Abstract
Embryogenesis is controlled by large gene-regulatory networks, which generate spatially and temporally refined patterns of gene expression. Here, we report the characteristics of the regulatory network orchestrating early mesodermal development in the fruitfly Drosophila, where the transcription factor Twist is both necessary and sufficient to drive development. Through the integration of chromatin immunoprecipitation followed by microarray analysis (ChIP-on-chip) experiments during discrete time periods with computational approaches, we identified >2000 Twist-bound cis-regulatory modules (CRMs) and almost 500 direct target genes. Unexpectedly, Twist regulates an almost complete cassette of genes required for cell proliferation in addition to genes essential for morophogenesis and cell migration. Twist targets almost 25% of all annotated Drosophila transcription factors, which may represent the entire set of regulators necessary for the early development of this system. By combining in vivo binding data from Twist, Mef2, Tinman, and Dorsal we have constructed an initial transcriptional network of early mesoderm development. The network topology reveals extensive combinatorial binding, feed-forward regulation, and complex logical outputs as prevalent features. In addition to binary activation and repression, we suggest that Twist binds to almost all mesodermal CRMs to provide the competence to integrate inputs from more specialized transcription factors.
Keywords: Twist, Dorsal, mesoderm, ChIP-on-chip, transcriptional network, temporal regulation
Embryonic development occurs through the progressive restriction of cell fates, from pluripotent fields of cells to complex organs and tissues. This process requires a directed progression through interlinked regulatory states, each defined by the total set of active transcription factors (TFs) (Davidson 2006). At each stage of development, the combined inputs of signaling and transcriptional networks regulate the expression of specific sets of genes that drive the transition to the next, often more specialized, state. Understanding how the underlying cis-regulatory networks produce temporal and spatial gene expression is therefore an essential step toward deciphering metazoan development.
Genetic studies have been very successful in identifying individual TFs, representing key network nodes for different developmental processes. However, the logics of only a limited number of developmental networks has been extensively studied using genetic and biochemical approaches, with careful dissection of individual cis-regulatory modules (CRMs) (Davidson et al. 2002; Schroeder et al. 2004; Inoue et al. 2005; Koide et al. 2005; Stathopoulos and Levine 2005). Due to the laborious nature of these experiments, it is very difficult to apply these approaches to global regulatory networks. Moreover, not all key components can be identified based on their phenotype alone, due to functional redundancy or pleiotropic effects. A number of recent studies have used different large-scale techniques to define developmental networks in a gene-centered approach, including RNA interference (RNAi)-based epistasis studies (Imai et al. 2006) and large-scale yeast-1 hybrid (Deplancke et al. 2006). Here, we focus on the identification of dynamically bound CRMs as a prerequisite for generating a transcriptional network during Drosophila embryogenesis, using in vivo TF-binding data generated by ChIP-on-chip analysis (chromatin immunoprecipiation followed by microarray analysis). While ChIP-on-chip has been used extensively to reveal network architecture under many conditions in yeast and mammalian tissue culture cells (Ren et al. 2000; Odom et al. 2004; Boyer et al. 2005), it has only been applied in limited cases within a developing embryo (Birch-Machin et al. 2005; Negre et al. 2006; Sandmann et al. 2006). To reveal the cis-regulatory code underlying early aspects of mesoderm development, we focused on the central regulator of this process in Drosophila melanogaster: the basic helix–loop–helix (bHLH) TF Twist.
The early Drosophila mesoderm is composed of a field of pluripotent cells (Beer et al. 1987; Farrell and Keshishian 1999), expressing the TF Twist (Leptin 1991; Baylies and Bate 1996). After gastrulation, these cells dissociate from each other, proliferate, and migrate dorsally, while remaining unspecified. Only at later stages does the mesoderm become subdivided into different tissue primordia, including the heart and gut muscle, marked by the restricted expression of key regulators essential for their development (Borkowski et al. 1995). As a consequence, these pluripotent cells need to express the appropriate TFs and signaling proteins to generate a permissive regulatory state, from which further specification can proceed down multiple developmental paths. How the system is primed for rapid activation, while simultaneously being held in an immature state until the appropriate time, is not understood.
The TF Twist is initially expressed within the presumptive mesoderm, where it acts as a dorsoventral (D–V) axis determinant with the TF Dorsal (stage 5) (Thisse et al. 1987; Ip et al. 1992b; Shirokawa and Courey 1997). Afterward, it drives the process of mesoderm gastrulation together with the TF Snail (stage 6). Within the unspecified mesoderm, Twist acts as a master regulator that is both essential and sufficient to initiate mesoderm development (stages 7–11) (Baylies and Bate 1996). Once the mesoderm primoridium is specified and differentiation begins, Twist expression is dramatically reduced and is only maintained in adult muscle precursors (Bate et al. 1991). How this single TF can regulate such a broad variety of different processes is poorly understood. Although many genes have been identified that are genetically downstream from twist (Casal and Leptin 1996; Furlong et al. 2001a; Stathopoulos et al. 2002) only 11 direct Twist targets are known to date (Supplementary Table 1). These include the TFs snail, tinman, and Mef2, as well as the microRNA mir-1, important downstream regulators of mesoderm development (Ip et al. 1992b; Lee et al. 1997; Yin et al. 1997; Cripps et al. 1998; Gajewski et al. 1998; Biemar et al. 2005; Sokol and Ambros 2005).
Does this limited set of target genes reflect a major part of Twist’s input into the mesodermal regulatory network? By directly regulating a restricted set of transcriptional regulators, which in turn independently control more downstream effectors, Twist could induce successive cascades of transcription indirectly. Alternatively, Twist may directly regulate a large number of as yet unidentified target genes, exerting immediate control over the properties of mesodermal cells.
To understand how Twist orchestrates the transcriptional program within pluripotent mesodermal cells, we have systematically identified Twist-bound CRMs and directly regulated target genes at two consecutive time periods in vivo. By combining ChIP-on-chip experiments with expression profiling of twist loss- and gain-of-function embryos, we discovered >2000 Twist-bound CRMs and verified the regulatory potential of 13 of these in transgenic reporter lines in vivo.
The requirement for twist at multiple stages of development is reflected by differential Twist binding to three temporal groups of CRMs, revealing dynamic changes in the regulatory network. The large number of newly identified direct target genes reveals surprising roles for Twist in the regulation of gene batteries essential for proliferation and transcriptional regulation. By incorporating in vivo binding data from three additional TFs, either upstream of or downstream from Twist, we generated an initial core network describing the transcriptional landscape during early mesoderm development. In contrast to the classical hierarchical role envisioned for a master regulator, Twist targets the majority of mesodermal CRMs and participates in extensive feed-forward regulation, thereby impinging directly on multiple network levels.
Results
A temporal map of Twist-bound enhancers and direct Twist target genes
ChIP-on-chip were performed at two consecutive developmental time periods: 2–4 h (stages 5–7) and 4–6 h (stages 8–9), covering the stages of gastrulation, mesoderm expansion, migration, and early subdivision into different primordia. For each time period, four independent ChIPs were performed using two different α-Twist antibodies to reduce possible off-target effects.
To systematically identify Twist-bound regions in an unbiased, global manner we designed a high-density microarray tiling across the Drosophila melanogaster genome with ∼380,000 60mer oligonucleotide probes. Twist binds to E-box motifs: As a degenerate E-box (CANNTG) is expected to occur every ∼256 base pairs (bp) in the Drosophila genome, we designed a 60mer oligonucleotide for each E-box motif within the nonrepetitive, noncoding regions of the genome (Supplemental Material). This design made no assumptions about the specificity of the E-box bound by Twist, yet ensured all putative E-boxes were covered and that each Twist-bound sequence was detected by at least two neighboring 60mers (see Supplemental Material for more details).
