Abstract
Heterochromatin plays an important role in transcriptional repression, for the correct segregation of chromosomes and in the maintenance of genome stability. Pericentric heterochromatin (PH) replication and formation have been proposed to occur in the pericentric heterochromatin duplication body (pHDB). A central question is how the underacetylated state of heterochromatic histone H4 tail is established and controlled, because it is a key event during PH replication and is essential to maintain the compacted and silenced state of these regions. Np95 is a cell cycle regulated and is a nuclear histone-binding protein that also recruits HDAC-1 to target promoters. It is essential for S phase and for embryonic formation and is implicated in chromosome stability. Here we show that Np95 is part of the pHDB, and its functional ablation causes a strong reduction in PH replication. Depletion of Np95 also causes a hyperacetylation of lysines 8, 12, and 16 of heterochromatin histone H4 and an increase of pericentromeric major satellite transcription, whose RNAs are key players for heterochromatin formation. We propose that Np95 is a new relevant protein involved in heterochromatin replication and formation.
INTRODUCTION
Heterochromatin represents a relevant fraction of most eukaryotic genomes (Perrod and Gasser, 2003) and it plays an important role for the regulation of transcriptional repression, the correct segregation of chromosomes and the maintenance of genome stability (Henikoff, 1990; Ekwall et al., 1997; Peters et al., 2001; Bitterman et al., 2003). It is mainly composed of repetitive DNA sequences, major (pericentromeric) and minor (centromeric) satellites, and its highly compacted and repressive environment (Elgin and Grewal, 2003) is mediated by the presence of specific epigenetic markers and the interaction with non-histone chromosomal proteins (Grewal and Moazed, 2003).
Although centromeric heterochromatin is mainly replicated in late S phase of the cell cycle (Maison and Almouzni, 2004), pericentric heterochromatin (PH) replication and formation are defined in mid-S phase (Bailis and Forsburg, 2003) in a specific operative unit: the pericentric heterochromatin duplication body (pHDB; Quivy et al., 2004). These replication “factories” are coincident with the horseshoe- or ring-like structures that form at the periphery of the PH core, evidenced by the characteristic nuclear DAPI spots in mouse cells, at the time of heterochromatin replication. According to this model, parental DNA is pulled out from the central core and is replicated in this unit. Once the DNA has been duplicated, newly synthesized nucleosomes are deposited and epigenetically modified to allow the formation of new heterochromatin domains (Quivy et al., 2004).
A crucial question during this process is how the underacetylation of heterochromatin is established and controlled (Grunstein, 1998), because from yeast to mammals the deacetylation of lysines 5, 8, 12, and 16 of histone H4 in these areas (Taddei et al., 1999) is essential to maintain the compacted and silenced condition of these regions (Agalioti et al., 2002). During heterochromatin replication newly synthesized H4 histones are deposited in a H4-K5 and K12 acetylated form. These residues are then hyperacetylated and maintained in this state for up to 20 min (Taddei et al., 1999). H4-K8 and K16, instead, are thought to be deposited already deacetylated and kept in this condition throughout the cell cycle. Many evidences link acetylation of H4-K8 and K16 with active transcription, whereas the deacetylated forms of these lysines are associated to silent chromatin (Agalioti et al., 2002). It has been recently demonstrated that RNAs produced by heterochromatic DNA satellite sequences are involved in heterochromatin formation in Schizosaccharomyces pombe and in mammalian cells (Reinhart and Bartel, 2002).
The expression of Np95 is apparently essential for entry from G1/G0 phase into S phase in fibroblast NIH-3T3 cells (Bonapace et al., 2002), but not in embryonic stem cells (Muto et al., 2002). Np95 KO (knockout) mice are lethal, probably because Np95 inactivation causes midgestational embryonic lethality (Muto et al., 2002). Np95 functions as a common component in the multiple response pathways against DNA damage and replication arrest and thereby contributes to genomic stability (Muto et al., 2002).
Np95 is a chromatin-binding protein that directly interacts with histones (with a H3 ≫ H1 > H2B hierarchy), both in vivo and in vitro, and is endowed with ubiquitin ligase activity (Citterio et al., 2004). It has been shown that ICBP90 binds to methylated DNA (Unoki et al., 2004) and to the methylated promoter of RB1 (Jeanblanc et al., 2005). Np95 and ICBP90 directly recruit HDAC-1 to target promoters (Unoki et al., 2004).
