Abstract
Disassembly of the nucleolus during mitosis is driven by phosphorylation of nucleolar proteins. RNA processing stops until completion of nucleolar reformation in G1 phase. Here, we describe the RNA methyltransferase NSUN2, a novel substrate of Aurora-B that contains an NOL1/NOP2/sun domain. NSUN2 was concentrated in the nucleolus during interphase and was distributed in the perichromosome and cytoplasm during mitosis. Aurora-B phosphorylated NSUN2 at Ser139. Nucleolar proteins NPM1/nucleophosmin/B23 and nucleolin/C23 were associated with NSUN2 during interphase. In mitotic cells, association between NPM1 and NSUN2 was inhibited, but NSUN2-S139A was constitutively associated with NPM1. The Aurora inhibitor Hesperadin induced association of NSUN2 with NPM1 even in mitosis, despite the silver staining nucleolar organizer region disassembly. In vitro methylation experiments revealed that the Aurora-B-phosphorylation and the phosphorylation-mimic mutation (S139E) suppressed methyltransferase activities of NSUN2. These results indicate that Aurora-B participates to regulate the assembly of nucleolar RNA-processing machinery and the RNA methyltransferase activity of NSUN2 via phosphorylation at Ser139 during mitosis.
INTRODUCTION
In higher eukaryotic cells, the nucleolus disassembles during each mitotic phase (for review, see Dimario, 2004). Nucleoli are generally composed of three morphologically distinct regions; the fibrillar centers containing RNA polymerase I and its associated transcription factors, a dense fibrillar component that surrounds the fibrillar centers, and a granular component that is the site of preribosome assembly and preribosomal particle synthesis (Stoykova et al., 1985). At the onset of mitosis, rDNA transcription is repressed by cyclin-dependent kinase (CDK) 1/Cdc2 cyclin-dependent kinase–Cyclin B-directed phosphorylation of components of the rDNA transcription machinery, and nucleolar disassembly is induced (Heix et al., 1998; Sirri et al., 2000). The RNA synthesis machinery for rDNA in the fibrillar centers remains associated with mitotic chromosomes in an inactive state at nucleolar-organizing regions (Stoykova et al., 1985). Other RNA processing components, such as fibrillarin, nucleolar protein Nop52, NPM1/nucleophosmin/B23/Numatrin/Nucleolar protein NO38, and nucleolin/C23/100-kDa nucleolar protein, are located at the periphery of chromosomes, away from nucleolar-organizing regions during mitosis (Jimenez-Garcia et al., 1994; Pinol-Roma, 1999; Savino et al., 1999; Dousset et al., 2000). CDK1-Cyclin B also phosphorylates NPM1 and nucleolin (Peter et al., 1990), and the disassembly is apparently driven by mitotic phosphorylation of nucleolar components. Inhibition of CDK1-Cyclin B during mitosis is sufficient to cause the resumption of rDNA transcription, but it is not sufficient to restore proper processing of RNA biogenesis or total rebuilding of the nucleolar processing machinery (Sirri et al., 2002). Thus, repression of the nucleolar assembly is spatiotemporally and multifactorially regulated.
Aurora-B is a conserved protein kinase essential for the segregation of eukaryotic chromosomes (Andrews et al., 2003; Carmena and Earnshaw, 2003; Yanagida, 2005), and it forms the mitotic passenger protein complex with inner centromere protein (INCENP), Survivin, and Borealin/Dasra (Carmena and Earnshaw, 2003; Gassmann et al., 2004; Sampath et al., 2004; Giet et al., 2005). In early mitosis, Aurora-B associates with centromeric heterochromatin and then moves to the midzone spindle in anaphase, to the cleavage furrow in telophase, and finally to both ends of the midbody in cytokinesis (Schumacher et al., 1998; Terada et al., 1998; Adams et al., 2001; Murata-Hori et al., 2002). Mitotic activation of Aurora-B is triggered by autophosphorylation, which is stimulated by association with and phosphorylation of its substrate INCENP. Survivin also stimulates Aurora-B activity by a direct interaction, and it is a substrate for Aurora-B (Wheatley et al., 2001). Borealin is also an Aurora-B substrate but does not stimulate Aurora-B activity. There are at least two different types of Aurora-B complex present during mitosis, suggesting that each complex targets different sites or contains different components (Gassmann et al., 2004; Giet et al., 2005). In addition, the mitotic passenger movements of Aurora-B are accompanied by continuous exchange with the surrounding cytoplasmic pool (Murata-Hori et al., 2002). The requirements for centromere or cleavage furrow targeting of Aurora-B and chromosomal and cytoskeletal Aurora-B substrates for each mitotic process are now being elucidated; however, the mitotic functions of Aurora-B may not be limited to cytokinesis or other processes of chromosome segregation.
Here, we report a novel substrate of Aurora-B, which contains an NOL1/NOP2/sun domain and is the second member of the NSUN family (NSUN2) of RNA methyltransferase. We have shown that NSUN2 has RNA methyltransferase activity that is repressed by Aurora-B–provoked phosphorylation and that the phosphorylation causes NSUN2 to dissociate from its nucleolar binding protein, NPM1. Phosphorylation of NSUN2 by Aurora-B seems to represent a novel mechanism for the regulation of nucleolar architecture and nucleic acids metabolism during mitosis.
MATERIALS AND METHODS
Synchronization of Cell Cycle
HeLa cells were seeded in a 10-cm dish at 2.5 × 105 cells/dish. After 2 d, cells were treated twice with 2.5 mM thymidine (at 8 and 16 h). When changed to fresh medium, the cells started to grow synchronously (time 0). The fidelity of synchronization was monitored by flow cytometry. After 6 h, 80–90% of the cell population was in S phase. The proportion of cells in mitosis peaked at 9–10 h. For mitotic inhibition, the cells were usually treated with 200 ng/ml nocodazole for 18 h.
Aurora Inhibitors
Hesperadin and ZM447439 were obtained from Boehringer Ingelheim (Ingelheim, Germany) and AstraZeneca Pharmaceuticals LP (Wilmington, DE), respectively. Synchronized HeLa cells were treated with 100 nM Hesperadin or 2 μM ZM447439 for 2 h (from hours 8–10).