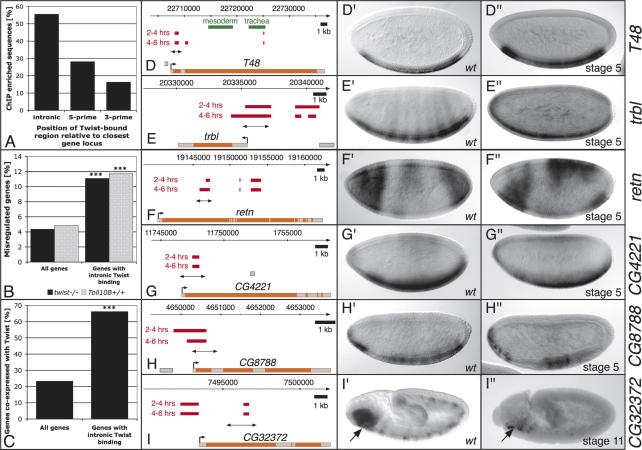
These experiments identified 2096 nonoverlapping genomic regions significantly bound by Twist within one or both developmental time periods. This set includes all known Twist-bound enhancers tested (Supplementary Table 1), except the eve-cardiac enhancer that is regulated outside the period of development assayed (Supplementary Fig. 1F). The majority of Twist-bound regions are found within introns of gene loci, rather than noncoding 5′ and 3′ regions (Fig. 1A). A similar positional bias was also observed for p53 (Wei et al. 2006) and Krüppel (Matyash et al. 2004), suggesting that introns close to the transcriptional start site represent hotspots for active CRMs. Intronic binding of Twist correlates significantly with the misregulation of these genes’ expression in twist loss-of-function mutant embryos (Fig. 1B) and their expression within the ventral blastoderm and mesoderm (Fig. 1C).
Figure 1.
Regions identified by Twist ChIP-on-chip represent functional enhancers in vivo. (A) Twist enhancers are more frequently located in intronic regions, as opposed to regions 5′ or 3′ relative to the nearest gene locus. Genes misexpressed in either twist loss-of-function or Toll10B gain-of-function mutants (B) or coexpressed with Twist (C) (information about expression patterns was compiled from the Berkeley Drosophila Genome Project in situ database, FlyBase, and published literature) are significantly enriched among genes with intronic Twist binding (right) compared with the whole Drosophila genome (left) (Fisher test, [***] p < 1e-10). (D–I) Schematic diagrams indicating the gene locus (exons in gray, introns in orange) together with Twist-enriched sequences (red bars, top: 2–4 h, bottom: 4–6 h) and the enhancer region tested (double-headed arrow). Known enhancers for T48 are indicated in green (Strutt and White 1994). (D′–I′) In situ hybridization of transcripts in wild-type embryos. H″ is reproduced from the Berkeley in situ database. (D″– I″) In situ hybridization of GFP transcripts in transgenic enhancer-GFP embryos assaying the regions indicated by double-headed arrows in D–I, respectively. All enhancers drive specific blastodermal or mesodermal expression reproducing (at least part of) the endogenous genes’ expression. All embryos are oriented with anterior facing left and the dorsal side up.
One of the major challenges for ChIP-on-chip studies is to accurately link the TF-bound enhancers to their appropriate target gene. Rather than simply taking the closest 5′ or 3′ gene, we took a more stringent approach and did not assign a Twist-bound region to a gene based on proximity alone. The results shown in Figure 1, B and C, demonstrate that Twist binds more frequently to gene loci genetically downstream from the TF and/or expressed in the same cells as the TF. We used these criteria to systematically match all 2096 Twist-bound regions (intronic or intergenic) to their likely targets, leading to a high-confidence gene assignment for 854 Twist-bound sequences (Supplementary Table 2; a detailed description of the automated gene assignment is available in the Supplemental Material). This increased the number of Twist direct targets from the previously known 11 to 494 genes. All Twist-bound regions and surrounding genes can be visualized and searched at http://furlonglab.embl.de.
Twist-bound regions function as CRMs in vivo
The RedFly database contains a comprehensive collection of previously described Drosophila enhancers, mainly characterized through single gene studies (Gallo et al. 2006). Of the 2096 Twist-bound regions, 143 overlap with known enhancers for 62 genes (Supplementary Table 2), confirming that these regions have regulatory potential in vivo. Twist was previously not known to bind to many of these enhancers; this overlap therefore provides strong evidence for a regulatory link between Twist and the 62 target genes (e.g., Abd-A, Abd-B, aop, Brd, slp1, and bap). To further examine the regulatory potential of Twist-bound regions, reporter constructs of new putative enhancer sequences were tested in transgenic animals. Six Twist-bound regions within or close to the following gene loci were assayed: T48, trbl, retn, CG4221, CG8788, and CG32372 (Fig. 1D–I). All regions proved sufficient to function as enhancers in vivo and could reproduce all or part of the endogenous spatio-temporal gene-expression pattern.
The T48, tribbles, retained, CG4221, and CG8788 enhancers initiate expression within the early blastoderm. The T48 module mirrors the expression of the endogenous gene within the presumptive mesoderm (Fig. 1D′, D″). The zygotic expression of tribbles is highly dynamic, which is reflected by the assayed CRM. This enhancer drives expression very transiently in the ventral blastoderm (Fig. 1E″) and quickly becomes ubiquitously expressed. The relatively small enhancer region for retained is activated in the anterior and posterior ventral blastoderm, where it is coexpressed with Twist (Fig. 1F″), and its expression extends into the dorsal blastoderm (Fig. 1F″). The CRMs for CG4221 and CG8788 initiate expression in the presumptive mesoderm (Fig. 1G″, H″), and continue to drive expression throughout the trunk mesoderm at later stages. The expression of the CG32372 module initiates after gastrulation in the head mesoderm, a domain that overlaps with twist expression (Fig. 1I″, arrow). It is interesting to note that Twist binds to multiple enhancer regions for many of these genes (Fig. 1D–I). This feature is also evident more globally: Almost 50% of Twist target genes have two or more Twist-bound enhancers, reflecting the complexity of their regulation (Supplementary Fig. 5).
In summary, these results demonstrate that our ChIP-on-chip experiments provide a sensitive and accurate global map of Twist-bound regulatory regions during key stages of early mesoderm development.
Twist activity is essential for target gene expression
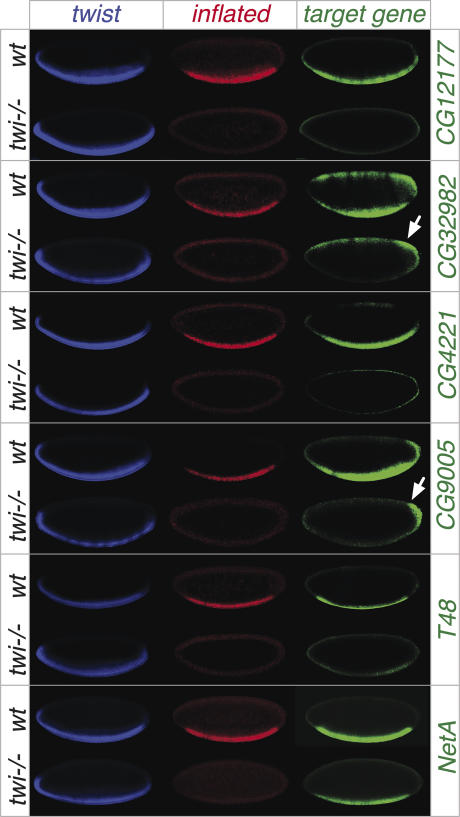
To assay the requirement of Twist function for target gene expression, we examined the expression of six novel direct targets in twist mutant embryos. These genes are expressed in the presumptive mesoderm prior to gastrualtion, and therefore at stages when the role of twist function can be assessed. Mesodermal cells are absent in twist mutant embryos later in development due to a block in gastrulation. Triple-fluorescent in situ hybridization was performed using probes directed against twist (blue channel; while twist1 is a protein-null allele, twist RNA is still expressed), inflated (red channel; this gene is dependent on twist for its expression and was used as a marker to distinguish homozygous mutant embryos from their siblings), and a probe directed against one of the six direct target genes (green channel). The spatial expression of all six targets overlaps with twist within the presumptive mesoderm (Fig. 2, cf. blue and green channels).
