability are unclear. Both abnormal fetal phenotypes (Dean et al. 1998) and lack of detectable abnormalities in postnatal animals (Humpherys et al. 2001) have been reported for derivatives of ES cells from fertilized embryos. For uniparental ES cell lines, parthenogenetic ES cell lines are more prone to lose imprinting-related gene expression and phenotype, while androgenetic ES cell lines exhibit higher fidelity of parent-of-origin characteristics (Mann 1992; Szabo and Mann 1994; Sturm et al. 1997; Hernandez et al. 2003), similar to our observations. Despite some variability observed in methylation of the Igf2r locus in one of the androgenetic ES lines and in gene expression patterns of androgenetic fetal liver cells, the phenotype of fetal (overgrowth, malformations, lethality) and postnatal (neonatal lethality, rib cartilage defects, polydactyly) (data not shown) chimeras generated with the androgenetic ES cell lines used in this study was consistent with the maintenance of functional outcomes of imprinting in androgenetic lines. Ultimately, for both paternal and maternal uniparental ES lines tested, we observed no difference in the effective repopulation of adult hematopoiesis. Although the significance of imprinting for hematopoiesis is undefined and we have only examined a subset of imprinted loci, our findings support the notion that there is considerable flexibility or tolerance in relation to variations in imprinted gene expression and methylation in the hematopoietic system of the adult.
The proposed use of uniparental (parthenogenetic) (Cibelli et al. 2002) ES cells for cell-based replacement therapies requires demonstration of immune compatibility and functionality while not conveying undesired pathologies. It has been verified that parthenogenetic ES cell lines can be generated with the full complement of major histocompatibility complex antigens (Kim et al. 2006), and the same should apply to androgenetic and gynogenetic lines. Our observations during long-term engraftment of androgenetic and gynogenetic cells in adults do not indicate malignancies or proliferation defects of uniparental cells. Adult cells, however, are typically not subject to the influence of fetal growth factors such as IGF-II that have been implicated with proliferation defects and malignant transformation in fetal fibroblasts (Hernandez et al. 2003). In adult gynogenetic chimeras, we have not detected a propensity for tumorigenesis (data not shown). Although the engrafted uniparental-derived hematopoietic cells appear normal, it cannot be excluded that the observed variant imprinted gene expression may cause problems in other transplanted tissues types. Characterization of the ability to safely transplant various cell types will be required to determine the broader therapeutic applicability of uniparental ES cell derivatives. Indirectly, this will be also valuable as a model to evaluate the role of genomic imprinting in adults.
Materials and methods
ES cell lines and chimeras
Animals were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania. AG embryos were produced by pronuclear transplantation (McGrath and Solter 1983) of the paternal pronuclei of zygotes from an intercross between C57BL/6NTac × C3H F1 females (Taconic; #B6C3F1; abbreviated B6C3) and eGFP transgenic C57BL/6-TgN (ACTbEGFP) 1Osb males (Jackson Laboratories; #003291; abbreviated B6Osb) (Okabe et al. 1997) into zygotes from a B6C3 × 129S1/SvImJ (Jackson Laboratories #002448; abbreviated 129S1) intercross, from which the maternal pronuclei had been removed. GG embryos were produced by transplantation of the maternal pronuclei of zygotes from a 129S1 × ICR (Taconic #ICR) intercross into zygotes from a B6Osb × ICR intercross, from which the paternal pronuclei had been removed. Embryos were cultured to the blastocyst stage in α-MEM (Sigma) supplemented with BSA (Pentex). Zona-free eGFP-positive blastocysts were placed on feeder fibroblasts, and ES cell lines were derived from outgrowths under standard conditions. N ES cell lines were derived from eGFP-positive blastocysts from a 129S1 × B6Osb intercross. Only uniparental embryos but not the donor zygotes could both be eGFP transgenic and express the A-form of GPI-1 that is distinct to the 129S1 strain (all other strains and outbred ICR males: GPI-1 bb), enabling unequivocal verification of the uniparental origin of ES cell lines. Parthenogenetic ES cells (B6129F1) were derived as described (Mann 2001), with the modification that oocytes were activated using 10 mM SrCl2.