Np95 expression starts at the G1/S boundary and lasts up to the end of mitosis. During mid-S phase, Muto and coworkers (Miura et al., 2001) have shown that Np95 is almost exclusively colocalized in the characteristic horseshoe-like structures with chromatin-bound proliferating cell nuclear antigen (PCNA), an auxiliary protein of DNA polymerase δ, whose chromatin-bound immunostaining is observed only in S-phase cells and whose patterns during the S-phase markedly changes in accord with the 5′-bromo-2′-deoxyuridine (BrdU) incorporation sites. Distinct localization of the two proteins, however, are evident in very early and late S phase, suggesting that Np95 is not directly involved in the replication machinery, like PCNA, but is presumably involved in other DNA replication-linked nuclear events (Miura et al., 2001). Thus, Np95 has many features of factors that bind to heterochromatin and are implicated in the replication process of this chromatin area. The experimental work presented in this article was undertaken to investigate the possible role of Np95 during heterochromatin replication.
MATERIALS AND METHODS
Cells
NIH-3T3 cells and a modified human embryonic kidney 293T cell were grown in DMEM supplemented with 10% fetal bovine serum (FBS).
DNA Transfection and RNA Interference
NIH-3T3 cells were transfected with vectors expressing Np95 myc tagged using FuGENE 6 (Roche, Indianapolis, IN) according to the manufacturer's instructions. For RNA interference, NIH3T3 cells were transfected with 20 nM small interfering RNA (siRNA) duplex using Oligofectamine (Invitrogen, Carlsbad, CA). Two rounds of transfection were done for all experiments. Cells were analyzed 24 h after the last transfection. siRNA oligos were from Ambion (Austin, TX) and the targeting sequences were as follows: RNA interference (RNAi) ctrl: AAAACGAGGCAGGAAAGGCGGTT; RNAi Np95: AACGCGGCTTCTGGTATGATGTT.
Replication Timing Analysis
NIH-3T3 cells were synchronized in G0 by growing them for 48 h in serum-starved medium: DMEM plus 5 μg/ml transferine and 5.2 ng/l selenium. FBS, 10%, was then added to stimulate cell cycle reentry. Cells were harvested at 8, 10, 12, 14, 16, 18, 20, and 22 h after release. Cells were pulse-labeled for 10 min with 10 μM BrdU (Sigma-Aldrich) just before harvesting. BrdU incorporation was then detected with anti-BrdU antibody (Becton Dickinson, Mountain View, CA). The different replication patterns (early, mid-, and late S phase) were assessed by analysis on number, size, and distribution of BrdU as previously published (Dimitrova and Berezney, 2002). In Figure 2 and 3 the quantitative assessment was performed on three independent experiments, in which at least 250 cells per experiment were counted.
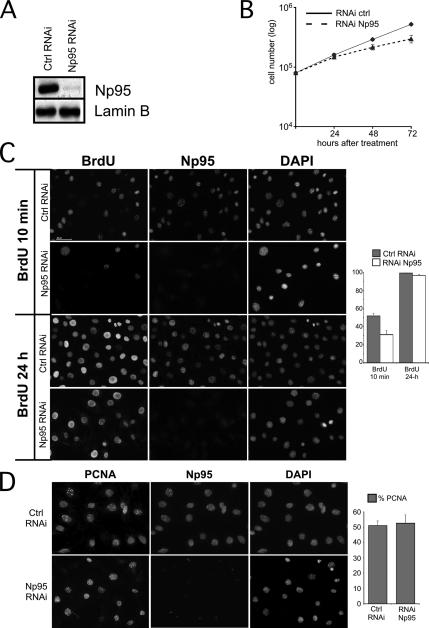
Figure 2.
Loss of Np95 leads to slow S-phase progression. (A) Whole-cell extracts of NIH-3T3 transfected with the RNAi oligo anti-Np95 (Np95 RNAi) or with the control oligo (ctrl RNAi) were analyzed by Western blot using antibody anti-Np95. An antibody anti-lamin B was used as a loading control. (B) Cell proliferation assay. NIH-3T3 cells were transfected with the RNAi oligo anti-Np95 (RNAi Np95) or with the control oligo (RNAi ctrl). Twenty-four hours after the second transfection, the cells were serum-starved for another 36 h to synchronize them in G0. At the indicated times after release, cells were harvested and counted with a Burker chamber. Each timing point corresponds to the average of three experiments. (C) Np95 depleted and control NIH-3T3 cells were pulsed for 10 min or 24 h with 10 μM BrdU and stained with antibodies anti-Np95 together with anti-BrdU. Nuclear counterstaining was visualized with DAPI. Representative pictures are shown. The plot indicates the percentage of cells positive for anti-BrdU. (D) NIH-3T3 cells treated as above were pulsed for 10 min with 10 μM BrdU and then stained with anti-Np95 together with anti-PCNA. Nuclear counterstaining was visualized with DAPI. Representative pictures are shown. The plot indicates the percentage of cells positive for anti-PCNA. Quantitative assessment in C and D was performed on three independent experiments, in which at least 250 cells per experiment were counted.
Figure 3.