Figure 2.
Twist activity is essential for the expression of many of its target genes. In situ hybridization of Twist targets in wild-type embryos (top region of panel) and twist mutant embryos (bottom). All embryos are at stage 5, with anterior to the left and dorsal to the top. The images show triple fluorescent in situ hybridization with probes against the following transcripts: twist (blue), inflated (red), and the target gene (green). As inflated is a target of Twist, lack of inflated expression serves as a functional marker for twist homozygous mutant embryos. Note, the presumptive mesoderm expression of CG12177, CG32982, CG4221, CG9005, and T48 is severely reduced or absent in twist mutant embryos. NetA expression appears largely unaffected.
Importantly, twist activity is essential for the expression of five out of six genes examined (Fig. 2). Note, for CG32982 and CG9005, residual expression remains outside the twist expression domain in the dorsal and posterior blastoderm, respectively (Fig. 2, arrow). These results, in combination with our in vivo binding data, indicate that Twist binding to a CRM is a prerequisite to activate target gene expression for a large percentage of its targets. The role of Twist binding to the NetA enhancer remains unclear (Fig. 2). Twist may act redundantly with other TFs, or alternatively may function in a more subtle manner to modulate the levels of expression.
Twist and Dorsal collaborate much more extensively than previously predicted
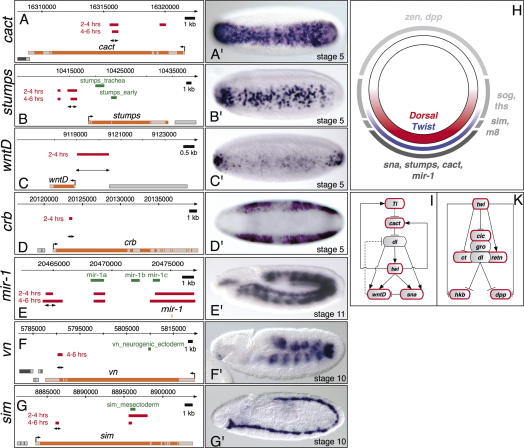
One of the earliest functions of Twist within the pregastrula embryo is the coregulation of D–V patterning with the NFκB ortholog Dorsal (Shirokawa and Courey 1997). Dorsal acts as a morphogen by regulating its target genes at (at least) three threshold concentrations along the D–V axis (Fig. 3H; for review, see Stathopoulos and Levine 2005). Type I-regulated Dorsal enhancers receive high levels of Dorsal, contain low-affinity Dorsal sites and drive expression in ventral mesodermal domains (e.g., sna, htl, twi). Type II enhancers receive intermediate levels of Dorsal and drive expression in mediolateral domains of different sizes (e.g., sim, brk, vn), while Type III enhancers receive low levels of Dorsal, contain high-affinity Dorsal sites, and can be either activated (sog, ths) or repressed (dpp, tld, zen) by Dorsal. This system has been studied so intensively that the level of knowledge is sufficient for quantitative modeling of cis-regulatory interactions (Zinzen et al. 2006). It was therefore of interest to determine whether our global analysis could reveal new insights into this process. Our data identified in vivo binding of Twist to both Type I and II Dorsal enhancers, as expected (Supplementary Fig. 1). The boundaries of Twist binding are in remarkable agreement with the limits of characterized minimal enhancers (e.g., htl, rho, and ths) (Supplementary Fig. 1G,L,R). More importantly, we were very surprised to identify new CRMs for several of these well-characterized genes (Fig. 3).
Figure 3.
Identification of novel enhancers for genes differentially expressed along the D–V axis. (A–G) Schematic diagrams indicating the target locus (exons in gray, introns in orange) together with Twist-enriched sequences (red bars, top: 2–4 h, bottom: 4–6 h), the location of known regulatory sequence (green bars), and the novel enhancer region tested (double-headed arrow). (A′–G′ In situ hybridization of lacZ transcripts in transgenic enhancer lines. (A′,B′) The enhancers for cactus (cact) and stumps drive expression in all or a subset of cells of the ventral mesoderm, respectively. The WntD enhancer displays repression in the central snail-expressing domain, and activation at the anterior and posterior poles (C′), while the crumbs (crb) enhancer is excluded from the entire ventral domain and is only activated in the ectoderm (D′). In addition to the recovery of known regulatory regions (E,G) novel enhancers were discovered for mir-1, vn, and sim (E′–G′). (H) Schematic overview of a transverse section through a stage 5 embryo. The nuclear Dorsal gradient (red) activates Twist expression (blue) on the ventral side of the embryo. Twist binds to CRMs associated with targets expressed in ventral (e.g., sna, stumps, cact, mir-1), lateral (m8, sim, ths, sog), and dorsal (zen, dpp) domains of the early embryo. (I) Twist regulates components of the Dorsal network; schematic model of the subcircuit leading to collaborative activation of targets in the ventral blastoderm. Twist binds to CRMs of all genes with a red border. Dashed lines indicate indirect regulation. (K) Twist also binds to CRMs regulating known components (red border) of the repressive complex associated with Dorsal.
Seven novel enhancers for D–V patterning genes reveal the regulatory complexity of Twist-bound CRMs: The cactus, stumps, and wntD enhancers drive expression in a domain overlapping Twist within the ventral blastoderm (Fig. 3A–C) and likely represent Type I enhancers. Cactus, an IκB ortholog, is expressed both maternally and zygotically (Kidd 1992) and sequesters Dorsal within the cytoplasm. While the regulation of zygotic cactus expression was previously not understood, our data reveals a Twist-bound CRM that is sufficient to drive expression in the presumptive mesoderm (Fig. 3A′). Twist also binds to a CRM of Toll (Supplementary Figs. 1, 3I). Although the function of cactus’ and Toll’s zygotic regulation remains unclear in Drosophila, positive feedback regulation of zygotic Toll-receptor expression is required to refine the Dorsal nuclear gradient in the flour beetle Tribolium castaneum (Chen et al. 2000).
The stumps CRM is expressed in a subset of Twist-expressing cells, yielding a salt and pepper pattern that may reflect the requirement for a second, partially redundant enhancer (e.g., the “stumps_early” enhancer) (Stathopoulos et al. 2004) to give robust expression (Fig. 3B′). The wntD CRM is highly expressed at the anterior and posterior poles of the ventral blastoderm, but is very weakly expressed within the central region (Fig. 3C′). This mirrors the transient expression of the endogenous gene at this stage of development (Fig. 1C in Ganguly et al. 2005). This single enhancer reflects the regulatory logic deduced from genetic studies: The inputs from Twist and Dorsal activate WntD, while Snail represses its transcription within the presumptive mesoderm (Fig. 3I; Ganguly et al. 2005). The CRM for crumbs reproduces the endogenous genes expression (Fig. 3D′). This 480-bp region can function as an enhancer in the ectoderm while acting as a silencer within the ventral blastoderm. This ventral repression is most likely due to direct input from Snail on this CRM (Leptin 1991). Therefore, even at the same stage of development, these four Twist-bound CRMs drive expression in different spatial patterns within a small population of cells. This complexity is clearly mediated by context-dependent integration of additional inputs. Three additional CRMs for mir-1 (Type I), vn, and sim (Type II Dorsal targets) drive expression later in development, reproducing part of the endogenous gene’s expression (Fig. 3E–G).
Unexpectedly, Twist also binds to characterized Dorsal Type III enhancers known to regulate dpp, ind, and ths (Supplementary Fig. 1C,I,R). Dorsal and its associated corepressors Cut, Retained, and Capicua recruit Groucho to repress dpp, confining its expression to the dorsal blastoderm (Valentine et al. 1998). The cobinding of Twist and Dorsal to Type III CRMs suggests that these factors may also collaborate in transcriptional repression. Interestingly, Twist binds to regulatory regions of all three Dorsal corepressors (Fig. 3K; Supplementary Table 2), providing another level at which Twist may modulate Dorsal-mediated repression. Overall, our exhaustive map of new CRMs for D–V patterning genes greatly extends our previous knowledge and will likely improve predictive models for this system.
