Abstract
Bacteria are able to switch between two mutually exclusive lifestyles, motile single cells and sedentary multicellular communities that colonize surfaces. These behavioral changes contribute to an increased fitness in structured environments and are controlled by the ubiquitous bacterial second messenger cyclic diguanosine monophosphate (c-di-GMP). In response to changing environments, fluctuating levels of c-di-GMP inversely regulate cell motility and cell surface adhesins. Although the synthesis and breakdown of c-di-GMP has been studied in detail, little is known about the downstream effector mechanisms. Using affinity chromatography, we have isolated several c-di-GMP-binding proteins from Caulobacter crescentus. One of these proteins, DgrA, is a PilZ homolog involved in mediating c-di-GMP-dependent control of C. crescentus cell motility. Biochemical and structural analysis of DgrA and homologs from C. crescentus, Salmonella typhimurium, and Pseudomonas aeruginosa demonstrated that this protein family represents a class of specific diguanylate receptors and suggested a general mechanism for c-di-GMP binding and signal transduction. Increased concentrations of c-di-GMP or DgrA blocked motility in C. crescentus by interfering with motor function rather than flagellar assembly. We present preliminary evidence implicating the flagellar motor protein FliL in DgrA-dependent cell motility control.
Keywords: diguanylate receptor, motility, PilZ, c-di-GMP, second messenger
Cyclic purine nucleotides are ubiquitous second messengers involved in cell signaling. They are produced through the action of growth factors, hormones, or neurotransmitters and elicit their response by acting on a range of downstream effector proteins such as protein kinases, transcription regulators, gated ion channels, or GTPase nucleotide exchange factors. Although cAMP is widespread through all kingdoms of life, cGMP seems to be restricted to signaling in eukaryotes. Recently, a third major cyclic nucleotide messenger, cyclic diguanosine monophosphate (c-di-GMP) has emerged as a ubiquitous signaling molecule in prokaryotes, where it antagonistically controls motility and virulence of planktonic cells on one hand and cell adhesion and persistence of multicellular communities on the other (1, 2) [supporting information (SI) Fig. 7]. C-di-GMP is synthesized from two molecules of GTP and degraded into the linear dinucleotide pGpG by the opposing activities of diguanylate cyclases (DGC) and c-di-GMP-specific phosphodiesterases (PDE). DGC and PDE activities are comprised in GGDEF and EAL domains, respectively (3–8), which represent two large families of output domains found in bacterial one- and two-component signal transduction systems (9, 10).
The molecular principles of c-di-GMP signaling have been studied in the model organism Caulobacter crescentus, where c-di-GMP coordinates the developmental transition from a motile swarmer cell to a surface-attached, replication-competent stalked cell. Both acquisition of flagellar motility in the predivisional cell and its replacement by an adhesive organelle later in development are controlled by c-di-GMP. TipF, an EAL domain protein, is required for an early step of flagellum assembly in the predivisional cell (11), whereas the diguanylate cyclase PleD is involved in flagellum ejection and subsequent steps in pole remodeling (3, 12–15). Similarly, the second messenger c-di-GMP regulates motility, adhesion factors, and biofilm formation in a wide variety of bacterial pathogens including Yersinia, Pseudomonas, Vibrio, and Salmonella (1, 2). C-di-GMP influences flagellar motility as a function of growth (16) or adaptation to surfaces (17), affects pilus assembly (18), and controls the production of surface structures such as fimbriae and exopolysaccaride matrices (19). The wide variety of cellular functions that are affected by c-di-GMP calls for multiple receptors and signaling mechanisms. However, little information is available on specific targets of c-di-GMP action. With the exception of a component of the cellulose synthase complex from Gluconacetobacter (20, 21) and the recent prediction of a candidate c-di-GMP-binding domain (22, 23), no c-di-GMP effector proteins have been reported. We have designed a biochemical approach to purify and characterize c-di-GMP effector molecules from C. crescentus crude cell extracts.
Results
Purification of c-di-GMP-Binding Proteins from C. crescentus.