Np95 depletion interferes with PH replication. NIH-3T3 cells were transfected with oligo against Np95 (Np95 RNAi) and a control (Ctrl RNAi). Twenty-four hours after treatment, the cells were pulsed for 10 min with 10 μM BrdU and then fixed onto glass slides and immunostained with anti-Np95 together with anti-BrdU. Representative pictures are shown. Arrows indicate the cells are replicating. The plot indicates percentage of cells in early (dark, bar), mid- (gray, bar), and late (white, bar) S phase. Quantitative assessment was performed on three independent experiments, in which at least 250 cells per experiment were counted.
Protein Extraction and Immunofluorescence
Proteins were extracted as described previously (Citterio et al., 2004). Immunofluorescence (IF) procedures were as described previously (Citterio et al., 2004). Antibodies used were as follows: rabbit polyclonals: anti-Np95 (Bonapace et al., 2002), anti-H4-acetyl-K8, -K12, and -K16 (Upstate Biotechnology, Lake Placid, NY); and anti-H2A-acetyl-K5 (AbCam, Cambridge, United Kingdom); mouse monoclonals: anti-Np95 (Fujimori et al., 1998), anti-BrdU (Becton Dickinson), and anti-PCNA PC10 (AbCam); and goat polyclonal anti-lamin B (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were as follows: Jackson Laboratories. Cells were analyzed with a fluorescent microscope (Olympus BX51, Melville, NY) equipped with 100× plan. Pictures were acquired with a color camera (Olympus DP50). Blots were digitalized with an Epson scan system (Epson expression 1600 Pro, Long Beach, CA). All images were managed with Adobe Photoshop (San Jose, CA).
RT-PCR Analysis
Total RNA was extracted from NIH3T3 cells with Trizol (Invitrogen) according to the manufacturer's instructions. RT-PCR procedures and the primers used for major and minor satellite were described previously (Lehnertz et al., 2003). RT-PCR analyses were performed with 18, 20, 22, 25, 27, and 30 cycles of amplification.
Picture Management
All pictures were managed with Adobe Photoshop and Canvas (Deneba Software, Miami, FL). Intensity profiles of Pictures 1 and 5 were done with ImageJ (National Institutes of Health, Bethesda, MD).
RESULTS
Np95 Targets Replicating Pericentromeric Heterochromatin and Is Part of the pHDB
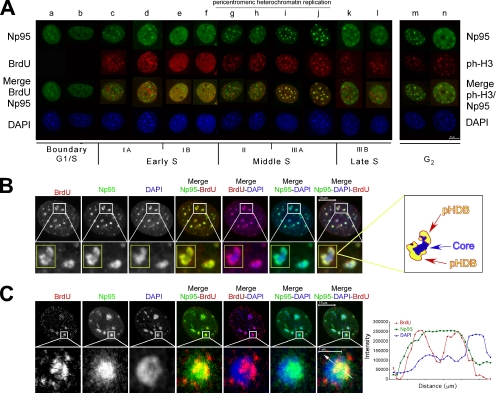
To gain insight into the function of Np95 during the progression through the cell cycle, we studied its subcellular localization during S and G2 phases. The spatiotemporal organization of DNA replication sites during S phase was assessed by a combined immunofluorescence analysis of the DAPI staining and of the pattern of BrdU incorporation. The distribution of nuclear DNA replication sites was classified into the five major types of patterns during S phase that have been shown to be identical in primary, immortalized, and transformed mammalian cells: IA and IB, early S and euchromatin replication; II and IIIA, middle S, pericentromeric heterochromatin replication; IIIB, late S, replication of centromeric heterochromatin and other structures (Ma et al., 1998; Dimitrova and Berezney, 2002). Serum-starved G0 synchronized NIH-3T3 cells were induced to re-enter the cycle by serum stimulation. At different times after cell cycle reentry, cells were pulse-labeled for 10 min with the nucleotide analogue BrdU, just before fixation. At the G1/S boundary (Figure 1A, lanes a and b) Np95 exhibits foci of staining over a more diffuse pattern. These foci are similar in number, size, and distribution to the pericentric heterochromatic foci typical of the DAPI staining (Figure 1A, lanes a and b). This pattern is conserved during early S (Figure 1A, lanes c–f). In middle S, the pattern changes and the bulk of Np95 staining localizes to the intense DAPI spots, overlapping with the characteristic BrdU ring-like structures corresponding to PH replication (Figure 1, A, lanes g–j, and B, insets and drawing). In these structures, Np95 and BrdU occupy the areas of less intense DAPI staining corresponding to a less compacted DNA (Quivy et al., 2004), as the confocal picture and the plot of the fluorescence intensity profiles of Figure 1C indicate (compare the blue line with respect to the green and the red ones). This is in agreement with the pHDB model that suggests that the ring-like structures located at the periphery of the dense DAPI spots correspond to the replication “factories,” made of the parental DNA pulled out from the central core and replicated (Quivy et al., 2004).