Twist targets functional modules required for diverse aspects of mesoderm development
Twist is not only required for D–V patterning. The 494 direct target genes are significantly enriched in functional groups of genes involved in cell communication, signal transduction, cell motility, and cell adhesion (Fig. 4A). Genes in these categories are essential for multiple aspects of development, including gastrulation and directed migration of mesodermal cells. Genetic studies have demonstrated a requirement for twist in these processes (Leptin and Grunewald 1990); however, the molecular mechanism remained ill-defined. Our data reveals Twist binding to CRMs for entire functional modules necessary for both gastrulation (Supplementary Table 3) and migration (the FGF pathway) (Fig. 4B).
Figure 4.
Twist targets functional cassettes required for diverse developmental processes. (A) Important developmental processes are overrepresented among direct Twist target genes: Comparing the fraction of direct Twist targets (gray) annotated with specific GO-slim terms with the frequency of these terms among all annotated FlyBase genes (black) reveals significant enrichment of several GO processes (EASE resampling analysis, [***] p < 0.001, [**] p < 0.01). (B,C) Numerous FGF pathway members and important regulators of cell cycle progression are direct Twist targets: manual pathway mapping of FGF signaling (B) and cell cycle regulation in Drosophila (C). Genes with a red border were identified as direct Twist targets.
The present study highlights a new direct connection between Twist and many key components involved in cell cycle progression and cell growth (Fig. 4C). Members of both the Cdk2/CyclinA/B and Cdk2/CyclinE complexes are targeted, as well as modifiers of their activity and genes involved in cytokinesis and replication. In many cases, Twist binds to several CRMs of these genes (e.g., cyclinE and E2f) (Supplementary Fig. 6), revealing the complexity of their regulation. This surprising link between Twist and the cell cycle is highly likely to be of regulatory significance; twist mutant embryos have proliferative defects that can be genetically separated from the block in mesoderm gastrulation (Arora and Nusslein-Volhard 1992).
These three functional groups of target genes (involved in morphogenesis, migration, and cell proliferation) have been defined as essential developmental network “plug-ins” (Davidson 2006). Twist orchestrates early mesoderm development by binding to CRMs of virtually all genes within functional groups essential for gastrulation, mesoderm proliferation, migration, and specification. In contrast, few CRMs for genes involved in terminal differentiation (e.g., sarcomere structure) are targeted by Twist.
Twist is a highly connected hub targeting a large repertoire of TFs
This global map of Twist-bound CRMs provides a first glimpse of Twist’s connectivity to the rest of the regulatory genome. Remarkably, TFs represent the largest group of Twist targets: Twist binds to CRMs of a striking 25% (113/454) of all sequence-specific Drosophila TFs (Supplementary Table 2). Among these are TFs essential for mesoderm development, including gap (hb, hkb, kr, kni), pair rule (eve, slp, opa, odd, prd, run), and segmentation genes (en, hh, ptc, wg), as well as homeotic genes (pb, Scr, Antp, Abd-A, Abd-B, Ubx). These classes of target genes implicate a new role for Twist in the establishment or maintenance of anterior–posterior patterning within the mesoderm in addition to its known role in D–V axis formation. Although the function of many of the remaining TFs is unknown, our data links these regulators to mesoderm development. The sheer number of TFs regulated by Twist does not support a simple hierarchical network, where Twist regulates a small set of TFs, which in turn control another layer of regulators, and so forth. Rather, our data suggests a model for Twist contributing to the regulation of the majority of TFs involved in every aspect of early mesoderm development.
Temporal enhancer occupancy by Twist reveals stage-specific coregulators
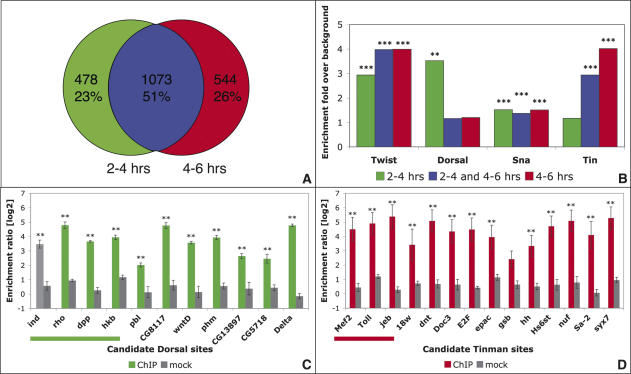
Although Twist is expressed during both developmental time periods assayed, it binds to CRMs in a temporally regulated manner. Approximately half of the enhancer regions are detected at both time periods, indicating continuous binding of Twist throughout these developmental stages (Fig. 5A). In contrast, 23% of Twist-CRMs are only bound in early development (2–4 h), while 26% are specific to later time periods (4–6 h). This dynamic occupancy reveals that the ability of Twist to bind to CRMs is tightly controlled beyond the mere presence of a suitable binding site, and is likely regulated by other TFs that aid or inhibit binding. To identify additional regulators that could differentiate between temporally bound CRMs, we searched for overrepresented sequence motifs, using two complementary computational approaches: statistical enrichment of position weight matrices (PWMs) for characterized TFs, and the de novo detection of overrepresented motifs (Supplemental Material).
Figure 5.
Twist occupies enhancers in a temporally regulated manner with Dorsal and Tinman. (A) Differentially enriched Twist-bound sequences: Roughly half of the CRMs are detected at both 2–4 h and 4–6 h (blue). About one-quarter of all detected regions are specific to either the early (green) or late (red) time periods. (B) Motifs for several characterized TFs are significantly enriched within Twist-bound CRMs: Scanning sequences bound specifically at 2–4 h (green), 4–6 h (red), or at both time periods (continuous, blue) with PWMs for known regulators in Drosophila revealed enrichment of Twist, Dorsal, Snail, and Tinman motifs within one or more of these temporal groups (Clover, [***] p < 0.001, [**] p < 0.01). (C) Dorsal co-occupies novel 2–4-h Twist-bound CRMs: ChIPs with two different α-Dorsal antisera (green, total n = 4) or preimmune serum (gray, n = 4) were performed at 2–4 h after egg laying and analyzed by quantitative real-time PCR using primers assaying predicted Dorsal sites within sequences enriched by Twist specifically at 2–4 h. Both known control sites (underlined, left) as well as all seven novel sites tested are significantly bound by Dorsal in vivo. The X-axis indicates the region tested; the Y-axis displays the level of enrichment as the ratio of enrichment using primers against the region of interest compared with primers covering a negative control region. (D) Tinman binds novel 4–6-h Twist-bound enhancers: Predicted Tinman-binding sites within continuously or late-bound sequences were assayed in α-Tinman (red) or mock (gray) ChIPs, using two independent antisera as described in C. All but one predicted site are significantly bound by Tinman in vivo. (**) p < 0.05, Wilcoxon rank test.
Twist and Snail consensus motifs are significantly overrepresented in all three groups of CRMs (Fig. 5B; Supplementary Table 4), indicating a potential for extensive cobinding between these two TFs. In contrast, Dorsal motifs are exclusively enriched in the early-bound CRMs, and not in the late group. While Tinman motifs are specifically overrepresented in the continuous and late-bound CRMs (Fig. 5B; Supplementary Table 4). A number of other motifs were also uncovered (Supplementary Fig. 7), including sites for potential Twist/Daughterless heterodimers, suggesting additional mechanisms to generate diverse outputs from Twist-CRMs.