The ability of ES cell-derived fetal liver cells to reconstitute irradiated adult recipients has been shown (Forrester et al. 1991). ES cell chimeras were produced by injection of ES cells into C57BL/6NTac (Taconic #B6, abbreviated B6) or B6C3 × B6 hybrid blastocysts, and embryo transfer into pseudopregnant ICR females. Fetuses were recovered at 13.5 dpc (AG) or at 14.5 dpc (GG and N ES). Uniparental and N ES cell lines were heterozygous for the alleles encoding the A and B electrophoretic forms of GPI-1, or homozygous for the A-encoding allele (AG ES line 3) (McLaughlin et al. 1997), and blastocysts were homozygous for the allele encoding the B form, permitting detection and quantification of ES cell-derived cells by GPI-1 isoenzyme electrophoresis. Standard curves for GPI-1 analysis were obtained by mixing peripheral blood from mice carrying different Gpi-1 alleles at known ratios.
Real-time RT–PCR
The eGFP-positive cell population from fetal livers from individual midgestation chimeras, and eGFP and CD3 double-positive cells from the spleens of recipients were collected using a FACSVantage Sort (BD Pharmingen). RNA was extracted using RNeasy columns (Qiagen). Eighty nanograms of total RNA were reverse-transcribed using Dynabeads (Dynal), resulting in bead-coupled cDNA libraries (Mann et al. 2003). Real-time PCR on Dynabead libraries was performed on a Roche LightCycler using LightCycler FastStart DNA Master SYBR Green I (Roche). The oligonucleotide sequences were Igf2r, 5′-TAGTTGCAGCT CTTTGCACG-3′ and 5′-ACAGCTCAAACCTGAAGCG-3′; p57Kip2/Cdkn1c, 5′-TTCAGATCTGACCTCAGACCC-3′ and 5′-AGTTCTCTTGCGCTTGGC-3′; Meg3/Gtl2, 5′-TTGCACA TTTCCTGTGGGAC-3′ and 5′-AAGCACCATGAGCCACTA GG-3′; Dlk-1, 5′-CTGGCGGTCAATATCATCTTCC-3′ and 5′-GAGGAAGGGGTTCTTAGATAGCG-3′; Igf2, 5′-CTAAGA CTTGGATCCCAGAACC-3′ and 5′-GTTCTTCTCCTTGGGT TCTTTC-3′; Peg3, 5′-TAGTCCTGTGAAGGTGTGGG-3′ and 5′-GTAGGGATGGGTTGATTTGG-3′; Ube3a, 5′-CACATA TGATGAAGCTACGA-3′ and 5′-CACACTCCCTTCATATT CC-3′; Impact, 5′-ACGTTTCCCCATTTTACAAG-3′ and 5′-CTCTACATATGATTTTCTCTAC-3′; U2af1-rs1, 5′-TAAG GCAGCACCACTTGGAC-3′ and 5′-TAAGGCAGCACCACT TGGAC-3′; and β-actin, 5′-GATATCGCTGCGCTGGTCG TC-3′ and 5′-ACGCAGCTCATTGTAGAAGGTGTGG-3′.
Fetal liver transplants
Single-cell suspensions of fetal livers from chimeras were injected into lethally irradiated (9.5 gy, Cesium 137 source) adult hybrid mice between B6 and 129S6/SvEv (B6129 Hybrid mice; Taconic #B6129; named B6129Sv; Gpi-1 alleles bc) mice via the lateral tail vein (0.6 × 106 to 3 × 106 fetal liver cells per recipient). For secondary reconstitutions, bone marrow harvested from tibiae and femora of primary recipients was injected into the lateral tail vein of lethally irradiated (9.5 gy) B6129Sv mice.
Flow cytometry and peripheral blood hematology
Cells were stained with phycoerythrin (PE), PE-Cy5, and biotin-conjugated monoclonal antibodies specific for lineage markers including CD4 (L3T4), CD8 (Ly-2), CD45R/B220, Ly-6G (Gr-1), Ter119/Ly-76, and IgM (Igh-6b). Biotinylated antibodies were detected using a secondary streptavidin-PE-Cy5 conjugate. All antibodies were obtained from BD Pharmingen. Cells were analyzed on a BD LSR. Peripheral blood hematology was performed as previously described (Bunting et al. 2002).