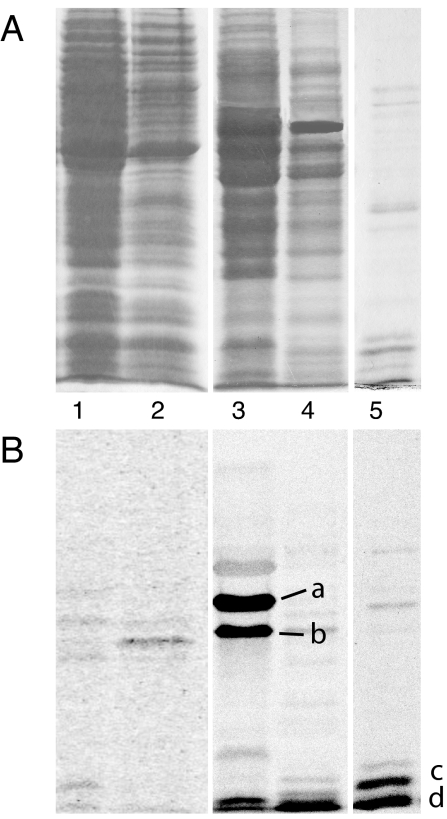
Based on the assumption that c-di-GMP signal transduction depends on specific receptor proteins, we designed a biochemical purification strategy to identify such components. C-di-GMP-binding proteins from C. crescentus were purified by two consecutive chromatography steps with Blue Sepharose CL-6B and affinity chromatography with GTP immobilized on epoxy-activated Sepharose 4B (Amersham Pharmacia, Piscataway, NJ). UV cross-linking with c-[33P]di-GMP was used to identify proteins with specific binding activity for c-di-GMP (see Materials and Methods and SI Table 2). Two binding proteins with apparent molecular masses of 47 and 36 kDa were detected in the 0.4–0.7 M NaCl eluate of the Blue Sepharose column (Fig. 1 A and B, lane 3), and the 0.7–0.9 M NaCl fraction contained several small c-di-GMP-binding proteins with apparent molecular masses of 8–12 kDa (Fig. 1 A and B, lanes 4 and 5). The latter fraction was dialyzed, concentrated, and separated on a GTP-epoxy-Sepharose 4B affinity column (Fig. 1 A and B, lane 4). One of these proteins (labeled c in Fig. 1B) was identified by tandem mass spectrometry (MS/MS) as the product of gene CC1599, a conserved hypothetical 12.5-kDa protein that we consequently renamed as diguanylate receptor A (DgrA). Sequence comparison disclosed DgrA as a member of the PilZ protein family, members of which have recently been proposed by bioinformatics to be c-di-GMP effector proteins (22).
Fig. 1.
Isolation of c-di-GMP-binding proteins from C. crescentus. (A) Coomassie blue-stained SDS/polyacrylamide gel with protein fractions used for UV cross-linking with c-[33P]di-GMP. Lane 1, 100.000 × g supernatant; lane 2, 60% ammonium sulfate precipitation; lane 3, 0.4–0.7 M NaCl eluate from Blue Sepharose; lane 4, 0.7–0.9 M NaCl eluate from Blue Sepharose; and lane 5, 125 mM NaCl eluate from GTP-Sepharose column. (B) Autoradiograph of SDS/polyacrylamide gel shown in A. C-di-GMP-binding proteins a, b, and d were identified by MS/MS. Protein c was identified by MS/MS as hypothetical protein CC1599 and was renamed DgrA.
DgrA Is a Diguanylate Receptor Protein.