Our data reveals Twist binding to almost all previously characterized Dorsal enhancers (Fig. 3; Supplementary Fig. 1). Twist and Dorsal are known to interact physically and to coregulate enhancers in the early, but not the late, time window of our experiment (Shirokawa and Courey 1997). We therefore hypothesized that Dorsal may be coregulating many of the newly discovered Twist CRMs, in keeping with the specific enrichment of Dorsal consensus motifs within these enhancers. To experimentally test Dorsal’s presence on predicted sites in vivo, we performed ChIP experiments at 2–4 h of development. Significant binding of Dorsal was detected by quantitative real-time PCR to all seven predicted sites tested (Fig. 5C). Similarly, as Tinman consensus sites were significantly enriched in 4–6-h CRMs, we tested the in vivo occupancy of predicted sites by Tinman at this stage of development. ChIP experiments detected significant binding of Tinman to 10 of 11 sites tested (Fig. 5D). Given the large number of early and late CRMs, the enrichment of these motifs highlights extensive combinatorial binding of Dorsal and Twist at 2–4 h, and Tinman and Twist at 4–6 h. A substantial part of Twist’s temporal specificity likely stems from its association with these upstream and downstream coregulators.
A core transcriptional network for early mesoderm development
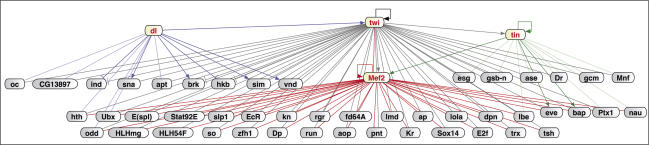
To delineate the combinatorial relationships between Twist and other TFs, we generated an initial transcriptional network for early mesoderm development. The temporal binding map for Twist was integrated with in vivo binding data for Mef2, Dorsal, and Tinman. Our previous study of Mef2-bound enhancers (Sandmann et al. 2006) offers the largest collection of regulatory regions bound at this stage of development to date. As it is difficult to visualize all 494 Twist target genes, we focused on TFs whose CRMs are cobound by two or more regulators during these stages of development (Fig. 6). Therefore, all links in this network represent direct connections to the same CRM at the same stages of development.
Figure 6.
A core transcriptional network for early mesoderm development. Through the integration of ChIP-on-chip data for Twist and Mef2 with in silico predictions, ChIPs, and literature searches for Dorsal and Tinman, we identified CRMs that are cobound by at least two TFs during the same stages of development. The regulatory connections for CRMs fulfilling these criteria are shown for all genes coding for TFs. Color code: Dorsal (dark blue, known regulation; light blue, predicted interaction), Twist (gray), Mef2 (red), and Tinman (dark green, known regulation; light green, predicted interaction). The direction of regulation, if known, is indicated by pointed or bar-ended arrows. Feed-forward loops and combinatorial regulation of downstream regulators are dominant features controlling early mesoderm development.
The resulting core network of 51 TFs is already relatively complex, with nine genes [nau, E(spl), eve, bap, Ubx, lbe, odd, hth, and Ptx1] being targeted by three out of the four examined regulators. The topology of the network provides several insights into how Twist functions to regulate multiple aspects of early mesoderm development. Extensive combinatorial binding and feed-forward regulation are abundant features. Dorsal activates twist, which in turn coregulates the majority of known direct Dorsal targets. This network motif is even more prominent within the mesoderm: Twist regulates the expression of Mef2 and tinman, and cobinds with these TFs to many of their targets’ enhancers. In fact, Twist co-occupies 42% of all Mef2-bound enhancers during early mesoderm development (Supplementary Table 2). Depending on the logical inputs from the two upstream regulators (transcriptional repression or activation), feed-forward loops can aid in cellular decision making by filtering out noisy regulatory inputs (Mangan et al. 2003) or control the timing of a transcriptional response (Penn et al. 2004; Mangan et al. 2006). For example, early gene expression in the mesoderm (e.g., activation of tin) depends on Twist alone, while transcription of other genes initiated at a later stage may require the input from both Twist and Tinman proteins.
Discussion
Through the integration of ChIP-on-chip analysis with expression profiling data during early stages of Drosophila development, we have identified >2000 Twist-bound regulatory regions and almost 500 direct target genes. This data, in combination with in vivo binding data for other TFs, lays the foundation of a transcriptional network describing early mesoderm development. The resulting network view reveals regulatory features that form the basis of Twist’s functional versatility.
Twist-bound CRMs correspond to silencers as well as enhancers of transcription
Our data revealed extensive Twist binding to characterized Dorsal enhancers and also, surprisingly, to Dorsal-regulated silencers (e.g., dpp; Supplementary Fig. 1C). Moreover, many of the new regulatory regions we identified for D–V patterning genes can function either as enhancers or integrated enhancer-silencer modules (e.g., WntD and crumbs) (Fig. 3). This ability of Twist to act within the context of silencers, as well as enhancers, may partially explain the widespread recruitment of Twist to many regulatory regions and its ability to regulate diverse developmental processes.
An attractive molecular explanation for this bifunctionality is the potential of Twist to form both homodimers and heterodimers. Twist homodimers drive gene activation in Drosophila, while Twist-Daughterless heterodimers are associated with transcriptional repression (Castanon et al. 2001). This model is supported by the significant overrepresentation of the Twist/Daughterless heterodimer consensus motif in both 2–4-h and 4–6-h CRMs (Supplementary Fig. 7). Direct protein– protein interactions between Twist and Dorsal (Shirokawa and Courey 1997) is an alternative mechanism for Twist’s incorporation into repressive complexes.
A network with unexpected topology governs early mesoderm development
Although the network generated in this study is far from complete, it represents the largest set of combinatorial-bound CRMs during these stages of development described to date, and therefore provides a comprehensive resource to decipher general regulatory principles. The resulting network topology was surprising. Instead of Twist regulating a restricted group of TFs, which in turn regulate a successive wave of transcription in a relay model, Twist directly impinges on CRMs for the vast majority of genes expressed in the early mesoderm.
The extent of combinatorial binding was also unanticipated. There is extensive cobinding of Twist and Dorsal to early 2–4-h CRMs. In fact, the presence of Dorsal binding may be a general prerequisite for Twist binding to enhancers specific for early development. The cooperative binding of Dorsal and Twist to the rho and sim CRMs supports this model (Ip et al. 1992a; Kasai et al. 1998; Markstein et al. 2004; Papatsenko and Levine 2005). At 4–6 h of development, the composition of TFs impinging on Twist-bound CRMs changes. Although genome wide ChIP-on-chip data is currently not available for Tinman, the significant overrepresentation of Tinman motifs in Twist-bound CRMs and the ability of Tinman to bind to the majority of sites tested (Fig. 5D) indicates prevalent combinatorial binding between these two TFs during 4–6 h of development. Comparing Twist-bound CRMs with a previously generated data set for Mef2 (Sandmann et al. 2006) revealed extensive cobinding to enhancers early in development (Fig. 6; Supplementary Table 2). Converging regulatory connections through combinatorial binding can produce diverse logical outputs, depending on the nature of the TFs. The cobinding of several pan-mesodermal TFs (Twist, Tinman, and Mef2) may ensure robust gene expression. While in other contexts—for example, the WntD-enhancer—the combined inputs of Twist and Snail (Fig. 2C) allow for spatial fine-tuning of gene expression.
The core network also revealed an abundance of feed-forward loops, providing directionality during early mesoderm development. This is prevalent with both upstream regulators of Twist (Dorsal and Twist) and downstream regulators (Tinman and Twist and Mef2 and Twist). This network motif will likely become even more widespread as additional ChIP-on-chip data becomes available. Twist targets an astounding number of TFs, which may represent an almost complete repertoire of TFs required for early mesoderm development. It is tempting to speculate that Twist participates in feed-forward regulation, with many of these factors through combinatorial binding to different subsets of the ∼2000 Twist-bound CRMs.
Temporal network dynamics reflect developmental progression
Both the composition and connectivity of regulatory networks describing developmental progression will naturally change over time. To capture dynamic changes within the early mesodermal network, we performed our experiments at consecutive time periods. Our data reveals temporally regulated binding of Twist to three classes of CRMs: early, continuous, and late. Similar temporally restricted enhancer occupancy has also been observed for other regulators with broad temporal expression, suggesting that this may be a general feature of developmental networks—e.g., MyoD (Bergstrom et al. 2002; Gaudet et al. 2004), PHA-4 (Bergstrom et al. 2002; Gaudet et al. 2004), and Mef2 (Sandmann et al. 2006).