Bisulfite sequencing
Genomic DNA isolated from bone marrow cells was digested with XhoI and treated with sodium bisulfite as described (Clark et al. 1994). Bisulfite-treated DNA was amplified by seminested PCR under standard conditions using ExTaq Hot Start Version (TaKaRa). Oligonucleotide sequences were 1401F (5′-TTTT GAATTATTATAAGGAA-3′) and 2159R (5′-ATCAAATATC CTCATAAATA-3′) for primary PCR, and 1401F and 1842R (5′-ACAACCCTAATCTTTACACA-3′) for secondary PCR. The amplified DNA fragment was subcloned into a pGEM-T Easy vector (Promega) for sequencing.
In vitro hematopoietic differentiation
Methylcellulose colony-forming assays in medium supplied with a cytokine cocktail (M3434; StemCell Technologies) were performed after 6 d of ES cell differentiation as described (Kennedy and Keller 2003).
For transplantation, ES cells were differentiated for 6 d, then transduced with MSCVHoxB4iGFP and cultured on OP9 stromal cells for 12 d as described (Kyba et al. 2002, 2003; Rideout et al. 2002). Differentiated cells were transplanted into lethally irradiated (9.5 gy), NK-depleted (i.p. injection of Anti-asialo GM1 [Wako] 24 h prior to transplant) B6129Sv mice.
Statistical analysis
One-way analysis of variance (ANOVA) and Kruskal-Wallis ANOVA on Ranks were applied using SigmaStat software.
Acknowledgments
We thank Michael Kyba for the MSCVhoxB4iGFP vector and for advice. We thank Ina Dobrinski, James Kehler, Satoshi Kurosaka, Jeff Mann, and Albrecht Müller for discussion and comments on data and versions of this manuscript; John Tobias for analysis of microarray data; Teresa Jordan for animal husbandry; and Marianne Friez for technical assistance. This work was supported by National Institutes of Health (NIH) Grants 1RO3 HD045291-01 (to K.J.M.); R01HL073738 (to K.D.B); the Stem Cell Research Foundation, the Marion Dilley and David George Jones Funds, and the Commonwealth and General Assembly of Pennsylvania (to K.J.M.); and the Wakayama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence of the Japan Science and Technology Corporation and a Grant-in-Aid for the 21st Century COE Program of the Japan Ministry of Education, Culture, Sports, Science and Technology (to H.K.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1524207
References
- Albrecht U., Sutcliffe J.S., Cattanach B.M., Beechey C.V., Armstrong D., Eichele G., Beaudet A.L., Sutcliffe J.S., Cattanach B.M., Beechey C.V., Armstrong D., Eichele G., Beaudet A.L., Cattanach B.M., Beechey C.V., Armstrong D., Eichele G., Beaudet A.L., Beechey C.V., Armstrong D., Eichele G., Beaudet A.L., Armstrong D., Eichele G., Beaudet A.L., Eichele G., Beaudet A.L., Beaudet A.L. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Allen N.D., Barton S.C., Hilton K., Norris M.L., Surani M.A., Barton S.C., Hilton K., Norris M.L., Surani M.A., Hilton K., Norris M.L., Surani M.A., Norris M.L., Surani M.A., Surani M.A. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- Allen N.D., Logan K., Lally G., Drage D.J., Norris M.L., Keverne E.B., Logan K., Lally G., Drage D.J., Norris M.L., Keverne E.B., Lally G., Drage D.J., Norris M.L., Keverne E.B., Drage D.J., Norris M.L., Keverne E.B., Norris M.L., Keverne E.B., Keverne E.B. Distribution of parthenogenetic cells in the mouse brain and their influence on brain development and behavior. Proc. Natl. Acad. Sci. 1995;92:10782–10786. doi: 10.1073/pnas.92.23.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton S.C., Surani M.A., Norris M.L., Surani M.A., Norris M.L., Norris M.L. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- Barton S.C., Ferguson-Smith A.C., Fundele R., Surani M.A., Ferguson-Smith A.C., Fundele R., Surani M.A., Fundele R., Surani M.A., Surani M.A. Influence of paternally imprinted genes on development. Development. 1991;113:679–687. doi: 10.1242/dev.113.2.679. [DOI] [PubMed] [Google Scholar]