Mammalian Expression Plasmids and Transfection
5′-Rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR) products were generated using primers corresponding to peptides lep56 and lep77-1 (Supplemental Figure S1). The full-length cDNA was isolated from the Okayama-Berg full-length cDNA library (produced from HeLa cells) by using the colony hybridization technique with the 5′-RACE PCR products. The obtained full-length cDNA was then sequenced. The coding region of this cDNA was subcloned into pcDNA3.1(−) for mammalian expression of tag-free NSUN2, generating plasmid pcDNA3.1(−)-NSUN2. The coding region was also subcloned into pcDNA3.1/His for mammalian expression of Xpress-NSUN2, generating plasmid pcDNA3.1-Xpress-NSUN2. The following mutant NSUN2 expression plasmids were also constructed: pcDNA3.1(−)-NSUN2-SA and pcDNA3.1-Xpress-NSUN2-SA, producing dephosphorylation-mimic NSUN2-S139A, in which Ser139 of NSUN2 was replaced with Ala by site-directed mutation at T410G; and pcDNA3.1(−)-NSUN2-SE and pcDNA3.1-Xpress-NSUN2-SE, producing phosphorylation-mimic NSUN2-S139E in which Ser139 of NSUN2 was replaced with Glu by site-directed mutation at T410G and C411A. Other mammalian expression vectors were also generated: pcDNA3-FLAG-Aurora-A-WT and -K/R (Tatsuka et al., 2005); pcDNA3-FLAG-Aurora-B-WT and -K/R (Kanda et al., 2005); pcDNA3-FLAG-Aurora-C-WT and -K/R (constructed from human Aurora-C cDNA isolated from the HeLa cDNA library by reverse transcription-PCR); and pEGFP-ΔCyclin B1 (constructed from the ΔCyclin B1 expression plasmid supplied by M. Brandeis, Department of Genetics, Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel). Transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Short Hairpin RNA (shRNA) Constructs
Oligonucleotides corresponding to Aurora or NSUN2 genes were synthesized as shown in Supplemental Figure S2. The oligonucleotides were then ligated into pSUPERIOR.PURO (OligoEngine). The mixture of each Aurora-shRNA was used to repress Aurora expression in HeLa cells. HeLa cell lines expressing each NSUN2-shRNA plasmid construct were established by puromycin selection (Supplemental Figure S3). We used clone 5 from NSUN2-shRNA 3 construct-transfected HeLa cells as HeLa-NSUN2-KD cells.
Antibodies
We raised a rabbit polyclonal antibody (αH3-P) against a phosphorylated synthetic Histone H3 peptide (ARKS*TGGKAPRKQL, where S* indicates the phosphorylated serine). Collected serum was affinity purified using the peptide. Rabbit polyclonal antibodies, αNSUN2 and αNSUN2-full, were raised against the C-terminal peptide (GCDPAGVHPPR) of NSUN2 and bacterially expressed full-length His-NSUN2, respectively. The bacterially expressed protein was isolated from the lysate of pRSET(C)-NSUN2-WT–expressing Escherichia coli cells by using a Talon nickel column (BD Biosciences, San Jose, CA) (Supplemental Figure S4). Other antibodies used in the experiments included the following: polyclonal rabbit anti-Cdc25A (Jinno et al., 1994) and anti-Human Aurora-C antibodies (38-9400; Zymed Laboratories, South San Francisco, CA); and monoclonal antibodies against FLAG (M5; Sigma-Aldrich, St. Louis, MO), Xpress (Invitrogen), α-tubulin (CLT-9002; Cedarlane Laboratories, Hornby, Ontario, Canada), enhanced green fluorescent protein (EGFP; JL-8; BD Biosciences), Aurora-A (clone 4; BD Biosciences), Aurora-B (Kanda et al., 2005), fibrillarin (38F3; Abcam, Cambridge, United Kingdom), NPM1 (B0556; Sigma-Aldrich), and nucleolin (sc-8031; Santa Cruz Biotechnology).
Immunoblot and Immunoprecipitation
Cells were lysed in TBSN buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM EGTA, 0.5 mM Na3VO4, 20 mM p-nitrophenyl phosphate, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Sigma-Aldrich], and 0.5% NP-40) on ice for 10 min. Cell lysates were then frozen in liquid nitrogen and thawed in water at 20°C. Thawed lysates were vortexed and centrifuged (15,000 rpm for 5 min), and the cleared extracted lysates were used for immunoblotting. For immunoprecipitation, the lysate (500 μg/tube) was incubated with rotation for 4 h at 4°C with antibody (10 μg)-coupled protein G-Sepharose beads. The beads were washed four times with TBSN buffer. The final volume was adjusted to 25 μl per tube, and the samples were processed for SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane for immunoblotting or were stained with Coomassie brilliant blue. For Edman degradation analysis, relevant areas of the gel were excised.
Indirect Immunofluorescence
Cells were fixed with methanol, blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS)(−) and were then incubated for 1 h with rabbit polyclonal antibody against NSUN2 (1:200), and a monoclonal antibody (mAb) against fibrillarin (1:500), NPM1 (1:200), or nucleolin (1:100). Cells were washed three times with PBS(−) containing 1% BSA and were then incubated for 1 h with secondary antibodies and 0.1 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride. Alexa 568-conjugated anti-rabbit or anti-mouse antibodies (1:1000; Invitrogen) and fluorescein isothiocyanate-conjugated anti-rabbit or anti-mouse antibodies (1:150; Chemicon International, Temecula, CA) were used as secondary antibodies. Stained cells were mounted with an anti-fade fluorescent mounting medium (Dako UK, Ely, Cambridgeshire, United Kingdom) and observed under a LSM510META laser scanning microscope (Carl Zeiss, Jena, Germany).
In Vitro Phosphorylation
Purified NSUN2 proteins were used as substrates for in vitro phosphorylation by GST-fused Aurora-B. The reaction was performed for 30 min at 25°C in 100 μl of a reaction mixture containing 25 mM Tris, pH 7.5, 2 mM MgCl2, 100 μM ATP, 100 μM [γ-32P]ATP, 0.1 μM calyculin A, 80 μg/ml glutathione S-transferase (GST)-Aurora-B, and 300 μg/ml NSUN2. Histone H3 was used as a positive control substrate. The reaction mixtures were processed for SDS-PAGE and were analyzed by autoradiography with x-ray film (Fuji, Tokyo, Japan).
Methyltransferase Assay
His-NSUN2 was phosphorylated by anti-Aurora-B antibody-immunoprecipitated Aurora-B (from nocodazole-treated HeLa cells) or anti-GST antibody-immunoprecipitated GST-Aurora-B. Both Aurora-B samples contain protein G-Sepharose beads. The Aurora-B kinase activity was monitored by the phosphorylation of H3 histone. Aurora-B-K/R was also used as control experiments. For MOCK kinase reaction, nonimmune IgG-immunoprecipitated sample and anti-GST antibody-immunoprecipitated GST were used. The kinase reacted NSUN2 proteins were used for the methyltransferase assay after removal of the beads containing Aurora-B. NSUN2-S139A and NSUN2-S139E proteins generated by the TNT in vitro transcription/translation (rabbit reticulocyte) system (Promega, Madison, WI) were also used for the methyltransferase assay. [35S]methionine (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) incorporation followed by SDS-PAGE and fluorography confirmed that a protein of the expected size was translated, and the NSUN2 protein amount was estimated by immunoblot.
In methyltransferase assay for DNA, lambda DNA (0.03 pmol) was reacted with the kinase-reacted His-NSUN2 (0.5 μM) in NE buffer 2 reaction buffer (New England Biolabs, Beverly, MA) containing 160 μM S-adenosylmethionine (SAM). Sss1 (New England Biolabs) and Dnmt1 (New England Biolabs) were used as positive and negative control, respectively. After reaction at 37°C for 1 h, the reacted lambda DNA was digested with BstU I (New England Biolabs) and analyzed by agarose gel electrophoresis.
In methyltransferase assay for hemimethylated DNA, hemimethylated mimic substrate poly(dI:dC) (Roche Diagnostics, Mannheim, Germany) (0.2 pmol) or tRNA (purified from E. coli; Sigma-Aldrich) (4 nmol) was reacted with the kinase reacted His-NSUN2 (0.5 μM) or TNT-produced NSUN2 (0.5 μM) in Dnmt1 reaction buffer (New England Biolabs) containing 6.5 μM S-adenosyl-L-[methyl-3H]methionine (specific activity 555 GBq/mmol; GE Healthcare) and RNasein (40 U/μl; Promega). After reaction at 37°C for 30 min (for His-NSUN2) or 5 min (for TNT-produced NSUN2), unincorporated labeled compound was removed by using MicroSpin G25 columns (GE Healthcare), and incorporated radioactivity was measured by liquid scintillation counting.