Figure 1.
During mid-S, Np95 targets pericentric heterochromatin and is part of pHDB. (A) Distribution of Np95 during S-phase. NIH-3T3 cells were synchronized in G0. At different time after release, cells were pulse-labeled with BrdU and then stained with anti-Np95 together with either anti-BrdU or anti-pH3 (specific for histone H3 phosphorylated at serine 10). Nuclear counterstaining was visualized with DAPI. (B) Np95 is part of pHDB. NIH-3T3 cells in mid-S phase were pulse-labeled with BrdU and then stained with anti-Np95 together with anti-BrdU. Nuclear counterstaining was visualized with DAPI. Representative pictures are shown. The insets correspond to magnifications of the areas indicated. On the right is a schematic representation of the pHDB of the inset. (C) NIH-3T3 cells in mid-S phase were stained as above and then analyzed by confocal microscopy. Representative pictures are shown. The insets correspond to magnifications of the areas indicated. The histogram shows the local intensity distribution (diagonal white lines through the images) of Np95 in green, BrdU in red, and DAPI in blue.
In very late S phase, Np95 and BrdU foci do not colocalize anymore (Figure 1A, lanes k and l), and Np95 regains the distribution pattern it had at the onset of S phase, which also persists in G2 (Figure 1A, lanes m and n; G2 phase is evidenced by an antibody against S10-phoshorylation of histone H3). During mid-S phase, Muto and coworkers (Miura et al., 2001) have shown that Np95 colocalizes with PCNA, whose chromatin-bound immunostaining is observed only in S-phase cells and whose patterns during the S phase markedly changes in accord with the BrdU incorporation sites.
Together with the results of Muto and coworkers (Miura et al., 2001) that show that Np95 colocalizes with PCNA in mid-S phase, these data suggest that Np95 is part of the pHDB and that it could have a role in heterochromatin replication. They also strengthen the idea that Np95 is involved in events linked to DNA replication, although it should not be directly part of the DNA duplication machinery itself.
Np95 Is Required for Normal S-Phase Progression
To address this question, we used RNAi to deplete Np95 from cells in vivo. Transfection of siRNA oligonucleotides against the mRNA encoding for Np95 (RNAi-Np95) strongly decreases the protein level in NIH-3T3 cells, although the knockdown is incomplete (Figure 2A).
RNAi against Np95 has a significant effect on the cell cycle. Loss of Np95 leads to decreased cell proliferation, as the growth curve in Figure 2B shows. To investigate if the observed effect on cell proliferation could be explained by alterations during S-phase, cells were pulsed-labeled for either 10 min or 24 h with BrdU and immunostained for BrdU and Np95. Counts were done on all the BrdU-incorporating cells of each field (either of the control-RNAi or Np95-RNAi), independently of the amount of Np95 inside the cell. After 10-min pulse, RNAi-control–treated cells displayed high levels of BrdU incorporation, whereas BrdU-labeled cells were more than 40% lower in Np95-depleted cells (Figure 2C, BrdU 10 min, and plot bar, BrdU 10 min). Labeling the cells with BrdU for 24 h abolished the differences in the number of incorporating cells (Figure 2C, BrdU 24 h, and plot bar, BrdU 24 h). Taken together, these data indicate that lower amounts of Np95 inside the cell perturb S phase and that DNA replication is slowed but not inhibited.
We then investigated if slowing of S phase was a consequence of a reduced ability of the replication machinery to bind chromatin. To this end, we tested if lower amounts of Np95 in the cells would influence the binding of PCNA to chromatin. As Figure 2D (picture and plot) shows, the levels of chromatin-bound PCNA in control and in Np95-depleted cells were equivalent. Thus, lower levels of Np95 inside the cell will not impede the formation of the replication complex.
On the whole, these results show that a reduced amount of Np95 affects S phase by slowing down DNA replication and suggest that the amount of Np95 could be a limiting factor for the progression of DNA duplication.