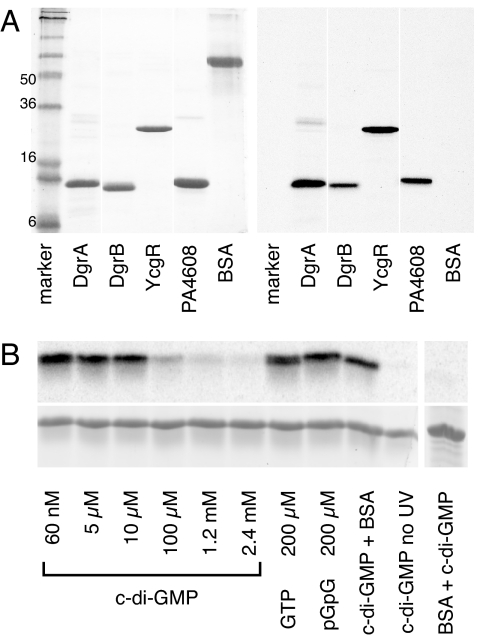
To confirm that the identified protein is a c-di-GMP receptor, dgrA, was subcloned into the expression vector pET-42b, and the recombinant His6-tagged protein was purified by nickel–nitrilotriacetic acid affinity chromatography. Like the semipurified protein from C. crescentus (Fig. 1B), the recombinant protein showed strong labeling upon UV cross-linking with c-[33P]di-GMP (Fig. 2A), confirming that DgrA is a c-di-GMP receptor protein. UV cross-linking experiments with DgrA in the presence of 60 nM 33P-labeled c-di-GMP and increasing concentrations of cold c-di-GMP, GTP (200 μM), or pGpG (200 μM) indicated that DgrA binds c-di-GMP with high affinity and specificity (Fig. 2B). C-di-GMP binds to DgrA in a noncovalent manner because no radiolabeled c-di-GMP was incorporated without UV irradiation (Fig. 2B). The dissociation constant for c-di-GMP of the recombinant DgrA was determined by using the UV cross-linking assay (Table 1). Saturated incorporation of radiolabeled c-di-GMP was already observed at 50 nM, indicating that the Kd of DgrA for c-di-GMP is <50 nM.
Fig. 2.
DgrA is a member of a new family of c-di-GMP-binding proteins. (A) UV cross-linking of purified His6-tagged diguanylate receptor proteins with c-[33P]di-GMP. The following proteins were used: DgrA (CC1599; C. crescentus), DgrB (CC3165; C. crescentus), PA4608 (P. aeruginosa), YcgR (S. typhimurium), and BSA (control). The Coomassie blue-stained gel (Left) and the autoradiograph (Right) are shown. (B) UV cross-linking of 10 μM DgrA in the presence of 60 nM 33P-labeled c-di-GMP. Autoradiograph (Upper) and Coomassie-stained gel (Lower). (Left) Samples were supplemented with increasing concentrations of nonlabeled nucleotides as indicated. (Right) Controls were carried out in the absence of UV irradiation or with BSA.
Table 1.
Binding constants of diguanylate receptor proteins determined by UV cross-linking with c-[33P]di-GMP
| Organism | Protein | Kd, nM | ΔKd* |
|---|---|---|---|
| C. crescentus | DgrB | 132 | 36 |
| DgrA | |||
| WT | <50 | 14 | |
| R11AR12A | ND | — | |
| D38A | 761 | 149 | |
| W75A | 6,200 | 496 | |
| S. typhimurium | YcgR | 182 | 29 |
| P. aeruginosa | PA4608 | <50 | 27 |
The protein concentration used for binding assay was 50 nM. ND, not detectable.
*S.E.
To test whether other members of the PilZ protein family also bind c-di-GMP, we analyzed several ortho- or paralogs of DgrA, including CC3165 (renamed as DgrB), YcgR from Salmonella typhimurium (24), and PA4608 from Pseudomonas aeruginosa (see Fig. 6). As shown in Fig. 2A, all four proteins were efficiently labeled with c-[33P]di-GMP upon UV cross-linking, whereas the control protein BSA did not incorporate c-di-GMP. The c-di-GMP-binding constants of DgrB, YcgR, and PA4608 were determined by performing UV cross-linking experiments with 50 nM receptor protein in the presence of increasing concentrations of 33P-labeled c-di-GMP (50–1,000 nM). All wild-type diguanylate receptor proteins exhibit a binding affinity in the nanomolar range (Table 1). Taken together, these data demonstrate that DgrA and its homologs containing a PilZ domain are members of a class of small diguanylate receptor proteins, which bind c-di-GMP, but not other guanine nucleotides, with high affinity. Thus, these proteins represent bona fide diguanylate receptor proteins and may be involved in the response of specific cell functions to fluctuating concentrations of c-di-GMP (2).
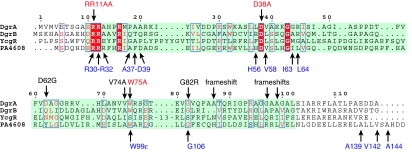
Fig. 6.