Ag-Nucleolar Organizer Region (NOR) Protein Silver Staining Method
To visualize the active nucleolus, selective silver staining for the Ag-NOR proteins (Robert-Fortel et al., 1993) was performed. Preparations were washed for 5 min in 50% ethanol and in deionized water. The slides were then covered with a freshly produced staining solution prepared by mixing equal volumes of the following solutions: 1) 0.5 g/ml AgNO3 (Merck, Darmstadt, Germany) in deionized water and 2) and 1 mg/ml gelatin dissolved in 2% (by volume) formic acid. The slides were incubated for 15–20 min at room temperature. After staining, slides were vigorously washed in distilled water, dehydrated in ethanol, and mounted in alcohol-Eukitt.
RESULTS
Identification of 100-kDa Mitotic Phosphorylated Protein
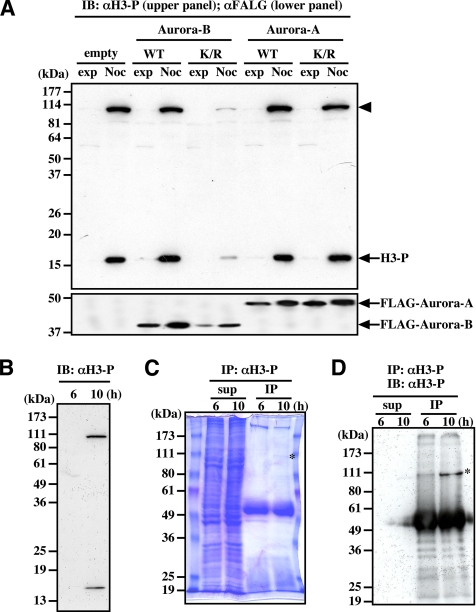
We generated a rabbit polyclonal antibody against a synthetic peptide corresponding to a phosphorylation site (Ser10) in Histone H3. The antibody (αH3-P) recognized mitotic phosphorylated Histone H3 (Figure 1A, H3-P) as well as an ∼100-kDa protein in mitotic cells (Figure 1A, arrowhead). Both this 100-kDa phosphorylated protein and phosphorylated Histone H3 were repressed by transfection with a kinase-negative form of Aurora-B (Aurora-B-K/R) but not by transfection with the kinase-negative form of Aurora-A (Aurora-A-K/R) (Figure 1A).
Figure 1.
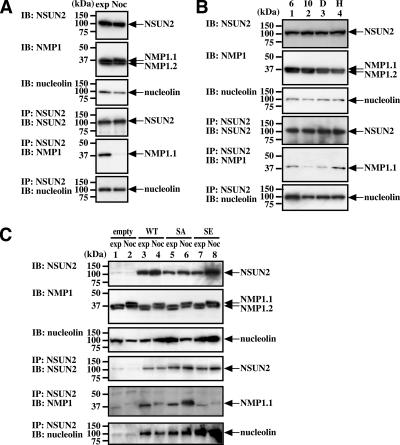
Antibody against Histone H3 phosphorylated at Ser10 binds to unknown 100-kDa protein. (A) The 100-kDa protein (arrowhead) is phosphorylated in cells treated for 18 h with nocodazole, and its phosphorylation is inhibited by the expression of Aurora-B-K/R but not Aurora-A-K/R. (B) The 100-kDa protein is phosphorylated during mitosis in synchronized cells. (C) Coomassie brilliant blue staining of the 100-kDa protein immunoprecipitated by αH3-P. Synchronized HeLa cells were collected at the indicated times, and the immunoprecipitated samples and the supernatant were separated by SDS-PAGE. (D) Immunoblot of the 100-kDa protein immunoprecipitated by αH3-P. Samples shown in C were analyzed by immunoblotting with αH3-P. In C and D, asterisks indicate the immunoprecipitated 100-kDa protein. IB, immunoblot; IP, immunoprecipitation; H3, Histone H3; WT, wild-type; K/R, kinase negative form; exp, exponentially growing; Noc, nocodazole treatment.
We used double-thymidine blocking and release to synchronize HeLa cells, and then we collected the interphase and mitotic cells 6 and 10 h after release, respectively (Figure 1B). After lysing the cells, we performed immunoprecipitation with the phosphorylated Histone H3 antibody, and the protein was visualized by staining with Coomassie brilliant blue (Figure 1C, asterisk). We confirmed that this 100-kDa band is recognized by αH3-P (Figure 1D, asterisk). The protein was extracted from the gel band, and partial amino acid sequences were obtained by Edman degradation. We determined the sequences of four peptide fragments (lep56, lep60, lep77-1, and lep77-2; Supplemental Figure S1, red letters). One fragment, lep60, was identical to a sequence in putative protein BC001041, but we did not find proteins homologous with the other three peptides. We then performed PCR with primers corresponding to peptides lep56 and lep77-1 (Supplemental Figure S1, underlined blue letters), followed by 5′-RACE. Thus, the PCR product was used to clone the full-length cDNA from a HeLa cDNA library (Supplemental Figure S1). This gene product contains an NOL1/NOP2/sun (NSUN) domain and is currently classified as the second member of the NSUN family (NSUN2).
NSUN2 Is Concentrated in Nucleoli
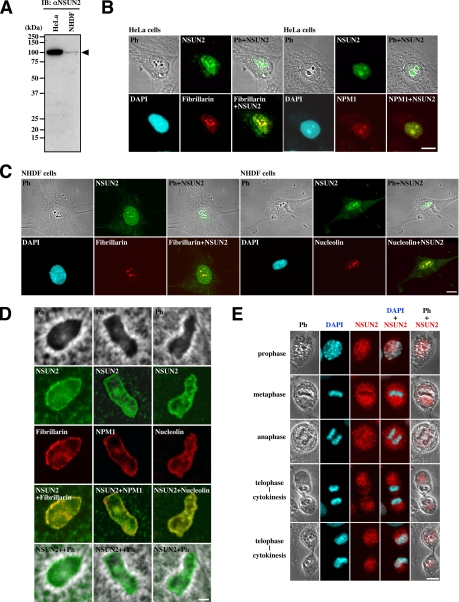
We raised a rabbit polyclonal antibody (αNSUN2) against the C-terminal peptide (GCDPAGVHPPR) of NSUN2. The affinity-purified antibody recognized a protein at the expected size (100 kDa) on immunoblots of lysates from both HeLa and normal human diploid fibroblasts (NHDFs) (Figure 2A). The levels of NSUN2 were markedly different between HeLa and NHDF cells (Figure 2A, arrowhead). Indirect immunofluorescence showed that NSUN2 is located in the nucleoplasm and is concentrated in the nucleoli in both cell lines (Figure 2, B and C). Simultaneous staining for nucleolar protein fibrillarin, NPM1, or nucleolin confirmed this nucleolar localization (Figure 2, B and C). When observed under higher magnification, NSUN2 and these nucleolar proteins were colocalized in the dense fibrillar component and the granular component, and none were significantly associated with the fibrillar centers (Figure 2D). The subnucleolar localization of NSUN2 was absent after RNase treatment (data not shown). During mitosis, NSUN2 is translocated into perichromosomal and cytoplasmic regions, accompanied by nucleolar disassembly, until the next G1 phase when nucleoli reassemble (Figure 2E). All immunoblot and immunofluorescence experiments shown in Figure 2 were confirmed using αNSUN2-full (data not shown).