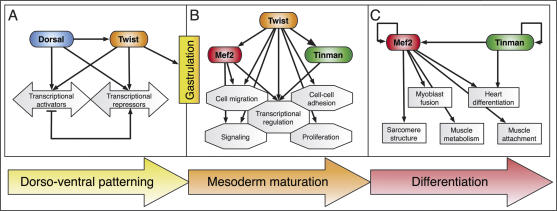
The temporal occupancy of specific CRMs by Twist reflects the development of this tissue (Fig. 7). At 2–4 h of development, Twist and Dorsal coregulate genes essential for D–V patterning (Fig. 7A; Supplementary Table 2). Twist also targets an almost complete set of genes essential for gastrulation and is required to progress to the next phase of development, mesoderm maturation (Fig. 7B). During this developmental window, the predominant target genes are part of functional modules essential for the cell migration, proliferation, patterning, and specification events occurring within the mesoderm at these stages. As expected for a TF essential for early aspects of mesoderm development, Twist does not bind to significant numbers of CRMs for genes involved in terminal differentiation. This is in sharp contrast to Mef2, which first co-occupies CRMs involved in early mesoderm development with Twist, and later selectively regulates an alternative group of CRMs driving genes involved in later aspects of differentiation; e.g., sarcomere structure or muscle attachment (Fig. 7C).
Figure 7.
Dynamic changes in the functional groups of genes targeted by Twist reflect developmental progression. (A) Before gastrulation, Twist collaborates with Dorsal to subdivide the embryo along the D–V axis and triggers target gene expression required for gastrulation. (B) Once the mesoderm has invaginated, Twist is associated with pan-mesodermal regulators (tin and Mef2) and CRMs regulating important “plug-ins” of development (Davidson 2006), providing the pluripotent cells with the competence to continue differentiation. (C) Twist expression is lost in cells activating gene batteries required for terminal differentiation (e.g., structural muscle proteins).
Conserved regulation of functional classes of genes by Twist
Integrating our data with genetic evidence from other species suggests that the regulation of several functional gene cassettes by Twist is conserved throughout evolution, from flies to man. These include (1) the FGF signaling pathway: Mutations in human FGF receptors phenocopy mutants in human twist (Htwist) (Wilkie and Morriss-Kay 2001). (2) Genes implicated in epithelial–mesenchymal transitions (EMTs): In mice and humans, Twist facilitates tumor metastasis through the promotion of EMTs (Yang et al. 2004). (3) Cell proliferation and apoptosis: Htwist has been classified as a potential oncogene, as it maintains tissue culture cells in a proliferative state. Interestingly, ectopic expression of Htwist in Drosophila also induces proliferation and inhibits p53-dependent apoptosis, indicating that the ability to regulate these processes is conserved (Gullaud et al. 2003). However, for each process, only a few Twist-regulated genes were known. Extrapolating from our findings in flies points toward a role for Twist in the direct regulation of entire gene modules required for each process in vertebrates.
An emerging model for Twist as a global competence factor for mesoderm development
Our results provide an initial global view of the transcriptional network describing early mesoderm development within the metazoan Drosophila. Twist resides at the top of this network and binds to CRMs for the vast majority of genes that need to be expressed during these stages. In many cases, Twist is essential and sufficient to drive expression of the target gene (e.g., Fig. 2; mir-1; Sokol and Ambros 2005). In other cases, however, the contribution of Twist remains unclear—e.g., crumbs (Leptin 1991) and NetA (Fig. 2). Rather than acting as a binary switch, Twist may act redundantly with other TFs. Alternatively, Twist may provide the competence for more specific TFs to bind to these CRMs; for example, by acting as a pioneer TF to facilitate chromatin remodeling (Berkes et al. 2004).
In species as diverse as flies, jellyfish, and mice, Twist is only expressed in mesodermal cells when they are in an immature state, and loss of twist expression correlates with the initiation of differentiation (Spring et al. 2000; Bialek et al. 2004). Moreover, overexpression of Twist-1 in mice is sufficient to block osteoblast differentiation (Bialek et al. 2004). We suggest that Twist provides the mesoderm with the competence to be pluripotent: first, by providing these cells with the components necessary to respond to inductive cues directing further specification; and second, by providing an almost universal repertoire of mesodermal CRMs with the competence to respond to other TFs. Once bound by Twist, these regulatory regions may be primed for activation by more specialized TFs, and thereby allow rapid developmental progression at the appropriate time.
Materials and methods
ChIP
ChIP was performed as described previously (Sandmann et al. 2006). In brief, for each time point studied, four independent chromatin samples were prepared from tightly staged 2-h wild-type embryo collections. Anti-Twist sera from two different rabbits were used to exclude serum-dependent biases: The first was raised against Twist’s amino acids 338–490. The second antiserum was a generous gift from Siegfried Roth. Only regions that were significantly enriched by both α-Twist antibodies, but not in mock experiments, were considered. Anti-Dorsal antisera were kindly provided by Michael Levine and Nicholas Gay. The first α-Tinman antiserum was raised against full-length Tinman (Furlong laboratory); the second α-Tinman serum was a generous gift from Manfred Frasch. In each experiment, an additional four mock immunoprecipitations with rabbit preimmunesera were performed. All ChIPs were assayed by quantitative real-time PCR (see Supplemental Material for details).
Expression profiling of twist loss-of-function mutants and gain-of-function embryos
The collection of the RNA samples has been described previously (Furlong et al. 2001a, b). The twistey53 allele was used to assay expression changes due to loss of twist function. The constitutively active Toll10B allele was used to produce embryos with ectopic Twist. Samples were amplified and hybridized to microarrays featuring at least one probe for each annotated gene in the Drosophila melanogaster genome as described previously (Sandmann et al. 2006). Four biological repeats were performed for each developmental time point. Dye swaps were included to account for possible dye biases.
Microarray hybridization and analysis
The raw data from the cDNA arrays was normalized by print-tip LOWESS normalization using the Tm4 analysis package (Saeed et al. 2003). To identify differentially expressed genes, the mutant samples were hybridized directly against stage-matched wild-type samples. A one-class SAM analysis (Tusher et al. 2001) was performed on four independent biological repeats for each time point. The following significance cutoffs were chosen: a log2 ratio of less than −0.5 and a q value <0.01 at two consecutive time points for the loss-of-function time course; log2 ratio of >0.7 and a q value <0.05 for the gain-of-function experiment.
The amplified Twist ChIPs were hybridized to custom-designed microarrays optimized for assaying all euchromatic E-box motifs outside repetitive or coding sequences (details available in Supplemental Material). The arrays were produced using maskless array synthesizer (MAS) technology (Nuwaysir et al. 2002). DNA labeling and hybridizations were performed as described previously (Stolc et al. 2004) and explained in detail in the Supplemental Material. To identify ChIP-enriched fragments, the experimental samples were hybridized against genomic DNA in a reference design. Dye swaps were included to account for possible dye biases. The oligonucleotide arrays were scanned at 5-μm resolution, and raw data was LOWESS normalized to remove (1) spatial and (2) concentration-dependent dye biases. Significantly enriched regions were identified using consecutive probe-level statistics combined with a hidden Markov model using Tilemap (Ji and Wong 2005).
Target gene assignments
The assignment of ChIP-enriched regions to a target gene was performed by integrating additional metadata for all surrounding genes (high-throughput in situ hybridization data, expression profiling, and the literature) using an automated scoring scheme (modified from Sandmann et al. 2006; see Supplemental Material for details).
Generation of transgenic reporter strains
Fragments within the following coordinates were cloned into the pH-stinger or pH-pelican vectors (Barolo et al. 2000) for germline transformation (coordinates based on genome release 4.2): chr2L:16,315,340–16,316,352 (cact), chr3L:7,495,261–7,497,085 (CG32372), chr3R:11,746,513–11,748,343 (CG4221), chr2R:4,650,254–4,650,882 (CG8788), chr3R:20,123,093–20,123,803 (crb), chr2R:19,145,341–19,147,650 (retn), chr3R:8,885,994– 8,886,691 (sim), chr3R:10,414,437–10,416,493 (stumps), chr3R:22,707,641–22,709,469 (T48), chr3L:20,335,011–20,337,399 (trbl), chr3L:5,790,824–5,791,419 (vn), chr3R:9,118,898–9,120,556 (wntD).