- Boediono A., Suzuki T., Li L.Y., Godke R.A., Suzuki T., Li L.Y., Godke R.A., Li L.Y., Godke R.A., Godke R.A. Offspring born from chimeras reconstructed from parthenogenetic and in vitro fertilized bovine embryos. Mol. Reprod. Dev. 1999;53:159–170. doi: 10.1002/(SICI)1098-2795(199906)53:2<159::AID-MRD5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Bunting K.D., Bradley H.L., Hawley T.S., Moriggl R., Sorrentino B.P., Ihle J.N., Bradley H.L., Hawley T.S., Moriggl R., Sorrentino B.P., Ihle J.N., Hawley T.S., Moriggl R., Sorrentino B.P., Ihle J.N., Moriggl R., Sorrentino B.P., Ihle J.N., Sorrentino B.P., Ihle J.N., Ihle J.N. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–487. doi: 10.1182/blood.v99.2.479. [DOI] [PubMed] [Google Scholar]
- Burns J.L., Jackson D.A., Hassan A.B., Jackson D.A., Hassan A.B., Hassan A.B. A view through the clouds of imprinting. FASEB J. 2001;15:1694–1703. doi: 10.1096/fj.010085rev. [DOI] [PubMed] [Google Scholar]
- Cattanach B.M., Beechey C.V., Peters J., Beechey C.V., Peters J., Peters J. Interactions between imprinting effects: Summary and review. Cytogenet. Genome Res. 2006;113:17–23. doi: 10.1159/000090810. [DOI] [PubMed] [Google Scholar]
- Cibelli J.B., Grant K.A., Chapman K.B., Cunniff K., Worst T., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Grant K.A., Chapman K.B., Cunniff K., Worst T., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Chapman K.B., Cunniff K., Worst T., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Cunniff K., Worst T., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Worst T., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Green H.L., Walker S.J., Gutin P.H., Vilner L., Tabar V., Walker S.J., Gutin P.H., Vilner L., Tabar V., Gutin P.H., Vilner L., Tabar V., Vilner L., Tabar V., Tabar V., et al. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- Clark S.J., Harrison J., Paul C.L., Frommer M., Harrison J., Paul C.L., Frommer M., Paul C.L., Frommer M., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W., Bowden L., Aitchison A., Klose J., Moore T., Meneses J.J., Reik W., Feil R., Bowden L., Aitchison A., Klose J., Moore T., Meneses J.J., Reik W., Feil R., Aitchison A., Klose J., Moore T., Meneses J.J., Reik W., Feil R., Klose J., Moore T., Meneses J.J., Reik W., Feil R., Moore T., Meneses J.J., Reik W., Feil R., Meneses J.J., Reik W., Feil R., Reik W., Feil R., Feil R. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: Association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J., Ludwig T., Fisher P., Leighton P.A., Tilghman S.M., Efstratiadis A., Ludwig T., Fisher P., Leighton P.A., Tilghman S.M., Efstratiadis A., Fisher P., Leighton P.A., Tilghman S.M., Efstratiadis A., Leighton P.A., Tilghman S.M., Efstratiadis A., Tilghman S.M., Efstratiadis A., Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes & Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester L.M., Bernstein A., Rossant J., Nagy A., Bernstein A., Rossant J., Nagy A., Rossant J., Nagy A., Nagy A. Long-term reconstitution of the mouse hematopoietic system by embryonic stem cell-derived fetal liver. Proc. Natl. Acad. Sci. 1991;88:7514–7517. doi: 10.1073/pnas.88.17.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fundele R., Norris M.L., Barton S.C., Reik W., Surani M.A., Norris M.L., Barton S.C., Reik W., Surani M.A., Barton S.C., Reik W., Surani M.A., Reik W., Surani M.A., Surani M.A. Systematic elimination of parthenogenetic cells in mouse chimeras. Development. 1989;106:29–35. doi: 10.1242/dev.106.1.29. [DOI] [PubMed] [Google Scholar]