Np95-negative Cells Accumulate in Early S Phase
We wondered if replicating RNAi-Np95 treated cells displayed a modified replication profile, i.e., if the amount of Np95 would be a limiting factor for the replication of specific chromatin areas. Because Np95 is part of the pHDB, we hypothesized that Np95 might affect replication and maturation of heterochromatin, leading to an altered distribution of replication foci in the cells. To verify this hypothesis, we pulsed-labeled control and Np95-RNAi–treated cells with BrdU for 10 min and analyzed the spatiotemporal organization of DNA replication sites to search for potential alterations in the replication pattern. Following the classification proposed by Berezney and coworkers (Dimitrova and Berezney, 2002), we counted the number of cells that displayed each of the five major types of patterns and divided them into three main categories: early replicating (IA and IB), middle replicating (II, IIIA), and late replicating (IIIB). Counts were done on all the BrdU-incorporating cells of each field (either of the control-RNAi or Np95-RNAi), independently of the amount of Np95 inside the cell. In Np95-RNAi–treated cells the number of early replicating cells is 28% higher, although both the number of middle and late replicating cells is almost 35% lower with respect to control-RNAi–treated cells (Figure 3). This result suggests that limiting amounts of Np95 inside a cell do not influence euchromatin replication, whereas they are critical for later stages of S phase when heterochromatin is duplicated. This prompted us to investigate a possible role of Np95 during heterochromatin replication.
Depletion of Np95 Causes Hyperacetylation of Lysine 8, 12, and 16 of Histone H4 Tail at PH
A crucial question with respect to heterochromatin replication is how the underacetylated state of specific histone H4 lysines is established and controlled (Grunstein, 1998), because from yeast to mammals deacetylated lysines (K) 5, 8, 12, and 16 of histone H4 are found at heterochromatin, and this condition is essential to maintain the compacted and silenced state of these regions (Chen and Townes, 2000; Agalioti et al., 2002). It is believed that histone H4 is deposited as a lysine 5 and 12 acetylated form (H4-AcK5 or H4-AcK12) and becomes deacetylated shortly after deposition. At late but not at early replicating foci, however, the immunofluorescence analysis of heterochromatin domains indicates that these domains are transiently enriched of the acetylated form of histone H4 in K5 and K12, which could be associated to the formation of new nucleosomes (Taddei et al., 1999). H4-K8 and -K16, instead, are never found in an acetylated form in heterochromatin during the cell cycle.
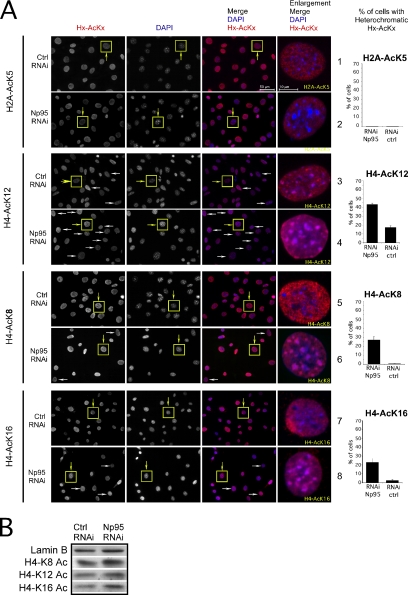
Np95 is a chromatin binding protein and both mouse and human (ICBP90) orthologues recruit HDAC-1 to target promoters (Unoki et al., 2004), whose activity has been proposed to be involved also in the deacetylation process of histone H4 (Annunziato and Hansen, 2000). Thus, we tested if Np95 could contribute to the specific H4 deacetylation event that occurs at H4 lysines in heterochromatin. To this end, we performed RNAi experiments to deplete Np95 from the cells and checked by immunofluorescence and Western blot the acetylation level of the NH2 tail lysines of H4. Figure 4A shows that the removal of Np95 more than doubles the number of cells that display an acetylation pattern of lysine 12 of histone H4 at the level of heterochromatin (Figure 4A, lanes 3 and 4, and inset; plot H4A-K12. The yellow boxed cells indicate the cells of the inset on the right; the white arrows indicate other representative cells). No modification of the patterns of a control acetylation marker (K5-H2A) is observed (Figure 4A, lanes 1 and 2, and inset, plot H2A-AcK5). The lack of a valid antibody for H4-AcK5 impeded us to check the behavior of this marker.
Figure 4.
Reduction of Np95 leads to specific enrichment of H4 acetylated at Lys 8, 12, and 16 at the periphery of heterochromatic dense DAPI dots. (A) Np95-depleted (Np95 RNAi) and control cells (Ctrl RNAi) were stained with anti-Np95, together with either anti-H2A-K5, -H4-K8, -H4-K12, and -H4-K16. Nuclear counterstaining was visualized with DAPI. Representative pictures are shown. The plots represent the percentage of cells that show H2A-AcK5, H4-AcK8, H4-AcK12, and H4-AcK16 acetylation on the periphery of heterochromatic dense DAPI dots. Quantitative assessment was performed on three independent experiments, in which at least 250 cells per experiment were counted. (B) Lysates of whole cells treated as above were analyzed by immunoblot with the indicate antibodies.
This result indicates that Np95 should control a key step for the deacetylation of lysine 12 of H4 in heterochromatin and prompted us to investigate if the reduction of Np95 in the cells could also have an effect on the acetylation of H4-K8 and K16 in the heterochromatic areas.