Sequence alignment of the c-di-GMP-binding proteins DgrA, DgrB, YcgR, and PA4608 according to the PilZ Pfam entry PF07238. The PilZ domain is highlighted in green. DgrA residues shown to be important for c-di-GMP binding and in vivo function (red) and the positions of intragenic dms suppressor mutations (black) are highlighted above the alignment. Residues of PA4608 with large chemical shift differences upon c-di-GMP binding (blue) are indicated below the alignment.
DgrA and DgrB Mediate c-di-GMP-Dependent Motility Control in C. crescentus.
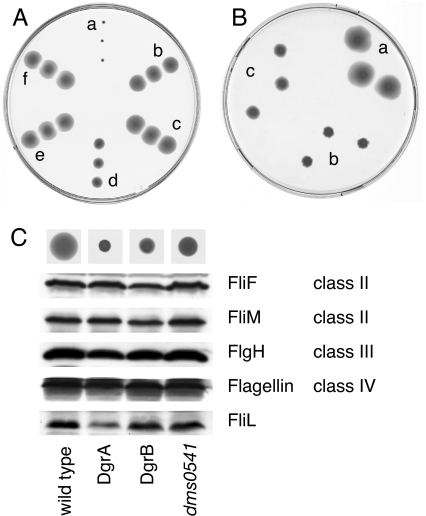
Low concentrations of c-di-GMP are generally associated with flagella- or pili-based motility of single planktonic cells, whereas increased concentrations of c-di-GMP promote multicellular traits and efficiently block cell motility (2). In agreement with this finding, C. crescentus cells are nonmotile in the presence of a plasmid-borne copy of dgcA, which encodes a highly active, soluble diguanylate cyclase (15) (Fig. 3A). Electron micrographs and immunoblot experiments showed that these cells were flagellated and expressed normal levels of flagellins (data not shown), arguing that increased c-di-GMP concentrations interfere with flagellar function rather than with the expression or assembly of flagellar components. To test whether motility control by c-di-GMP involves dgrA or dgrB, single and double in-frame deletion mutants were generated by using a two-step homologous recombination procedure (see SI Materials and Methods). In contrast to C. crescentus wild-type, ΔdgrA and ΔdgrB mutants were motile even in the presence of the dgcA plasmid (Fig. 3A). Motility was not the result of a reduction of the c-di-GMP concentration because cellular levels of c-di-GMP in these mutants were indistinguishably high (data not shown). At low cellular concentrations of c-di-GMP, motility phenotypes were not significantly altered in the deletion mutants (data not shown), indicating that DgrA and DgrB affect cell motility primarily at conditions where the level of c-di-GMP is elevated. Together, these data suggested that the c-di-GMP-binding proteins DgrA and DgrB are part of a signal transduction pathway that interferes with flagellar function in response to high concentrations of c-di-GMP. In agreement with this suggestion, overexpression of dgrA or dgrB from a plasmid efficiently blocked motility on swarmer plates (Fig. 3B) and in liquid media as observed microscopically (data not shown).
Fig. 3.
DgrA and DgrB are involved in motility control by c-di-GMP. Motility behavior of C. crescentus wild-type strain CB15 and mutants is shown on semisolid agar plates. Three different colonies from independent conjugation experiments are shown. (A) The following strains containing plasmid pUJ142::dgcA or control plasmid pUJ142 were analyzed: CB15/pUJ142::dgcA (a), CB15ΔdgrA/pUJ142::dgcA (b), CB15dgrAW75A/pUJ142::dgcA (c), CB15ΔdgrB/pUJ142::dgcA (d), CB15ΔdgrAΔdgrB/pUJ142::dgcA (e), and CB15/pUJ142 (f). (B) Overexpression of dgrA or dgrB from the lactose promoter (Plac) repressed C. crescentus motility. (a) CB15/pBBR (vector control). (b) CB15/pBBR::dgrA. (c) CB15/pBBR::dgrB. (C) Levels of class II, class III, and class IV structural components of the C. crescentus flagellum were determined by immunoblot analysis for the following strains: CB15/pBBR (wild-type), CB15/pBBR::dgrA (DgrA), CB15/pBBR::dgrB (DgrB), and the extragenic diguanylate receptor motility suppressors CB15dms0541 pBBR::dgrA (dms0541). The motility behavior of each strain is shown on top of the graph.