- Fundele R.H., Norris M.L., Barton S.C., Fehlau M., Howlett S.K., Mills W.E., Surani M.A., Norris M.L., Barton S.C., Fehlau M., Howlett S.K., Mills W.E., Surani M.A., Barton S.C., Fehlau M., Howlett S.K., Mills W.E., Surani M.A., Fehlau M., Howlett S.K., Mills W.E., Surani M.A., Howlett S.K., Mills W.E., Surani M.A., Mills W.E., Surani M.A., Surani M.A. Temporal and spatial selection against parthenogenetic cells during development of fetal chimeras. Development. 1990;108:203–211. doi: 10.1242/dev.108.1.203. [DOI] [PubMed] [Google Scholar]
- Fundele R., Barton S.C., Christ B., Krause R., Surani M.A., Barton S.C., Christ B., Krause R., Surani M.A., Christ B., Krause R., Surani M.A., Krause R., Surani M.A., Surani M.A. Distribution of androgenetic cells in fetal mouse chimeras. Rouxs Arch. Dev. Biol. 1995;204:484–493. doi: 10.1007/BF00360856. [DOI] [PubMed] [Google Scholar]
- Hernandez L., Kozlov S., Piras G., Stewart C.L., Kozlov S., Piras G., Stewart C.L., Piras G., Stewart C.L., Stewart C.L. Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc. Natl. Acad. Sci. 2003;100:13344–13349. doi: 10.1073/pnas.2234026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W.K., Takeuchi S., Frantzen M.A., Hoelzer D., Koeffler H.P., Takeuchi S., Frantzen M.A., Hoelzer D., Koeffler H.P., Frantzen M.A., Hoelzer D., Koeffler H.P., Hoelzer D., Koeffler H.P., Koeffler H.P. Loss of genomic imprinting of insulin-like growth factor 2 is strongly associated with cellular proliferation in normal hematopoietic cells. Exp. Hematol. 2002;30:318–323. doi: 10.1016/s0301-472x(01)00797-4. [DOI] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S., Monk M., Hardy K., Handyside A., Hunter S., Monk M., Handyside A., Hunter S., Monk M., Hunter S., Monk M., Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Humpherys D., Eggan K., Akutsu H., Hochedlinger K., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R., Eggan K., Akutsu H., Hochedlinger K., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R., Akutsu H., Hochedlinger K., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R., Hochedlinger K., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R., Biniszkiewicz D., Yanagimachi R., Jaenisch R., Yanagimachi R., Jaenisch R., Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Jagerbauer E.M., Fraser A., Herbst E.W., Kothary R., Fundele R., Fraser A., Herbst E.W., Kothary R., Fundele R., Herbst E.W., Kothary R., Fundele R., Kothary R., Fundele R., Fundele R. Parthenogenetic stem cells in postnatal mouse chimeras. Development. 1992;116:95–102. doi: 10.1242/dev.116.1.95. [DOI] [PubMed] [Google Scholar]
- Kaufman M.H., Barton S.C., Surani M.A., Barton S.C., Surani M.A., Surani M.A. Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature. 1977;265:53–55. doi: 10.1038/265053a0. [DOI] [PubMed] [Google Scholar]
- Kennedy M., Keller G.M., Keller G.M. Hematopoietic commitment of ES cells in culture. Methods Enzymol. 2003;365:39–59. doi: 10.1016/s0076-6879(03)65003-2. [DOI] [PubMed] [Google Scholar]
- Kim K., Lerou P., Yabuuchi A., Lengerke C., Ng K., West J., Kirby A., Daly M.J., Daley G.Q., Lerou P., Yabuuchi A., Lengerke C., Ng K., West J., Kirby A., Daly M.J., Daley G.Q., Yabuuchi A., Lengerke C., Ng K., West J., Kirby A., Daly M.J., Daley G.Q., Lengerke C., Ng K., West J., Kirby A., Daly M.J., Daley G.Q., Ng K., West J., Kirby A., Daly M.J., Daley G.Q., West J., Kirby A., Daly M.J., Daley G.Q., Kirby A., Daly M.J., Daley G.Q., Daly M.J., Daley G.Q., Daley G.Q. Histocompatible embryonic stem cells by parthenogenesis. Science. 2006 doi: 10.1126/science.1133542. DOI: 10.1126/science.1133542. [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C., Daley G.Q., Perlingeiro R.C., Daley G.Q., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C., Daley G.Q., Perlingeiro R.C., Daley G.Q., Daley G.Q. Development of hematopoietic repopulating cells from embryonic stem cells. Methods Enzymol. 2003;365:114–129. doi: 10.1016/s0076-6879(03)65008-1. [DOI] [PubMed] [Google Scholar]