To this end we performed RNAi experiments to deplete Np95 from the cells, and we observed a strong acetylation on lysine 8 and 16 of H4, at the level of the heterochromatin DAPI spots in NIH-3T3 cells. Although acetylation in these lysines in the heterochromatic areas of control cells is virtually absent at any replication timing, in Np95-depleted cells more than 20% of the cells are heavily acetylated at the level of the dense DAPI regions (Figure 4A, lanes 5–8, and inset; plots H4-AcK8 and H4-AcK16). Confocal central sections of cells in RNAi-Np95 experiments show that the acetylation is concentrated prevalently at the periphery of the DAPI, as indicated by the fluorescence intensity profiles taken along the arrow on two sequentially DAPI spots inside the cell in the indicated images and plotted as a function of distance from the start point along the arrow (Figure 5, insets and intensity profiles. Compare the intensity profiles of H4-AcK8 and AcK16, red line, and DAPI, blue line). Western blot experiments performed on total extract from RNAi-Np95 and controls indicate that there are no variations in the overall amount of H4 acetylation (Figure 4B), suggesting that Np95 has a specific action only on the acetylation of heterochromatic H4 histones.
Figure 5.
Confocal analyses of distribution histone H4 acetylated at Lys 8 and 16 in Np95-depleted cells. Np95-depleted cells (Np95 RNAi) were stained with anti-Np95 antibody, together with antibodies against either acetylated H2A-K5, H4-K8, and H4-K16 and then analyzed by confocal microscopy. Nuclear counterstaining was visualized with DAPI. Representative pictures are shown (right panel). The histogram shows the local (diagonal white lines through the images) intensity distribution of Np95 in green, H4 acetylated in red, and DAPI in blue (left panel).
These results suggest a role for Np95 in the regulation of the acetylation of the key lysines of histone H4 in heterochromatin.
Depletion of Np95 Up-regulates Major Satellite Transcripts
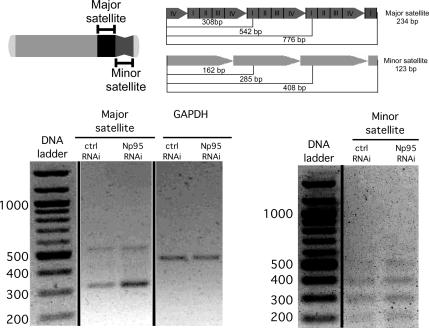
In S. pombe and in higher eukaryotic cells (Peters et al., 2001; Hall et al., 2002; Lehnertz et al., 2003) repeated sequences of heterochromatin are not completely silenced, but small RNA molecules transcribed from centromeric and pericentromeric regions appear to be necessary to initiate repressive chromatin modifications. The deacetylation of H4-K8 and H4-K16 of histone H4 has been shown to be required for the silencing of major satellite sequences (Nicol and Jeppesen, 1996). Thus, we investigated if Np95 could contribute to the negative regulation of these repetitive DNA sequences. To this end, we analyzed the transcriptional level of major and minor satellites, in control and Np95-depleted NIH-3T3 cells. Semiquantitative RT-PCR on total RNA shows that depletion of Np95 causes a 3–4-fold increase in the transcriptional expression of the major, but not of the minor satellites (Figure 6 and Supplementary Information). This suggests that Np95 exerts a transcriptional control on the DNA satellites of pericentromeric and not of centromeric regions of heterochromatin.
Figure 6.
Np95 depletion increases the transcription expression of major satellites. Top, schematic representation of major (pericentric heterochromatin) and minor satellite (centromeric heterochromatin) repeats on a mitotic mouse chromosome. Bottom, relative RNA levels for major satellites, minor satellites, and GAPDH was determined by RT-PCR analyses on cDNA derived from total RNA obtained from Np95-depleted (Np95 RNAi) and control cells (Ctrl RNAi). The results of 18 cycles of amplification are shown. The negative digital image of agarose gel analysis is shown.
We conclude that these data argue in favor of a direct action of Np95 on the control of the deacetylation of histone H4 tails and of the silencing of major satellite sequences.
DISCUSSION
In this article, we provide evidence that Np95 is a new protein involved in replication through PH. The results presented here show that Np95 is part of this unit and are consistent with a role of Np95 during replication of PH for several reasons. First, Np95 is specifically enriched in heterochromatin only at the time of PH replication in mid-S phase. Second, Np95 is part of the pHDB for it colocalizes with BrdU (this article) and with PCNA (Miura et al., 2001) in the ring-like structures that correspond to PH replication. Third, it colocalizes with active replication foci during PH DNA replication (mid-S phase) and not during the replication of centromeric heterochromatin and other chromatin structures (late S phase). Fourth, but most important, functional ablation of Np95 reduces both mid- and late S-phase replication more than 35% with respect to controls, in a 10-min BrdU pulse experiment.