Figure 2.
Intracellular localization of NSUN2. (A) HeLa cells express higher levels of NSUN2 (arrowhead) than NHDF cells during the exponential growth phase. (B) In interphase HeLa cells, NSUN2 is localized in the nucleoplasm and concentrated in the nucleolus. (C) In interphase NHDF cells, NSUN2 is localized in nucleoplasm and concentrated in the nucleolus. (D) In the nucleolus, NSUN2 is localized in the dense fibrillar and granular components and is not significantly associated with the fibrillar centers. (E) NSUN2 is localized in the perichromosomal and cytoplasmic regions during mitosis and is translocated into the nucleoplasm during cytokinesis. Bar, 10 μm (B and C) and 1 μm (D).
NSUN2 Is Phosphorylated at Ser139 by Aurora-B during Mitosis
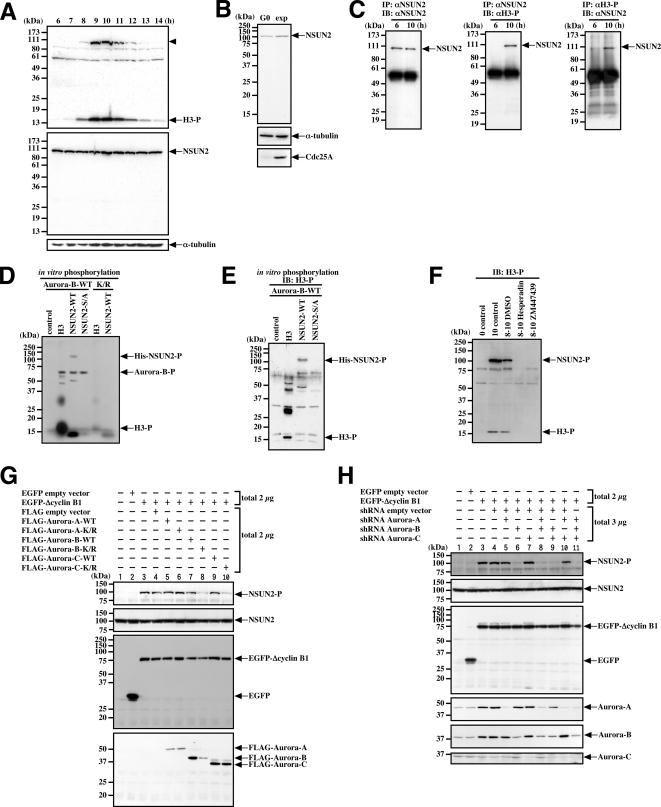
Using double-thymidine blocked and released synchronized HeLa cells, we identified a phosphorylated 100-kDa protein, presumably NSUN2, that occurs during mitosis (Figure 3A, arrowhead). The levels of NSUN2 in HeLa cells, however, remain stable during the cell cycle (Figure 3A). Similarly, the levels of NSUN2 are unchanged in NHDFs, even when serum starved (Figure 3B). Thus, NSUN2 is constitutively expressed in both cancerous HeLa cells and normal NHDFs, although the expression levels are markedly different.
Figure 3.
NSUN2 is phosphorylated during mitosis on Ser139 by Aurora-B. (A) NSUN2 is constitutively expressed and possibly phosphorylated during mitosis. The samples were collected at the indicated times in synchronized HeLa cells. The top, middle, and bottom panels show immunoblots (IB) using αH3-P, αNSUN2, and anti-α-tubulin antibody, respectively. Arrowhead indicates the phosphorylated 100-kDa protein, presumably phosphorylated by NSUN2. (B) NSUN2 is expressed even in the quiescent state. NHDF cells were cultured under exponential (exp) growth conditions or in 0.2% fetal bovine serum for 48 h (G0). (C) NSUN2 is phosphorylated during mitosis but not during interphase in synchronized HeLa cells. (D) NSUN is phosphorylated by Aurora-B in vitro. (E) NSUN2 phosphorylated in vitro by Aurora-B is recognized by αH3-P. The samples shown in D were analyzed by immunoblotting using αH3-P. (F) Aurora inhibitors Hesperadin and ZM447439 repress NSUN2 and Histone H3 phosphorylation during mitosis in synchronized HeLa cells. Eight hours after release, the cells were treated with inhibitors for 2 h. The control lysate was collected immediately after release. (G) Aurora-B-K/R and Aurora-C-K/R repress the phosphorylation of NSUN2 during mitosis in ΔCyclin B1-arrested cells. HeLa cells were transfected with the indicated pair of plasmids, and after 18 h, cells were lysed and analyzed by immunoblotting by using αH3-P, αNSUN2, anti-EGFP antibody, or anti-FLAG antibody. (H) Aurora-B-shRNA represses the phosphorylation of NSUN2 during mitosis in ΔCyclin B1-arrested cells. HeLa cells were transfected with the indicated pair of plasmids, and after treatment for 48 h with puromycin, cells were lysed and analyzed by immunoblotting αH3-P, αNSUN2, anti-EGFP antibody, or anti-Aurora-A, -B, or -C antibodies. H3, Histone H3; H3-P, phosphorylated Histone H3; NSUN2-P, phosphorylated NSUN2; Aurora-B-P, phosphorylated Aurora-B; IB, immunoblot; IP, immunoprecipitation.
To confirm whether NSUN2 is phosphorylated during mitosis, we prepared lysates from interphase (6 h after release) and mitotic (10 h after release) cells, immunoprecipitated proteins with αH3-P or αNSUN2, and then performed immunoblotting with the opposite antibody. Immunoprecipitated NSUN2 was recognized by αH3-P (Figure 3C, middle). In addition, NSUN2 was one of the proteins immunoprecipitated by αH3-P (Figure 3C, right).
Antibody αH3-P was raised against a synthetic phosphorylated peptide corresponding to the Ser10 phosphorylation site in Histone H3 (ARKS*TGGKAPRKQL, where S* represents phosphorylated serine). We detected the same RKS* sequence around the 139 site (Supplemental Figure S1). This suggests that NSUN2 is phosphorylated at Ser139. To confirm this observation, we used site-directed mutagenesis to replace Ser139 of NSUN2 with Ala. We then produced full-length wild-type and Ser139Ala mutants of NSUN2 (NSUN2-WT and NSUN2-S139A, respectively) in E. coli. The purified proteins (Supplemental Figure S4) were used in an in vitro kinase assay with either N-terminal GST-fused recombinant Aurora-B-WT or Aurora-B-K/R. We found that NSUN2 was phosphorylated by Aurora-B-WT but not by Aurora-B-K/R (Figure 3D). In addition, NSUN2-S139A was not phosphorylated by Aurora-B-WT (Figure 3D). We also confirmed that the in vitro phosphorylated band for His-tagged NSUN2 was recognized by αH3-P (Figure 3E). Thus, it seems that Ser139 on NSUN2 is phosphorylated by Aurora-B.