- Latham K.E. Stage-specific and cell type-specific aspects of genomic imprinting effects in mammals. Differentiation. 1995;59:269–282. doi: 10.1046/j.1432-0436.1996.5950269.x. [DOI] [PubMed] [Google Scholar]
- Mann J.R. Properties of androgenetic and parthenogenetic mouse embryonic stem cell lines; are genetic imprints conserved? Semin. Dev. Biol. 1992;3:77–85. [Google Scholar]
- Mann J.R. Deriving and propagating mouse embryonic stem cell lines for studying genomic imprinting. Methods Mol. Biol. 2001;181:21–39. doi: 10.1385/1-59259-211-2:21. [DOI] [PubMed] [Google Scholar]
- Mann J.R., Stewart C.L., Stewart C.L. Development to term of mouse androgenetic aggregation chimeras. Development. 1991;113:1325–1333. doi: 10.1242/dev.113.4.1325. [DOI] [PubMed] [Google Scholar]
- Mann J.R., Gadi I., Harbison M.L., Abbondanzo S.J., Stewart C.L., Gadi I., Harbison M.L., Abbondanzo S.J., Stewart C.L., Harbison M.L., Abbondanzo S.J., Stewart C.L., Abbondanzo S.J., Stewart C.L., Stewart C.L. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: Implications for genetic imprinting. Cell. 1990;62:251–260. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- Mann M.R., Chung Y.G., Nolen L.D., Verona R.I., Latham K.E., Bartolomei M.S., Chung Y.G., Nolen L.D., Verona R.I., Latham K.E., Bartolomei M.S., Nolen L.D., Verona R.I., Latham K.E., Bartolomei M.S., Verona R.I., Latham K.E., Bartolomei M.S., Latham K.E., Bartolomei M.S., Bartolomei M.S. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Thompson J.S., Edwards M.C., Bartletta J.M., Grundy P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Thompson J.S., Edwards M.C., Bartletta J.M., Grundy P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Edwards M.C., Bartletta J.M., Grundy P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Bartletta J.M., Grundy P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Grundy P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Kalikin L.M., Harper J.W., Elledge S.J., Feinberg A.P., Harper J.W., Elledge S.J., Feinberg A.P., Elledge S.J., Feinberg A.P., Feinberg A.P. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc. Natl. Acad. Sci. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier F., Fontanellas A., Salesse S., Taine L., Landriau S., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Fontanellas A., Salesse S., Taine L., Landriau S., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Salesse S., Taine L., Landriau S., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Taine L., Landriau S., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Landriau S., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Moreau-Gaudry F., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Reiffers J., Peault B., Di Santo J.P., de Verneuil H., Peault B., Di Santo J.P., de Verneuil H., Di Santo J.P., de Verneuil H., de Verneuil H. A novel immunodeficient mouse model—RAG2 × common cytokine receptor γ chain double mutants—requiring exogenous cytokine administration for human hematopoietic stem cell engraftment. J. Interferon. Cytokine Res. 1999;19:533–541. doi: 10.1089/107999099313983. [DOI] [PubMed] [Google Scholar]
- McGrath J., Solter D., Solter D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion. Science. 1983;220:1300–1302. doi: 10.1126/science.6857250. [DOI] [PubMed] [Google Scholar]