Analysis of c-di-GMP Binding to the Diguanylate Receptor by NMR Spectroscopy.
The available NMR structure and resonance assignments of the DgrA homolog PA4608 from P. aeruginosa (25) [Protein Data Bank (PDB) ID code 1YWU; Biological Magnetic Resonance Bank (BMRB) ID code 6514]‡ provided an opportunity to characterize the ligand-binding site on a molecular level and to investigate the structural consequences of ligand binding. PA4608 carrying an N-terminal His6 tag was produced in uniformly 15N- and 13C-labeled form for NMR spectroscopy. The 1H and 15N chemical shifts observed for pure PA4608 were in good agreement with those reported in BMRB entry 6514. When c-di-GMP was added to the protein, 1H-15N heteronuclear sequential quantum correlation spectra (HSQC) changed dramatically (SI Fig. 8). Free and ligand-bound PA4608 were in slow exchange on the NMR chemical shift time scale, and titration curves were in agreement with a Kd in the sub-μM range (data not shown). To assign resonances of the PA4608*c-di-GMP complex, exchange (EXSY) spectra were recorded on a roughly 3:1 mixture of free and c-di-GMP-bound PA4608 at 313 K. Standard triple-resonance NMR spectra recorded on PA4608 saturated with c-di-GMP were used to complete the backbone resonance assignments. No resonances were observed for residues Met3–His12 (His6 tag), His22, Phe33–Ile36, Gly73, Ile91, Glu125, Leu128, and Asp130–Leu138. These residues are probably flexible on a microsecond to millisecond time scale, and peaks are broadened beyond detection because of intermediate chemical exchange. Secondary 13Cα and 13Cβ shifts (26) showed that the secondary structure of PA4608 remained essentially unchanged after ligand binding (SI Fig. 9).
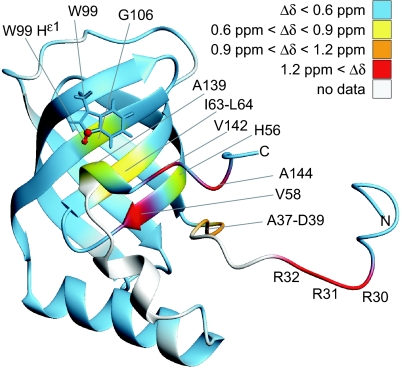
To localize the ligand-binding site on the protein surface, backbone amide 1H and 15N chemical shifts of the PA4608*c-di-GMP complex were compared with those of the free protein, and the differences were mapped on the structure of the free protein (see Fig. 5 and SI Fig. 8). Large shift differences were found on one face of the β-barrel (around Val58, Ile63), in the C terminus (Val142, Ala144), and in the N terminus (Arg30–Asp39). We conclude that c-di-GMP binds to the outside of the β-barrel close to Val58 and that the termini, which are partially flexible in the apo form, fold around the bound ligand. Presumably the side chain N-H group of Trp99 forms a hydrogen bond with the ligand because the 15Nε1 and 1Hε1 resonances strongly shift toward higher chemical shifts by 8.24 and 1.66 ppm, respectively.
Fig. 5.
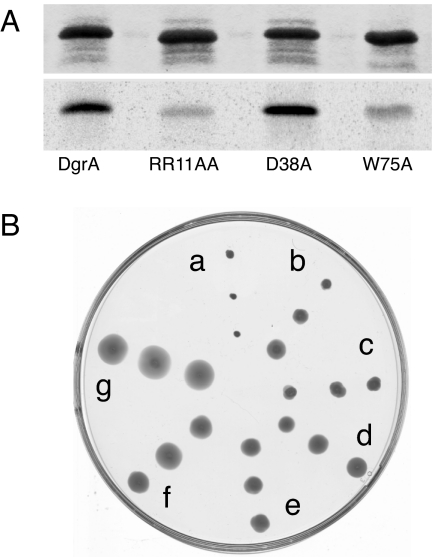
C-di-GMP binding and motility control of DgrA mutants. (A) UV cross-linking of different DgrA mutant proteins with c-[33P]di-GMP. Coomassie blue-stained SDS/PAGE (Upper) and autoradiograph (Lower) with purified wild-type and mutant DgrA proteins (10 μM) are shown. (B) Motility behavior of C. crescentus wild-type CB15 overexpressing different dgrA alleles: CB15/pBBR::dgrA (a), CB15/pBBR::dgrAR11AR12A (b), CB15/pBBR::dgrAD38A (c), CB15/pBBR::dgrAV74A (d), CB15/pBBR::dgrAR11AR12AV74A (e), CB15/pBBR::dgrAW75A (f), and CB15/pBBR (vector control) (g). Three different colonies from independent conjugation experiments are shown.