We next expressed NSUN2 proteins containing N-terminal Xpress tags in HeLa cells and synchronized the cells in mitosis by treatment with nocodazole. Immunoprecipitation with anti-Xpress mAb (αXpress) followed by immunoblotting with αH3-P detected NSUN2-WT but not NSUN2-S139A (Supplemental Figure S5). Together with our other results, this suggests that αH3-P recognizes NSUN2 phosphorylated during mitosis at Ser139 as well as Histone H3 phosphorylated at Ser10. This phosphorylation was repressed by Aurora-B-K/R (Figure 1A). In addition, a 2-h treatment with Hesperadin or ZM447439 to inhibit Aurora-B repressed mitotic phosphorylation of NSUN2 in the synchronized HeLa cells (Figure 3F).
Because Aurora-B plays a central role in spindle checkpoint regulation, the repression of NSUN2 phosphorylation by Aurora-B-K/R or the inhibitors may be due to an inability to accumulate mitotic cells caused by the absence of spindle checkpoint regulation. To determine whether this is the case, we used a destruction-defective Cyclin B (ΔCyclin B1), which induces mitotic arrest, even in cells expressing Aurora-B-K/R or an Aurora-B shRNA, or treatment with an Aurora-B inhibitor (data not shown). Cotransfection of HeLa cells with ΔCyclin B1 and Aurora-B-K/R repressed mitotic phosphorylation of NSUN2 (Figure 3G, lane 8). Similar repression was observed in cells cotransfected with the expression vectors for ΔCyclin B1 and Aurora-C-K/R (Figure 3G, lane 10). In contrast, in the Aurora shRNA experiments, only the Aurora-B shRNA was able to inhibit NSUN2 phosphorylation in the presence of ΔCyclin B1 expression (Figure 3H, lanes 6, 8, 9, and 11). Based on these results, we concluded that NSUN2 is phosphorylated by Aurora-B.
Conservation of Aurora-B Phosphorylation Site of NSUN2 in Vertebrates
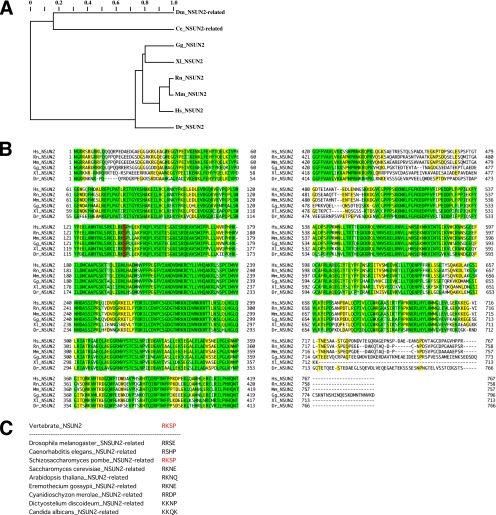
The human NSUN2 gene belongs to the methyltransferase superfamily and is located on chromosome 5p15.31. Saccharomyces cerevisiae Ncl1/Trm4 is related to human NSUN2 (∼35% similarity). We also detected a remote similarity between NSUN2 and predicted proteins of Drosophila melanogaster and Caenorhabditis elegans (Figure 4A, Dm and Ce, respectively). Among vertebrates, NSUN2 is highly conserved (Figure 4B). The Aurora-B phosphorylation site at Ser139, which lies within the NOL1/NOP2/sun domain (residues 102-427 in human NSUN2), is also conserved among vertebrates (Figure 4B, red letters). In contrast, nonvertebrates other than Schizosaccharomyces pombe lack the putative Aurora-B phosphorylation site in this domain (Figure 4C).
Figure 4.
NSUN2 is conserved among vertebrates and is distantly related to proteins in other species. (A) Tentative evolutionary tree for the vertebrate NSUN2 family and distantly related proteins in insects and nematodes. (B) Multiple sequence alignment for the vertebrate NSUN2 family. Colors indicate the extent of similarity: yellow, >60%; and green, 100%. The conserved Aurora-B phosphorylation site is indicated in red. (C) Nonconservation of the Aurora-B phosphorylation site among the NSUN2-related proteins in nonvertebrates.
NSUN2 Is Associated with Nucleolar Proteins NPM1 and Nucleolin during Interphase, but Phosphorylated NSUN2 Disassociates from NPM1 during Mitosis
Indirect immunofluorescence by using monoclonal antibodies against NPM1 and nucleolin confirmed that NSUN2 is colocalized with NPM1 and nucleolin in nucleoli, particularly at the dense fibrillar and granular components (Figure 2, B–D). In agreement with this, NPM1 and nucleolin coprecipitated with NSUN2 in interphase HeLa cells (Figure 5A, lanes exp). In the immunoblot for NPM1, there were two bands (NPM1.1 and NPM1.2), both of which migrated more slowly (i.e., shifted to a higher molecular weight) in nocodazole-treated cells (Figure 5A, IB: NPM1 and C, IB: NPM1). Such a shift in migration of the bands was not observed in synchronized mitotic cells (Figure 5B, IB: NPM1). These results suggested that nocodazole induces hyperphosphorylation of both NPM1 proteins. Regardless, only the top band (NPM1.1) was immunoprecipitated with αNSUN2 (Figure 5A, IP: NSUN2; IB: NPM1). In addition, in the immunoprecipitation experiments, association between NSUN2 and NPM1 was not observed in nocodazole-treated mitotic cells, whereas the association of NSUN2 with nucleolin was unchanged (Figure 5A, IP: NSUN2, IB: NPM1 and IP: NSUN2, IB: nucleolin). We confirmed these data both by immunoblot using the supernatant of immunoprecipitated samples and by immunoprecipitation using the opposite combination of antibodies (IP: NPM1 or nucleolin; IB: NSUN2) (data not shown). Thus, NSUN2 phosphorylated by Aurora-B during mitosis does not seem to be associated with NPM1.
Figure 5.
NSUN2 associates with NPM1 and nucleolin, but this association is reduced during mitosis. (A) Association between NSUN2 and NPM1 is reduced in nocodazole-treated cells. Exponentially growing and nocodazole-treated HeLa cells were analyzed by immunoprecipitation. (B) The association between NSUN2 and NPM1 is reduced during mitosis in synchronized cells. This reduction is inhibited by Hesperadin. Eight hours after release, synchronized HeLa cells were treated for 2 h with Hesperadin or dimethyl sulfoxide (DMSO). Samples were collected at 6 (lane 1) and 10 h (lane 2) after release (after 2 h with DMSO (lane 3) or Hesperadin (lane 4)). (C) NSUN2-S139A, which mimics unphosphorylated NSUN2, is associated with NPM1 even in mitotic cells, but the association of NPM1 with NSUN2-S139E, which mimics constitutively phosphorylated NSUN2, is reduced. HeLa-NSUN2-KD cells were transfected with a plasmid for expressing NSUN2-WT, NSUN2- SA, or NSUN2-SE and were then treated for 18 h with or without nocodazole. IB, immunoblot; IP, immunoprecipitation; WT, wild-type; SA, NSUN2-S139A; SE, NSUN2-S139E; exp, exponentially growing; Noc, nocodazole treatment.