Histological techniques
In situ hybridizations were performed as described previously (Furlong et al. 2001a).
Identification of overrepresented known TF sites
Significantly overrepresented PWMs were identified in a two-step procedure: Motifs for 104 known transcriptional regulators from Drosophila melanogaster were obtained from FlyReg (Bergman et al. 2005), Transfac (Matys et al. 2006), Jaspar (Vlieghe et al. 2006), or the literature. First, the Clover tool (Frith et al. 2004) was used to identify motifs significantly enriched between Twist-bound and control sequences with a cutoff of p < 0.05. Second, for the set of significant PWMs, different similarity thresholds were evaluated using Patser (Hertz and Stormo 1999) and only motifs achieving at least a 1.5-fold enrichment, while simultaneously reporting hits within >10% of the enriched sequences, were considered to be of interest (see Supplemental Material for details).
Prediction of novel Dorsal and Tinman sites
Dorsal and Tinman sites were predicted using Patser with a score cutoff of 8.5 (Dorsal) and 10.5 (Tinman). See Supplementary Table 4 for individual predictions.
Accession numbers
Details about the microarrays designed and used in this study have been deposited in the ArrayExpress database under the following accession numbers: A-MEXP-588 (E-box array), A-MEXP-278 (cDNA microarray), and A-MEXP-314 (INDAC oligonucleotide microarray).
Raw microarray data can be accessed using the following accession numbers: E-TABM-162 (twist−/− loss-of-function time course hybridized to cDNA microarrays), E-TABM-161 (twist−/− loss-of-function time course hybridized to INDAC oligonucleotide microarrays), E-TABM-160 (Toll10B gain-of-function time course hybridized to INDAC oligonucleotide microarrays), E-TABM-159 (Twist ChIP-on-chip data). Alternatively, all normalized data can be downloaded from http://furlonglab.embl.de.
Acknowledgments
We are very grateful to Detlev Arendt and Jürg Müller for critically reading the manuscript. We thank all members of the Furlong laboratory, especially Julien Gagneur, Janus Jakobsen, Martina Braun, and Hilary Gustafson. We are grateful to Lars Jensen and Peer Bork for help with target gene assignments. We thank Anne-Mari Voie and the MyoRes transgenic platform for embryo injections. This work was supported by funding from EMBL and from the EU 6th framework MyoRes grant.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1509007
References
- Arora K., Nusslein-Volhard C., Nusslein-Volhard C. Altered mitotic domains reveal fate map changes in Drosophila embryos mutant for zygotic dorsoventral patterning genes. Development. 1992;114:1003–1024. doi: 10.1242/dev.114.4.1003. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L.A., Posakony J.W., Carver L.A., Posakony J.W., Posakony J.W. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bate M., Rushton E., Currie D.A., Rushton E., Currie D.A., Currie D.A. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- Baylies M.K., Bate M., Bate M. twist: A myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- Beer J., Technau G.M., Campos-Ortega J.A., Technau G.M., Campos-Ortega J.A., Campos-Ortega J.A. Lineage analysis of transplanted individual cells in embryos of Drosophila melanogaster. Roux’s Arch. Dev. Biol. 1987;196:222–230. doi: 10.1007/BF00376346. [DOI] [PubMed] [Google Scholar]
- Bergman C.M., Carlson J.W., Celniker S.E., Carlson J.W., Celniker S.E., Celniker S.E. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J., Perry R.L., Rudnicki M.A., Tapscott S.J., Rudnicki M.A., Tapscott S.J., Tapscott S.J. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J., Seaver K.J., Knoepfler P.S., Tapscott S.J., Knoepfler P.S., Tapscott S.J., Tapscott S.J. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Wu H., Yu K., Ornitz D.M., Olson E.N., Yu K., Ornitz D.M., Olson E.N., Ornitz D.M., Olson E.N., Olson E.N., et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Biemar F., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Sementchenko V., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin I., Gao S., Huen D., McGirr R., White R.A., Russell S., Gao S., Huen D., McGirr R., White R.A., Russell S., Huen D., McGirr R., White R.A., Russell S., McGirr R., White R.A., Russell S., White R.A., Russell S., Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowski O.M., Brown N.H., Bate M., Brown N.H., Bate M., Bate M. Anterior–posterior subdivision and the diversification of the mesoderm in Drosophila. Development. 1995;121:4183–4193. doi: 10.1242/dev.121.12.4183. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J., Leptin M., Leptin M. Identification of novel genes in Drosophila reveals the complex regulation of early gene activity in the mesoderm. Proc. Natl. Acad. Sci. 1996;93:10327–10332. doi: 10.1073/pnas.93.19.10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon I., Von Stetina S., Kass J., Baylies M.K., Von Stetina S., Kass J., Baylies M.K., Kass J., Baylies M.K., Baylies M.K. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- Chen G., Handel K., Roth S., Handel K., Roth S., Roth S. The maternal NF-κB/dorsal gradient of Tribolium castaneum: Dynamics of early dorsoventral patterning in a short-germ beetle. Development. 2000;127:5145–5156. doi: 10.1242/dev.127.23.5145. [DOI] [PubMed] [Google Scholar]
- Cripps R.M., Black B.L., Zhao B., Lien C.L., Schulz R.A., Olson E.N., Black B.L., Zhao B., Lien C.L., Schulz R.A., Olson E.N., Zhao B., Lien C.L., Schulz R.A., Olson E.N., Lien C.L., Schulz R.A., Olson E.N., Schulz R.A., Olson E.N., Olson E.N. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophilamyogenesis. Genes & Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H. The regulatory genome. Academic Press; Burlington, MA: 2006. [Google Scholar]
- Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Amore G., Hinman V., Arenas-Mena C., Hinman V., Arenas-Mena C., Arenas-Mena C., et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Reece-Hoyes J.S., Hope I.A., Hope I.A., et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Farrell E.R., Keshishian H., Keshishian H. Laser ablation of persistent twist cells in Drosophila: Muscle precursor fate is not segmentally restricted. Development. 1999;126:273–280. doi: 10.1242/dev.126.2.273. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Fu Y., Yu L., Chen J.F., Hansen U., Weng Z., Fu Y., Yu L., Chen J.F., Hansen U., Weng Z., Yu L., Chen J.F., Hansen U., Weng Z., Chen J.F., Hansen U., Weng Z., Hansen U., Weng Z., Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong E.E., Andersen E.C., Null B., White K.P., Scott M.P., Andersen E.C., Null B., White K.P., Scott M.P., Null B., White K.P., Scott M.P., White K.P., Scott M.P., Scott M.P. Patterns of gene expression during Drosophila mesoderm development. Science. 2001a;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- Furlong E.E., Profitt D., Scott M.P., Profitt D., Scott M.P., Scott M.P. Automated sorting of live transgenic embryos. Nat. Biotechnol. 2001b;19:153–156. doi: 10.1038/84422. [DOI] [PubMed] [Google Scholar]
- Gajewski K., Kim Y., Choi C.Y., Schulz R.A., Kim Y., Choi C.Y., Schulz R.A., Choi C.Y., Schulz R.A., Schulz R.A. Combinatorial control of Drosophila mef2 gene expression in cardiac and somatic muscle cell lineages. Dev. Genes Evol. 1998;208:382–392. doi: 10.1007/s004270050194. [DOI] [PubMed] [Google Scholar]
- Gallo S.M., Li L., Hu Z., Halfon M.S., Li L., Hu Z., Halfon M.S., Hu Z., Halfon M.S., Halfon M.S. REDfly: A regulatory element database for Drosophila. Bioinformatics. 2006;22:381–383. doi: 10.1093/bioinformatics/bti794. [DOI] [PubMed] [Google Scholar]
- Ganguly A., Jiang J., Ip Y.T., Jiang J., Ip Y.T., Ip Y.T. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132:3419–3429. doi: 10.1242/dev.01903. [DOI] [PubMed] [Google Scholar]
- Gaudet J., Muttumu S., Horner M., Mango S.E., Muttumu S., Horner M., Mango S.E., Horner M., Mango S.E., Mango S.E. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullaud M., Delanoue R., Silber J., Delanoue R., Silber J., Silber J. A Drosophila model to study the functions of TWIST orthologs in apoptosis and proliferation. Cell Death Differ. 2003;10:641–651. doi: 10.1038/sj.cdd.4401222. [DOI] [PubMed] [Google Scholar]