Because of their distinct chemical shifts (>10.7 ppm), the H1 imino hydrogens of guanine in c-di-GMP could be identified once the assignment of protein backbone 1HN and tryptophan 1Hε1 resonances had been completed. Because four separate H1 resonances of about equal intensity are observed for c-di-GMP in complex with PA4608 and each molecule of c-di-GMP contains two guanine bases, c-di-GMP binds to PA4608 as a dimer. Consistent with the ligand-binding site outlined above, two of these H1 imino resonances show intermolecular NOEs to Leu64 and Trp99 (SI Fig. 10).
Amide 15N T1 and T2 relaxation times and heteronuclear {1H}-15N NOEs were measured at 293 K for free and c-di-GMP-bound PA4608 (data not shown). Isotropic rotational correlation times (τc) were determined from these data with the program TENSOR (27) as 11.3 and 12.3 ns for free and ligand-bound protein, respectively. These τc are in reasonable agreement with values expected for monomeric apo-PA4608 (16.7 kDa, 9.8 ns) and c-di-GMP-bound PA4608 (18.1 kDa, 10.6 ns). Thus, PA4608 is a monomer before and after ligand binding.
C-di-GMP-Binding Mutants of DgrA Are Unable to Control Motility.
Alignments of the amino acid sequences of PA4608, DgrA, DgrB, and YcgR revealed that the key residues that were postulated based on NMR data to be involved in c-di-GMP binding to PA4608 are conserved among other diguanylate receptor proteins (see Fig. 6). To probe the c-di-GMP-binding site of DgrA and to define the minimal requirements for c-di-GMP binding, residues Arg11, Arg12, Asp38, and Trp75 were replaced with Ala, and the mutant proteins were analyzed for c-di-GMP binding. Mutants R11A/R12A and W75A strongly reduce c-di-GMP binding, whereas mutant D38A is still able to bind c-di-GMP (see Fig. 5A). In agreement with this result, the binding constant for the D38A mutant was marginally increased to 740 nM, whereas the Kd for the W75A mutant (6.4 μM) was increased 100- to 1,000-fold compared with wild type (Table 1). Binding of c-di-GMP was completely abolished in the R11A/R12A mutant. To analyze the importance of c-di-GMP binding for DgrA-mediated signaling, the dgrA mutant alleles were tested for functionality in vivo. As indicated above, overexpression of wild-type dgrA renders cells nonmotile (Fig. 4B; see also Fig. 6B). In contrast, overexpression of dgrAD38A, dgrAR11AR12A, or dgrAW75A only partially affected motility (Fig. 5B). In particular, changing Trp75 to Ala almost completely abolished the ability of DgrA to block motility under these conditions (Fig. 5B). Also, when the dgrAW75A mutant allele was expressed in single copy from its original chromosomal locus, cells were fully motile even in the presence of the dgcA plasmid, arguing that DgrAW75A can no longer control motility in response to increased c-di-GMP levels (Fig. 3). We isolated suppressors that alleviated the dgrA-mediated motility block (see Materials and Methods). One of the intragenic dms (diguanylate receptor motility suppressors) mutations mapped to Val74, in the immediate vicinity of the Trp residue critical for c-di-GMP binding (Figs. 5B and 6). Other intragenic dms mutations (Asp62, Gly82) mapped to conserved residues of DgrA, emphasizing the functional importance of these residues (Fig. 6). In conclusion, these results support the view that ligand binding is essential for the regulatory function of the diguanylate receptor and suggest that DgrA blocks motility in its c-di-GMP-bound state.
Fig. 4.