To confirm these findings, we examined the effects of 1) the Aurora-B inhibitor Hesperadin and 2) the NSUN2 mutants NSUN2-S139A, which cannot be phosphorylated, and NSUN2-S139E, which mimics NSUN2 phosphorylated on Ser139. As in the nocodazole-treated mitotic cells (Figure 5A), there was a reduced association between NSUN2 and NPM1 in synchronized cells during mitosis (10 h after release; Figure 5B, lanes 10). In contrast, a 2-h treatment with Hesperadin prevented the reduction in NPM1–NSUN2 association (Figure 5B, lanes H).
We next examined the effects of the NSUN2 mutants. HeLa cells were transfected with plasmids expressing Xpress-tagged NSUN2-WT, NSUN2-S139A, or NSUN2-S139E. Although Xpress-tagged NSUN2-WT was phosphorylated in mitotic cells (Supplemental Figure S5C), its association with NPM1 was not detected, even in interphase cells (data not shown).
We speculated that the long N-terminal His-Xpress polypeptide disrupts the NSUN2 structure necessary for association with NPM1. We also examined tag-free NSUN2 expressed in NSUN2 knockdown (KD) cells. HeLa cells were transfected with the vector for expressing NSUN2 shRNA, and stable clones with low NSUN2 expression (HeLa-NSUN2-KD cells) were established (Supplemental Figure S3). HeLa-NSUN2-KD cells were transfected with expression plasmids for NSUN2-WT, NSUN2-S139A, or NSUN2-S139E (without tag). When NSUN2-WT was expressed in HeLa-NSUN2-KD cells, the association of NSUN2 with NPM1 and nucleolin was detected in interphase cells (Figure 5C, lane 3). The association of NSUN2 with NPM1 was reduced in mitotic cells, but the association with nucleolin was not (Figure 5C, lane 4). In contrast, the phosphorylation site mutant NSUN2-S139A was associated with NPM1, even during mitosis (Figure 5C, lanes 5 and 6). Finally, the phosphorylation-mimic mutant NSUN2-S139E was associated with nucleolin, but not NPM1 (Figure 5C, lanes 8 and 7, respectively). These data indicate that Aurora-B-mediated phosphorylation of NSUN2 at Ser139 during mitosis directs the dissociation of NPM1 from the NSUN2-nucleolin complex.
Aurora-B-provoked Phosphorylation Affects Methyltransferase Activities of NSUN2
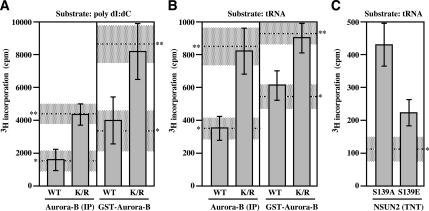
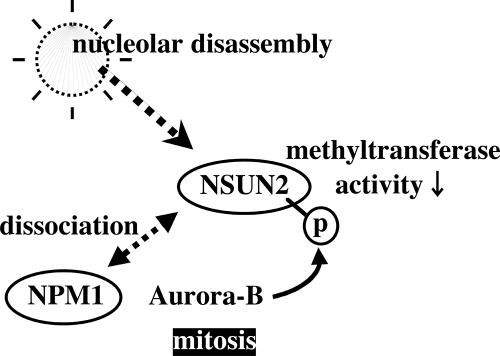
Next, we tested whether phosphorylation status of NSUN2 provoked by Aurora-B affects the methyltransferase activity. NSUN2 contains an NOL1/NOP2/sun domain and relates to yeast protein Ncl1, which is a methyl transferase that generates m5C in RNA. As expected, NSUN2 has no methyltransferase activity in DNA that is not methylated on either strand (Supplemental Figure S6). In contrast, NSUN2 has a methyltransferase activity in hemimethylated DNA. When poly(dI:dC) was used as substrate, NSUN2 stimulated the incorporation of tritirated methyl group from methyl donor SAM into the substrate (Figure 6A, **). In addition, NSUN2 has methyltransferase activity in RNA. When tRNA was used as substrate, the activity was also found (Figure 6B, **). The phosphorylation of NSUN2 provoked by either nocodazole-treated HeLa cells-derived immunoprecipitated Aurora-B or GST-Aurora-B suppressed NSUN2 methyltransferase activities (Figure 6, A and B). Such a suppression was not found when Aurora-B-K/R was used (Figure 6, A and B). The effect of phosphorylation in NSUN2 was also confirmed by using NSUN2-S139A and NSUN2-S139E proteins generated by the TNT in vitro transcription/translation (rabbit reticulocyte) system (Figure 6C). Therefore, as schematically illustrated in Figure 7, we propose a model that the NSUN2 is negatively regulated by Aurora-B-provoked phosphorylation during mitosis.
Figure 6.
Phosphorylation status of NSUN2 affects its methyltransferase activities. (A) [3H]methylation of poly(dI:dC) DNA upon incubation for 30 min with purified NSUN2 after phosphorylation reaction with immunoprecipitated Aurora-B or GST-Aurora-B. (B) [3H]methylation of tRNA upon incubation for 30 min with purified NSUN2 after phosphorylation reaction with immunoprecipitated Aurora-B or GST-Aurora-B. (C) [3H]methylation of tRNA upon incubation for 5 min with TNT-produced NSUN2-S139A and NSUN2-S139E. Error bar, SD (n = 3)., basal incorporation level (shadow is SD range)., incorporation level in a reaction mixture with purified NSUN2 following MOCK phosphorylation reaction (shadow is SD range).
Figure 7.
Schematic model of NSUN2 regulation by Aurora-B.
Nucleolar Disassembly–Reassembly Processes under Aurora-B Inhibition
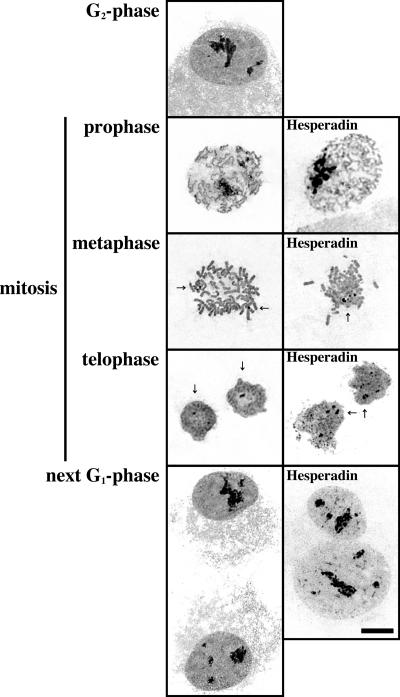
During mitosis, NSUN2 is phosphorylated by Aurora-B to suppress its methyltransferase activities and the association of NSUN2 with NPM1 is inhibited. The inhibition of Aurora- B kinase activity by Hesperadin induces constitutive association of NSUN2 with NPM1 during mitosis. Moreover, the constitutively dephosphorylated form of NSUN2-S139A is continuously associated with NPM1 during mitosis. Finally, to test whether mitotic nucleolar disassembly is affected by NUSN2 phosphorylation induced by Aurora-B, the disassembly process was monitored by Ag-NOR. Nucleolar disassembly was clearly observed by the silver staining (Figure 8, left). The Ag-NOR was also detected in mitotic chromosomes of HeLa cells (Figure 8, arrow). In Hesperadin-treated mitotic cells, the nucleolus was completely disassembled, although the number and size of Ag-NORs were larger than in nontreated cells during mitosis (Figure 8). Aurora-B inhibition by Hesperadin induced binuclear cells as a result of cytokinesis error. In the next G1 cells, the disassembled nucleolus was reassembled, even when Hesperadin was continuously present in the culture medium (Figure 8, bottom right). Under Hesperadin-induced Aurora-B inhibition, the disassembly–reassembly processes of Ag-NOR are likely to be intact. Therefore, Aurora-B affects both the assembly of NSUN2 and its associated protein NPM1 and NSUN2 activities, but it does not affect Ag-NOR organization.