- Hertz G.Z., Stormo G.D., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Imai K.S., Levine M., Satoh N., Satou Y., Levine M., Satoh N., Satou Y., Satoh N., Satou Y., Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Inoue T., Wang M., Ririe T.O., Fernandes J.S., Sternberg P.W., Wang M., Ririe T.O., Fernandes J.S., Sternberg P.W., Ririe T.O., Fernandes J.S., Sternberg P.W., Fernandes J.S., Sternberg P.W., Sternberg P.W. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip Y.T., Park R.E., Kosman D., Bier E., Levine M., Park R.E., Kosman D., Bier E., Levine M., Kosman D., Bier E., Levine M., Bier E., Levine M., Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992a;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Park R.E., Kosman D., Yazdanbakhsh K., Levine M., Park R.E., Kosman D., Yazdanbakhsh K., Levine M., Kosman D., Yazdanbakhsh K., Levine M., Yazdanbakhsh K., Levine M., Levine M. dorsal–twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes & Dev. 1992b;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Ji H., Wong W.H., Wong W.H. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Kasai Y., Stahl S., Crews S., Stahl S., Crews S., Crews S. Specification of the Drosophila CNS midline cell lineage: Direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Expr. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- Kidd S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell. 1992;71:623–635. doi: 10.1016/0092-8674(92)90596-5. [DOI] [PubMed] [Google Scholar]
- Koide T., Hayata T., Cho K.W., Hayata T., Cho K.W., Cho K.W. Xenopus as a model system to study transcriptional regulatory networks. Proc. Natl. Acad. Sci. 2005;102:4943–4948. doi: 10.1073/pnas.0408125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Park T., Schulz R.A., Kim Y., Park T., Schulz R.A., Kim Y., Schulz R.A., Kim Y., Kim Y. Twist-mediated activation of the NK-4 homeobox gene in the visceral mesoderm of Drosophila requires two distinct clusters of E-box regulatory elements. J. Biol. Chem. 1997;272:17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes & Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- Leptin M., Grunewald B., Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110:73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- Mangan S., Zaslaver A., Alon U., Zaslaver A., Alon U., Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Mangan S., Itzkovitz S., Zaslaver A., Alon U., Itzkovitz S., Zaslaver A., Alon U., Zaslaver A., Alon U., Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J. Mol. Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Markstein M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Zinzen R., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Markstein P., Yee K.P., Erives A., Stathopoulos A., Levine M., Yee K.P., Erives A., Stathopoulos A., Levine M., Erives A., Stathopoulos A., Levine M., Stathopoulos A., Levine M., Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Matyash A., Chung H.R., Jackle H., Chung H.R., Jackle H., Jackle H. Genome-wide mapping of in vivo targets of the Drosophila transcription factor Kruppel. J. Biol. Chem. 2004;279:30689–30696. doi: 10.1074/jbc.M403345200. [DOI] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Reuter I., Chekmenev D., Krull M., Hornischer K., Chekmenev D., Krull M., Hornischer K., Krull M., Hornischer K., Hornischer K., et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Lavrov S., Bellis M., White K.P., Cavalli G., Bellis M., White K.P., Cavalli G., White K.P., Cavalli G., Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Richmond T., Gorski T., Berg J.P., Ballin J., Gorski T., Berg J.P., Ballin J., Berg J.P., Ballin J., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Schreiber J., Rolfe P.A., Gifford D.K., Rolfe P.A., Gifford D.K., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D., Levine M., Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn B.H., Bergstrom D.A., Dilworth F.J., Bengal E., Tapscott S.J., Bergstrom D.A., Dilworth F.J., Bengal E., Tapscott S.J., Dilworth F.J., Bengal E., Tapscott S.J., Bengal E., Tapscott S.J., Tapscott S.J. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes & Dev. 2004;18:2348–2353. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Braisted J., Klapa M., Currier T., Thiagarajan M., Klapa M., Currier T., Thiagarajan M., Currier T., Thiagarajan M., Thiagarajan M., et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sandmann T., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Eichenlaub M.P., Bork P., Furlong E.E., Bork P., Furlong E.E., Furlong E.E. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Schroeder M.D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Rajewsky N., Siggia E.D., Gaul U., Siggia E.D., Gaul U., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokawa J.M., Courey A.J., Courey A.J. A direct contact between the dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol. Cell. Biol. 1997;17:3345–3355. doi: 10.1128/mcb.17.6.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring J., Yanze N., Middel A.M., Stierwald M., Groger H., Schmid V., Yanze N., Middel A.M., Stierwald M., Groger H., Schmid V., Middel A.M., Stierwald M., Groger H., Schmid V., Stierwald M., Groger H., Schmid V., Groger H., Schmid V., Schmid V. The mesoderm specification factor twist in the life cycle of jellyfish. Dev. Biol. 2000;228:363–375. doi: 10.1006/dbio.2000.9956. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M., Levine M. Genomic regulatory networks and animal development. Dev. Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M., Van Drenth M., Erives A., Markstein M., Levine M., Erives A., Markstein M., Levine M., Markstein M., Levine M., Levine M. Whole-genome analysis of dorsal–ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Tam B., Ronshaugen M., Frasch M., Levine M., Tam B., Ronshaugen M., Frasch M., Levine M., Ronshaugen M., Frasch M., Levine M., Frasch M., Levine M., Levine M. pyramusand thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes & Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Hua S., Herreman T., Tongprasit W., Barbano P.E., Herreman T., Tongprasit W., Barbano P.E., Tongprasit W., Barbano P.E., Barbano P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Strutt D.I., White R.A., White R.A. Characterisation of T48, a target of homeotic gene regulation in Drosophila embryogenesis. Mech. Dev. 1994;46:27–39. doi: 10.1016/0925-4773(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Thisse B., el Messal M., Perrin-Schmitt F., el Messal M., Perrin-Schmitt F., Perrin-Schmitt F. The twist gene: Isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine S.A., Chen G., Shandala T., Fernandez J., Mische S., Saint R., Courey A.J., Chen G., Shandala T., Fernandez J., Mische S., Saint R., Courey A.J., Shandala T., Fernandez J., Mische S., Saint R., Courey A.J., Fernandez J., Mische S., Saint R., Courey A.J., Mische S., Saint R., Courey A.J., Saint R., Courey A.J., Courey A.J. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol. Cell. Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Wasserman W.W., van Roy F., Lenhard B., van Roy F., Lenhard B., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Shahab A., Yong H.C., Fu Y., Weng Z., Yong H.C., Fu Y., Weng Z., Fu Y., Weng Z., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wilkie A.O., Morriss-Kay G.M., Morriss-Kay G.M. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Savagner P., Gitelman I., Richardson A., Weinberg R.A., Gitelman I., Richardson A., Weinberg R.A., Richardson A., Weinberg R.A., Weinberg R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yin Z., Xu X.L., Frasch M., Xu X.L., Frasch M., Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R.P., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Zinzen R.P., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Stark A., Kellis M., Zhang H., Young R.A., Levine M., Kellis M., Zhang H., Young R.A., Levine M., Zhang H., Young R.A., Levine M., Young R.A., Levine M., Levine M. Whole-genome ChIP–chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes & Dev. 2007 doi: 10.1101/gad.1509607. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R.P., Senger K., Levine M., Papatsenko D., Senger K., Levine M., Papatsenko D., Levine M., Papatsenko D., Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]