Combined amide 1H and 15N shift differences (Δδ) between PA4608 in its free and ligand-bound form. Shift differences are color-coded on the structure of free PA4608 (PDB 1YWU, model 12). Combined chemical shift differences were calculated as . These data are also shown in SI Fig. 8. Residue Trp99 is shown as sticks, and Nε1 and Hε1 are shown in red to highlight the large Δδ value (1.67 ppm) for these atoms.
Motility Control by DgrA Correlates with Cellular Levels of the FliL Motor Protein.
Immunoblot analysis revealed that overexpression of dgrA or dgrB blocks motility without interfering with the expression of known class II, class III, or class IV components of the flagellar hierarchy (Fig. 3C). Because the expression of each class of genes depends on the successful expression and assembly of components of the preceding class of the hierarchy (28), this result suggested that flagella are assembled normally in cells overexpressing dgrA or dgrB. In agreement with this suggestion, flagella were readily observed by electron microscopy in these nonmotile cells (data not shown). The only flagellar protein whose concentration was severely affected in cells overexpressing dgrA was FliL (Fig. 3C). The C. crescentus fliL gene is not part of the flagellar hierarchy, and its product is not assembled into the flagellar structure (29). However, fliL is required for flagellar rotation (29). To examine whether reduced FliL levels are linked to motility, we screened the pool of dms mutants (see above) for extragenic suppressors (see Materials and Methods). From a total of 120 independently isolated motile suppressors, only one mapped to the chromosome. This suppressor mutation (dms0541), which mapped to gene CC3587 coding for the ribosomal protein S1, not only restored motility but also reestablished normal levels of FliL (Fig. 3C).
Discussion
Motility control by c-di-GMP is implemented through gene expression, organelle assembly, or motor function (2). In C. crescentus, increased cellular concentrations of c-di-GMP block motility by interfering with motor function rather than by altering expression or assembly of structural components of the flagellum (13). How are increased levels of c-di-GMP sensed and how is this information transmitted to the flagellar motor? The data presented here show that DgrA and DgrB are high affinity receptors for c-di-GMP that, in a ligand-bound form, interfere with the flagellar motor either directly or indirectly. Motor control by DgrA-like proteins is not unique to Caulobacter. Escherichia coli H-NS mutants lack flagella because the expression of the flagellar master control operon flhCD is reduced. Ectopic expression of flhCD restores flagellation but leaves the motors partially paralyzed (24). Under these conditions, flagellar function can be restored either by a mutation in ycgR, coding for the E. coli DgrA homolog, or by providing multiple copies of yhjH, which encodes a presumable c-di-GMP-specific phosphodiesterase (24). Together with our data demonstrating that the Salmonella YcgR protein specifically binds c-di-GMP, this finding suggests that in C. crescentus and in enteric bacteria flagellar motor function might be controlled by c-di-GMP through similar mechanisms.
But how would DgrA or YcgR interfere with the function of the flagellum? Our data propose the FliL protein as a candidate for such a role. FliL was the only flagellar protein that showed significantly reduced levels in nonmotile cells overexpressing dgrA. In C. crescentus the FliL protein is not part of the flagellar structure but is required for flagellar rotation (29). Intriguingly, fliL mutant strains exhibit a motility phenotype identical to that of cells that have high levels of c-di-GMP or overexpress dgrA (29). Because the expression of fliM, the gene located immediately downstream from fliL in the same operon (30), was not affected by DgrA, FliL changes must be the result of altered translation or protein stability. An extragenic suppressor mutation that restored motility under these conditions also reestablished normal FliL concentrations, indicating that the two phenotypes are linked. The simplest model that is in agreement with these results predicts that DgrA, upon binding of c-di-GMP, represses FliL by a so far unknown mechanism, and through this mechanism blocks motor function. The extragenic suppressor mutation restoring FliL levels was mapped to the coding region of rpsA (ribosomal protein S1). RpsA enhances translation initiation by binding to mRNA regions upstream from the Shine–Dalgarno sequence and by tethering the mRNAs on the 30S subunit of the ribosome (31–33). How DgrA and its ligand c-di-GMP modulate FliL levels is a subject for future investigation. Recently, FliL was reported to be involved in surface sensing and virulence gene expression in the urinary tract pathogen Proteus mirabilis (34). Thus, it is possible that FliL has a more general role in controlling the switch between a planktonic and a surface-associated lifestyle.