- McGrath J., Solter D., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McLaughlin K.J., Kochanowski H., Solter D., Schwarzkopf G., Szabo P.E., Mann J.R., Kochanowski H., Solter D., Schwarzkopf G., Szabo P.E., Mann J.R., Solter D., Schwarzkopf G., Szabo P.E., Mann J.R., Schwarzkopf G., Szabo P.E., Mann J.R., Szabo P.E., Mann J.R., Mann J.R. Roles of the imprinted gene Igf2 and paternal duplication of distal chromosome 7 in the perinatal abnormalities of androgenetic mouse chimeras. Development. 1997;124:4897–4904. doi: 10.1242/dev.124.23.4897. [DOI] [PubMed] [Google Scholar]
- Morison I.M., Eccles M.R., Reeve A.E., Eccles M.R., Reeve A.E., Reeve A.E. Imprinting of insulin-like growth factor 2 is modulated during hematopoiesis. Blood. 2000;96:3023–3028. [PubMed] [Google Scholar]
- Morrison S.J., Hemmati H.D., Wandycz A.M., Weissman I.L., Hemmati H.D., Wandycz A.M., Weissman I.L., Wandycz A.M., Weissman I.L., Weissman I.L. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Sass M., Markkula M., Sass M., Markkula M., Markkula M. Systematic non-uniform distribution of parthenogenetic cells in adult mouse chimaeras. Development. 1989;106:321–324. doi: 10.1242/dev.106.2.321. [DOI] [PubMed] [Google Scholar]
- Narasimha M., Barton S.C., Surani M.A., Barton S.C., Surani M.A., Surani M.A. The role of the paternal genome in the development of the mouse germ line. Curr. Biol. 1997;7:881–884. doi: 10.1016/s0960-9822(06)00377-0. [DOI] [PubMed] [Google Scholar]
- Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y., Ikawa M., Kominami K., Nakanishi T., Nishimune Y., Kominami K., Nakanishi T., Nishimune Y., Nakanishi T., Nishimune Y., Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Paldi A., Nagy A., Markkula M., Barna I., Dezso L., Nagy A., Markkula M., Barna I., Dezso L., Markkula M., Barna I., Dezso L., Barna I., Dezso L., Dezso L. Postnatal development of parthenogenetic in equilibrium with fertilized mouse aggregation chimeras. Development. 1989;105:115–118. doi: 10.1242/dev.105.1.115. [DOI] [PubMed] [Google Scholar]
- Rideout W.M., Hochedlinger K., Kyba M., Daley G.Q., Jaenisch R., Hochedlinger K., Kyba M., Daley G.Q., Jaenisch R., Kyba M., Daley G.Q., Jaenisch R., Daley G.Q., Jaenisch R., Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- Robertson E.J., Kaufman M.H., Bradley A., Evans M.J., Kaufman M.H., Bradley A., Evans M.J., Bradley A., Evans M.J., Evans M.J. Isolation, properties, and karyotype analysis of pluripotential (EK) cell lines from normal and parthenogenetic embryos. In: Silver L.M., et al., editors. Teratocarcinomal stem cells, Cold Spring Harbor Conferences on Cell Proliferation. Vol. 10. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. pp. 647–663. [Google Scholar]
- Sanchez-Pernaute R., Studer L., Ferrari D., Perrier A., Lee H., Vinuela A., Isacson O., Studer L., Ferrari D., Perrier A., Lee H., Vinuela A., Isacson O., Ferrari D., Perrier A., Lee H., Vinuela A., Isacson O., Perrier A., Lee H., Vinuela A., Isacson O., Lee H., Vinuela A., Isacson O., Vinuela A., Isacson O., Isacson O. Long-term survival of dopamine neurons derived from parthenogenetic primate embryonic stem cells (cyno-1) after transplantation. Stem Cells. 2005;23:914–922. doi: 10.1634/stemcells.2004-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.M., Garfield A.S., Ward A., Garfield A.S., Ward A., Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet. Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu. Rev. Genet. 1988;22:127–146. doi: 10.1146/annurev.ge.22.120188.001015. [DOI] [PubMed] [Google Scholar]
- Stevens L.C. Totipotent cells of parthenogenetic origin in a chimaeric mouse. Nature. 1978;276:266–267. doi: 10.1038/276266a0. [DOI] [PubMed] [Google Scholar]