A strong reduction of Np95 inside the cells does not completely inhibit DNA replication nor hamper the binding of PCNA to the DNA, indicating that the effect of the reduction of Np95 is more likely to be a slowing down rather than a block of DNA replication and that the impairment of heterochromatin replication does not depend on the stability of the replication machinery. Np95, moreover, does not seem to be a common part of the DNA synthesizing machinery. Thus, the slowing down effect on heterochromatin replication in RNAi experiments must be a consequence of DNA replication–linked nuclear events not directly connected to the replication machinery.
Np95 is a histone-binding protein involved in histone epigenetic modifications. The results we obtained, together with previously published data, are consistent with a model in which Np95 binds to histone H3 (Citterio et al., 2004), recruits HDAC-1 (Unoki et al., 2004), and deacetylates the tails of heterochromatic histone H4 (this article). The marked increase of heterochromatic H4Ac-K12 we observe in the absence of Np95 reinforces the thought that Np95 has a role in heterochromatin replication. H4-K12, in fact, has been shown to be acetylated in heterochromatic areas only during histone deposition and for a time window of 20 min afterward (Taddei et al., 1999). Interestingly, we also observe an acetylation of H4-K8 and H4-K16 at the periphery of the DAPI spots at the time of heterochromatin replication with a distribution that recalls the ring-like one of Np95, suggesting that Np95 should control deacetylation of NH2 lysines of H4 at the level of the pHDB. At this stage of the study, however, we cannot distinguish if the acetylation of H4 lysines occurs at the time of heterochromatin replication or in other moments of the cell cycle, nor we can define if this event involves the newly deposited histones, the parental ones, or both. Importantly, heterochromatic acetylation of H4-K12 has been observed only at the time of heterochromatin replication, and an acetylated form of lysine 8 of H4 has been found in the chromatin assembly complex (CAC), which contains the three subunits of CAF-1 and that is a key intermediate for the de novo nucleosome assembly pathway as it is able to promote DNA replication–dependent chromatin assembly (Verreault et al., 1996). A very short time might exist, therefore, in which newly deposited histone H4 tails are acetylated at K8 and K16, and the high concentration of Np95 in the pHDB unit, right at the time of heterochromatin replication, guarantees an efficient recruitment of HDAC-1 for the deacetylation of these residues. Further experiments will be necessary to conclusively prove this hypothesis.
However, H32/H42 tetramers lacking the amino-terminal domains of both histones can be stably bound to CAF-1, which can efficiently promote their assembly in vitro during SV40 DNA replication (Shibahara et al., 2000). This indicates that the acetylation-deacetylation of newly synthesized H4 is not a limiting step for histone deposition. Thus, the impairment of heterochromatin replication in RNAi-Np95 experiments cannot be due to a histone deposition failure as a consequence of the modified acetylation pattern of NH2 tail lysines of H4.
We show here that an increased level of the RNA of repetitive pericentromeric major satellite sequences in the absence of Np95. This can be interpreted as a direct consequence of the deacetylation of histone H4 NH2 tails, as the deacetylation of both H4AcK8 and H4AcK16 is required for gene silencing, including the transcriptional repression of heterochromatic satellite DNA sequences, and is necessary for proper chromatin maturation in a postreplicative phase. Biamonti and coworkers (Rizzi et al., 2004) have shown that heat-shock stress bodies, formed on the pericentromeric heterochromatic regions of specific human chromosomes, are enriched in histone H4AcK8 and K16 isoforms and that heat shock triggers the transient accumulation of heterogeneous RNA molecules containing the subclass of satellite III sequences found in human PH. Small RNA produced by satellite sequences are involved in the RNAi machinery that is required for heterochromatin formation (Wassenegger, 2005). Deacetylation of H4 in PH, therefore, could be involved in establishing and maintaining the silent state of heterochromatin after replication, and this could be a first event during heterochromatin formation. The impairment of satellite silencing blocks many processes associated with the formation of heterochromatin, including DNA and histone methylation (Kanellopoulou et al., 2005). Np95 has a ubiquitin ligase activity with a H3 specificity in vitro and “ex-vivo” (Citterio et al., 2004). An interesting possibility is that Np95 could bind to chromatin, deacetylate the key lysines of H4, and ubiquitinate H3, and these could be epigenetic marks needed for the addition of further PH epigenetic marks, such as H3-K9 methylation. Experiments are in progress to address this issue.
Thus, Np95 might be implicated in the process that controls a very initial postreplicative step that leads to the establishment of silent PH, a precondition for the formation of this chromatin district. No variations are observed in the RNA level of minor satellites, which correspond to centromeric heterochromatin, once more suggesting a role of Np95 only for PH. We cannot yet distinguish if the increased transcription of pericentromeric repeats is due to an enhanced transcriptional rate, to an increased stability of the RNA owing to the inhibition of enzymes involved in the RNAi process (Dicer, for example; Wassenegger, 2005), or both. Nevertheless, in both cases we would expect an impairment of PH formation.