Figure 8.
Aurora-B kinase activity does not affect nucleolar Ag-NOR disassembly-reassembly processes during mitosis. Ag-NOR silver staining images visualizing the nucleolus in interphase nuclei and NOR on mitotic chromosomes in Hesperadin-treated or nontreated synchronized HeLa cells. Arrow, Ag-NOR.
DISCUSSION
NSUN2 is Phosphorylated during Mitosis by Aurora-B
NSUN2 is a novel 100-kDa substrate of Aurora-B conserved in all vertebrates examined, with more distant relatives in D. melanogaster and C. elegans. The site of NSUN2 phosphorylated by Aurora-B is also conserved among vertebrates but not in D. melanogaster and C. elegans.
NSUN2 was recently identified in a large-scale screening of nuclear phosphoproteins in HeLa cells by mass spectroscopic analysis of tryptic peptides (Beausoleil et al., 2004). This study identified three phosphorylation sites on NSUN2 (Ser456, Ser593, and Ser743). It is possible that Ser139 was not identified as one of the phosphorylation sites due to technical limitations of the large-scale screening by using trypsin. Furthermore, because the cells were not synchronized, mitosis-specific phosphorylation would have been diluted. Whether the three identified sites are constitutively or periodically phosphorylated during the cell cycle was not determined. Together, the current results and those of Beausoleil et al. (2004) indicate that phosphorylation of NSUN2 on Ser139 is mediated by Aurora-B and that this is one of many sites phosphorylated in vivo.
Like Aurora-B, Aurora-C is a mitotic passenger protein that is active during mitosis (Li et al., 2004; Sasai et al., 2004; Dutertre et al., 2005). We also found that kinase-negative Aurora-C-K/R represses mitotic phosphorylation of NSUN2 due to overlapping repression of Aurora-B; however, we found that the level of Aurora-C was very low in HeLa cells and that repression of expression by an shRNA does not affect the phosphorylation of NSUN2 at Ser139 in mitotic cells. Therefore, Aurora-C may not make a substantial contribution to the phosphorylation of NSUN2, and Aurora-B may be the protein kinase critical for its phosphorylation at Ser139 during mitosis.
NSUN2 Is a Nucleolar Protein That Has RNA Methyltransferase Activity
NSUN2 is a novel mammalian RNA methyltransferase that was independently discovered in a study of myc-induced proliferation mediators in epithelial cells (Frye and Watt, 2006). The original working name of the protein in our laboratory was substrate of AIM-1/Aurora-B kinase (SAKI; accession no. AB255451), whereas that reported by Frye and Watt (2006) was myc-induced SUN-domain-containing protein (MISU; accession no. DQ490066). The protein is now designated “NSUN2.” NSUN2 contains an NOL1/NOP2/sun domain. This domain is found in archaeal, bacterial, and eukaryotic proteins. In the archaea and bacteria, the majority of proteins possessing this domain have an S-adenosyl methionine binding-domain and are related to E. coli Sun/Fmu protein, which catalyzes 5-methylation of C967 within 16S rRNA (Foster et al., 2003). In S. cerevisiae, the nucleolar protein Nop2 is known to be a probable RNA m5C methyltransferase (Katz et al., 2003) and is essential for processing and maturation of rRNA (de Beus et al., 1994; Hong et al., 1997). Yeast Nop2 is thought to be closely related to human NOL1/proliferation-associated nucleolar antigen p120 (Fonagy et al., 1989; de Beus et al., 1994). Yeast contain Ncl1, another protein related to Nop2 (Wu et al., 1998), but unlike Nop2, Ncl1 is not essential for cell survival. Yeast Ncl1, also called as Trm4, has tRNA m5C methyltransferase activity, and it probably catalyzes the methylation of other RNA molecules (Motorin and Grosjean, 1999). Like the relationship between yeast Nop2 and human NOL1, there may be substantial identity between yeast Ncl1 and human NSUN2.
Similar to the case of yeast Ncl1, repression of NSUN2 expression in HeLa cells by shRNA did not affect cell growth properties such as doubling time and the ability of anchorage-independent growth (data not shown). Thus, like Ncl1, NSUN2 is probably nonessential for growth. Interestingly, disruption of Ncl1 in yeast increases the sensitivity to rRNA-targeting antibiotics, such as paromomycin (Wu et al., 1998). In addition, under disruption of the tRNA methyltransferase Trm8, disruption of Ncl1 greatly reduces tRNA stability (Alexandrov et al., 2006). The evolutionary conservation of NSUN2 suggests that it plays important biological roles other than the nonessential methylation of RNAs. This is also supported by the fact that NSUN2 expression is markedly elevated in cancer cells (Figure 2A; our unpublished data).
Here, we showed that NSUN2 methylated hemimethylated DNA, poly(dI:dC), as well as tRNA. This raises the possibility that NSUN2 is implicated in alterations of not only RNA methylation patterns but also genomic methylation patterns. Unlike prokaryotic restriction DNA methyltransferases that have innate sequence specificity, eukaryotic DNA cytosine methyltransferases exhibit a preference for hemimethylated substrates with indirect sequence recognition. Mammalian DNA methyltransferase-2 (Dnmt2) methylates tRNA, suggesting eukaryotic DNA methyltransferases were derived from ancestral RNA methyltransferases rather than prokaryotic restriction DNA methyltransferases (Goll et al., 2006). Thus, eukaryotic RNA or DNA methyltransferases possibly have broad substrate specificity in nucleic acids, even though their sequence and their organization of catalytic motifs are characteristic of RNA or DNA.
Aurora-B Regulates NSUN2-NPM1 Association and NSUN2 Methyltransferase Activities
During interphase, EGFP-Aurora-B is placed in nucleoplasm but is not located in nucleolus (our unpublished data). During prophase, EGFP-Aurora-B rapidly spreads throughout the chromatin (Murata-Hori et al., 2002). Consequently, Histone H3 is phosphorylated on Ser10 (Crosio et al., 2002). Simultaneously, NSUN2 is rapidly phosphorylated by Aurora-B. NSUN2 phosphorylation probably occurs in parallel with nucleolar disassembly during mitosis. CDK1-Cyclin B is a major regulator of this disassembly but is not the sole driving factor (Sirri et al., 2002). When ribosome disassembles, nucleolar proteins may be phosphorylated by multiple kinases at multiple sites. NPM1 contains four or five CDK1-Cyclin B phosphorylation sites (Thr4, Thr199, Thr219, Thr234, and Thr237) (Peter et al., 1990; Okuwaki et al., 2002), and the interaction of NPM1 with nucleolin is inhibited during mitosis (Liu and Yung, 1999). Here, we have found that NSUN2 is associated with nucleolar proteins NPM1 and nucleolin and that the interaction of NSUN2 with NPM1 is inhibited during mitosis. This dissociation is driven by Aurora- B-provoked phosphorylation of NSUN2. Thus, Aurora-B is also a mitotic kinase that participates to nucleolar disassembly-reassembly processes.