- Stevens L.C., Varnum D.S., Eicher E.M., Varnum D.S., Eicher E.M., Eicher E.M. Viable chimaeras produced from normal and parthenogenetic mouse embryos. Nature. 1977;269:515–517. doi: 10.1038/269515a0. [DOI] [PubMed] [Google Scholar]
- Strain L., Warner J.P., Johnston T., Bonthron D.T., Warner J.P., Johnston T., Bonthron D.T., Johnston T., Bonthron D.T., Bonthron D.T. A human parthenogenetic chimaera. Nat. Genet. 1995;11:164–169. doi: 10.1038/ng1095-164. [DOI] [PubMed] [Google Scholar]
- Sturm K.S., Berger C.N., Zhou S.X., Dunwoodie S.L., Tan S., Tam P.P., Berger C.N., Zhou S.X., Dunwoodie S.L., Tan S., Tam P.P., Zhou S.X., Dunwoodie S.L., Tan S., Tam P.P., Dunwoodie S.L., Tan S., Tam P.P., Tan S., Tam P.P., Tam P.P. Unrestricted lineage differentiation of parthenogenetic ES cells. Dev. Genes Evol. 1997;206:377–388. doi: 10.1007/s004270050067. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Barton S.C., Barton S.C. Development of gynogenetic eggs in the mouse: Implications for parthenogenetic embryos. Science. 1983;222:1034–1036. doi: 10.1126/science.6648518. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Barton S.C., Kaufman M.H., Barton S.C., Kaufman M.H., Kaufman M.H. Development to term of chimaeras between diploid parthenogenetic and fertilised embryos. Nature. 1977;270:601–603. doi: 10.1038/270601a0. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Barton S.C., Norris M.L., Barton S.C., Norris M.L., Norris M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Surani M.A.H., Barton S.C., Norris M.L., Barton S.C., Norris M.L., Norris M.L. Influence of parental chromosomes on spatial specificity in androgenetic ↔ parthenogenetic chimaeras in the mouse. Nature. 1987;326:395–397. doi: 10.1038/326395a0. [DOI] [PubMed] [Google Scholar]
- Szabo P., Mann J.R., Mann J.R. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development. 1994;120:1651–1660. doi: 10.1242/dev.120.6.1651. [DOI] [PubMed] [Google Scholar]
- Szabo P.E., Mann J.R., Mann J.R. Allele-specific expression and total expression levels of imprinted genes during early mouse development: Implications for imprinting mechanisms. Genes & Dev. 1995a;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- Szabo P.E., Mann J.R., Mann J.R. Biallelic expression of imprinted genes in the mouse germ line: Implications for erasure, establishment, and mechanisms of genomic imprinting. Genes & Dev. 1995b;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- Van Zant G., Holland B.P., Eldridge P.W., Chen J.J., Holland B.P., Eldridge P.W., Chen J.J., Eldridge P.W., Chen J.J., Chen J.J. Genotype-restricted growth and aging patterns in hematopoietic stem cell populations of allophenic mice. J. Exp. Med. 1990;171:1547–1565. doi: 10.1084/jem.171.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zant G., Scott-Micus K., Thompson B.P., Fleischman R.A., Perkins S., Scott-Micus K., Thompson B.P., Fleischman R.A., Perkins S., Thompson B.P., Fleischman R.A., Perkins S., Fleischman R.A., Perkins S., Perkins S. Stem cell quiescence/activation is reversible by serial transplantation and is independent of stromal cell genotype in mouse aggregation chimeras. Exp. Hematol. 1992;20:470–475. [PubMed] [Google Scholar]
- Vorwerk P., Wex H., Bessert C., Hohmann B., Schmidt U., Mittler U., Wex H., Bessert C., Hohmann B., Schmidt U., Mittler U., Bessert C., Hohmann B., Schmidt U., Mittler U., Hohmann B., Schmidt U., Mittler U., Schmidt U., Mittler U., Mittler U. Loss of imprinting of IGF-II gene in children with acute lymphoblastic leukemia. Leuk. Res. 2003;27:807–812. doi: 10.1016/s0145-2126(03)00014-6. [DOI] [PubMed] [Google Scholar]
- Wood A.J., Oakey R.J., Oakey R.J. Genomic imprinting in mammals: Emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]