Proper replication and formation of PH are important for the correct segregation of chromosomes and for the maintenance of genome stability. In yeast, heterochromatin is specifically required for cohesion between sister centromeres (Bernard et al., 2001). In murine cells, forced accumulation of 120-nt centromeric transcripts leads to defects in chromosome segregation and sister-chromatid cohesion, changes in hallmark centromeric epigenetic markers, and mislocalization of centromere-associated proteins essential for centromere function (Bouzinba-Segard et al., 2006). Although more experiments are needed to deeply investigate the role of major satellites in chromosome segregation and stability, our results can, at least partially, explain the higher genome instability that has been seen in Np95 +/− and −/− ES mouse cells (Muto et al., 2002).
Preliminary experiments conducted to study the effect of overexpression of Np95 in the cells (Papait, Pistou, Cogliati, Pecoraro, Babbio, and Bonapace, unpublished results) show that high levels of this protein inside the cells cause profound modifications of PH. Because Np95 and ICBP90 have been found overexpressed in numerous cancers, higher levels of this protein inside the cell could affect proper chromosome organization and thereby chromosome stability and segregation.
Although our results do not formally prove that the deacetylation of H4 lysines is responsible for the observed reduction of heterochromatin replication, a conceivable hypothesis is that the absence of Np95 is preventing the initial deacetylation step necessary for the correct formation of PH and that this event would impair and retard the transfer of replicated, but yet unformed (or at least not yet deacetylated), heterochromatin from the pHDB back into the heterochromatic core, as proposed by the pHDB model (Quivy et al., 2004).
In conclusion, Np95 is a new important protein for PH replication, and it has a key role in the regulation of the deacetylation of lysine residues of the NH2 tails of histone H4 and in the transcriptional control of heterochromatic major satellites, a process that occurs during heterochromatin replication. Further experiments will be needed to investigate more deeply how the regulation of the deacetylation of H4 tails and the transcriptional control of major satellites by Np95 are involved in heterochromatin replication and formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Pier Paolo Di Fiore for continuous support and encouragement and Prof. Gianfranco Badaracco for reviewing the manuscript. This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) to I.M.B. and from Fondazione Cariplo. R.P. was supported by a fellowship from AIRC.
Abbreviations used:
- PH
pericentric heterochromatin
- pHDB
pericentric heterochromatin duplication body
- RNAi
RNA interference
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0874) on December 20, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Annunziato A. T., Hansen J. C. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis J. M., Forsburg S. L. It's all in the timing: linking S phase to chromatin structure and chromosome dynamics. Cell Cycle. 2003;2:303–306. [PubMed] [Google Scholar]
- Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., Allshire R. C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bitterman K. J., Medvedik O., Sinclair D. A. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace I. M., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P. P. Np95 is regulated by E1A. during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzinba-Segard H., Guais A., Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Y., Townes T. M. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl. Acad. Sci. USA. 2000;97:377–382. doi: 10.1073/pnas.97.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P. P., Bonapace I. M. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. S., Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson T., Turner B. M., Cranston G., Allshire R. C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Grewal S. I. Heterochromatin: silence is golden. Curr. Biol. 2003;13:R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Fujimori A., Matsuda Y., Takemoto Y., Hashimoto Y., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., Grewal S. I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990;6:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- Jeanblanc M., Mousli M., Hopfner R., Bathami K., Martinet N., Abbady A. Q., Siffert J. C., Mathieu E., Muller C. D., Bronner C. The retinoblastoma gene and its product are targeted by ICBP90, a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S. A., Kung A. L., Ganesan S., Drapkin R., Jenuwein T., Livingston D. M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Ma H., Samarabandu J., Devdhar R. S., Acharya R., Cheng P. C., Meng C., Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Miura M., Watanabe H., Sasaki T., Tatsumi K., Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp. Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- Muto M., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Nicol L., Jeppesen P. Chromatin organization in the homogeneously staining regions of a methotrexate-resistant mouse cell line: interspersion of inactive and active chromatin domains distinguished by acetylation of histone H4. J. Cell Sci. 1996;109(Pt 9):2221–2228. doi: 10.1242/jcs.109.9.2221. [DOI] [PubMed] [Google Scholar]
- Perrod S., Gasser S. M. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell. Mol. Life Sci. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Quivy J. P., Roche D., Kirschner D., Tagami H., Nakatani Y., Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Bartel D. P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K., Verreault A., Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Roche D., Sibarita J. B., Turner B. M., Almouzni G. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M., Nishidate T., Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.