The C-terminal region of NPM1.1, which has a 35-amino acid C-terminal extension, was shown to bind RNA during interphase and to lose this ability upon phosphorylation during mitosis (Okuwaki et al., 2002). Our results show that NSUN2 binds only to NPM1.1, indicating that the interaction must be mediated by the C-terminal region of NPM1.1. We found that NPM1.1 can associate with NSUN2 but that these proteins dissociate during mitosis when NSUN2 is phosphorylated on Ser139 by Aurora-B. This dissociation may also occur in concert with CDK1 phosphorylation of NPM1.1. However, this dissociation is primarily regulated by Aurora-B-provoked phosphorylation of NSUN2, because the Aurora-B kinase inhibitor Hesperadin is able to inhibit this dissociation during mitosis.
Inhibition of CDK1 leads to resumption of rDNA transcription, but it is not sufficient to build a functioning rRNA processing complex in mitotic cells (Sirri et al., 2002). Inhibition of Aurora-B is also not sufficient to activate constitutively rRNA processing in mitosis. Silver staining to visualize the nucleolus confirms that Hesperadin is not able to completely inhibit nucleolar disassembly. The Ag-NOR is not sufficiently spread in Hesperadin-treated mitotic chromosomes, but the number and the size of NOR are not drastically altered. It is possible that this is due to impaired chromosome organization induced by Aurora-B inhibition. Thus, in Aurora-B–inhibited cells, Ag-NOR nucleolar disassembly–reassembly processes are likely to be intact.
The methyltransferase activities of NSUN2 are suppressed by Aurora-B–provoked phosphorylation. During normal mitosis, NSUN2 should be inactive due to its phosphorylation by Aurora-B. It remains uncertain what are precise target nucleic acids of NSUN2 in the cells, although NSUN2 may have broad in vivo substrate specificities including DNA and RNAs such as rRNA, tRNA or microRNAs. The elucidation of the in vivo targets of NSUN2 helps us to understand the physiological effects of the phosphorylation of NSUN2 provoked by Aurora-B.
In summary, Aurora-B regulates not only chromosome segregation but also nucleolar protein function during mitosis. Our findings presented here possibly create a new aspect to investigate how nucleic acid modifications are regulated in mitotic cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. M. Brandeis for providing Cyclin B1 plasmids and Drs. Takahide Ota and Yasuhiko Terada for helpful comments. This work was supported by a grant from the Two Cell Co. Ltd (H17-SAKI).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-1021) on January 10, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams R. R., Maiato H., Earnshaw W. C., Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., Grayhack E. J., Phizicky E. M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Andrews P. D., Knatko E., Moore W. J., Swedlow J. R. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 2003;15:672–683. doi: 10.1016/j.ceb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J.E., Villen J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G. M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C. D., Sassone-Corsi P. Mitotic phosphorylation of histone H3, spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beus E., Brockenbrough J. S., Hong B., Aris J. P. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimario P. J. Cell and molecular biology of nucleolar assembly and disassembly. Int. Rev. Cytol. 2004;239:99–178. doi: 10.1016/S0074-7696(04)39003-0. [DOI] [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S., Hamard-Peron E., Cremet J. Y., Thomas Y., Prigent C. The absence of p53 aggravates polyploidy and centrosome number abnormality induced by Aurora-C overexpression. Cell Cycle. 2005;4:1783–1787. doi: 10.4161/cc.4.12.2172. [DOI] [PubMed] [Google Scholar]
- Fonagy A., Henning D., Jhiang S., Haidar M., Busch R. K., Larson R., Valdez B., Busch H. Cloning of the cDNA and sequence of the human proliferating-cell nucleolar protein P120. Cancer Commun. 1989;1:243–251. [PubMed] [Google Scholar]
- Foster P. G., Nunes C. R., Greene P., Moustakas D., Stroud R. M. The first structure of an RNA m5C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Structure. 2003;11:1609–1620. doi: 10.1016/j.str.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Frye M., Watt F. M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A. J., Ruchaud S., Hudson D. F., Honda R., Nigg E. A., Gerloff D. L., Earnshaw W. C. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Petretti C., Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Goll M. G., Kirpekar F., Maggert K. A., Yoder J. A., Hsieh C. L., Zhang X., Golic K. G., Jacobsen S. E., Bestor T. H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Heix J., Vente A., Voit R., Budde A., Michaelidis T. M., Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B., Brockenbrough J. S., Wu P., Aris J. P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L. F., Segura-Valdez M. L., Ochs R. L., Rothblum L. I., Hannan R., Spector D. L. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S., Suto K., Nagata A., Igarashi M., Kanaoka Y., Nojima H., Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A., Kawai H., Suto S., Kitajima S., Sato S., Takata T., Tatsuka M. Aurora-B/AIM-1 kinase activity is involved in Ras-mediated cell transformation. Oncogene. 2005;24:7266–7272. doi: 10.1038/sj.onc.1208884. [DOI] [PubMed] [Google Scholar]
- Katz J. E., Dlakic M., Clarke S. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell Proteomics. 2003;2:525–540. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- Li X., Sakashita G., Matsuzaki H., Sugimoto K., Kimura K., Hanaoka F., Taniguchi H., Furukawa K., Urano T. Direct association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C. J. Biol. Chem. 2004;279:47201–47211. doi: 10.1074/jbc.M403029200. [DOI] [PubMed] [Google Scholar]
- Liu H. T., Yung B. Y. In vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett. 1999;144:45–54. doi: 10.1016/s0304-3835(99)00184-6. [DOI] [PubMed] [Google Scholar]
- Motorin Y., Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori M., Tatsuka M., Wang Y. L. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 2002;13:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M., Tsujimoto M., Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Nakagawa J., Doree M., Labbe J. C., Nigg E. A. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Fortel I., Junera H. R., Geraud G., Hernandez-Verdun D. Three-dimensional organization of the ribosomal genes and Ag-NOR proteins during interphase and mitosis in PtK1 cells studied by confocal microscopy. Chromosoma. 1993;102:146–157. doi: 10.1007/BF00387729. [DOI] [PubMed] [Google Scholar]
- Sampath S. C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. The chromosomal passenger complex is required for chromatin- induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sasai K., et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil. Cytoskeleton. 2004;59:249–263. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- Savino T. M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Schumacher J. M., Golden A., Donovan P. J. AIR-2, An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Hernandez-Verdun D., Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 2002;156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Roussel P., Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 2000;148:259–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A. S., Dabeva M. D., Dimova R. N., Hadjiolov A. A. Ribosome biogenesis and nucleolar ultrastructure in neuronal and oligodendroglial rat brain cells. J. Neurochem. 1985;45:1667–1676. doi: 10.1111/j.1471-4159.1985.tb10521.x. [DOI] [PubMed] [Google Scholar]
- Tatsuka M., Sato S., Kitajima S., Suto S., Kawai H., Miyauchi M., Ogawa I., Maeda M., Ota T., Takata T. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene. 2005;24:1122–1127. doi: 10.1038/sj.onc.1208293. [DOI] [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., Otsu M. AIM-1, a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S. P., Carvalho A., Vagnarelli P., Earnshaw W. C. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Wu P., Brockenbrough J. S., Paddy M. R., Aris J. P. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Basic mechanism of eukaryotic chromosome segregation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005;360:609–621. doi: 10.1098/rstb.2004.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.