A bioinformatics study originally proposed that the PilZ domain is a specific c-di-GMP-binding module (22). This proposal was recently substantiated by the demonstration that YcgR, a PilZ protein from E. coli, is able to bind c-di-GMP (23). Here we presented genetic, biochemical, and structural evidence that further validates this hypothesis, and we propose a model for ligand binding and activation of proteins containing a PilZ domain. NMR studies with the DgrA homolog PA4608 showed that a dimer of c-di-GMP binds to a well defined binding site on the surface of the β-barrel (Fig. 4). Large chemical shift differences between free and ligand-bound PA4608, which indicate changes in the local environment, were also observed in both termini of the protein, with the largest differences observed for residues Arg30–Arg32, Val142, and Ala144. These regions are structurally ill defined in the absence of ligand (25) and are probably flexible. The observed chemical shift differences indicate that these regions come in direct contact with the ligand after complex formation. The N-terminal part of PA4608 contains three consecutive Arg residues, which are conserved in most PilZ domains (22) (Fig. 6). Arg side chains are likely to be involved in hydrogen bonds or in electrostatic or π stacking interactions with c-di-GMP, as in the allosteric binding site of the diguanylate cyclases PleD and DgcA (15, 35). Furthermore, it is conceivable that the positively charged head groups of Arg are sufficient for transient binding to the phosphate groups of c-di-GMP and that their position on the flexible N terminus increases the ligand capture radius of the protein, as in the “fly-casting mechanism” proposed in ref. 36. Alternatively, the observed folding of previously flexible parts of the protein may be responsible for communication of the c-di-GMP signal to downstream elements, either by forming new interaction surfaces or by determining the relative position of neighboring domains. Similarly, the chemical shift differences of the C-terminal part of PA4608 could be explained by a specific role in ligand binding. However, the fact that residues Val142 and Ala144, which showed the largest chemical shift differences, are not conserved, argues against this possibility. Several of the motile dgrA loss-of-function suppressors that were isolated had frameshift mutations in the very C terminus of DgrA (Fig. 6), suggesting that this part of the protein is critical for its in vivo function. One possibility is that the C terminus contributes to the specific readout mechanism of this protein family. Upon c-di-GMP binding to the β-barrel surface, the C terminus could be untied to interact with downstream components. In accordance with such a view, the very C terminus of the P. aeruginosa PilZ protein has recently been proposed to interact with the PilF protein required for type 4 pilus assembly (37). To complement our picture of the c-di-GMP circuitry, future studies will have to focus on interaction partners of DgrA and related PilZ domain proteins.
It is intriguing that genetic and biochemical studies of the C. crescentus DgrA protein and structural analysis of PA4608 from P. aeruginosa identified the same set of key amino acids involved in c-di-GMP binding (Fig. 6). This finding is a strong indication that these proteins bind c-di-GMP in a similar way and suggests that they may share a common signaling mechanism. Based on these results, we postulate that most or all PilZ domain proteins function as diguanylate receptor proteins.
Materials and Methods
Strains, Plasmids, and Media.
E. coli strains were grown in Luria broth. C. crescentus strains were grown in complex peptone yeast extract (38) supplemented with antibiotics, where necessary. For the exact procedure of strain and plasmid construction, see SI Materials and Methods.
UV Cross-Linking with C-[33P]di-GMP and Isolation of DgrA.
Procedures for enzymatic production of c-[33P]di-GMP and UV cross-linking with c-[33P]di-GMP were published earlier (6, 15). For a detailed protocol used for the isolation of DgrA, see SI Materials and Methods.
Preparation of Isotope-Labeled Protein, NMR Samples, and NMR Spectroscopy.
The detailed procedures for overexpression and 13C,15N labeling of PA4608 are described in SI Materials and Methods. NMR samples (Shigemi microtubes) were prepared as 0.8 mM U-13C,15N-labeled protein in 300 μl of 95% H2O/5% D2O/250 mM NaCl/10 mM DTT/1 mM NaN3/10 mM Tris at pH 7.1. C-di-GMP was added at suitable molar ratios from a 7.7 mM stock solution. NMR spectra were recorded on Bruker (Billerica, MA) DRX 600 and 800 MHz spectrometers at 293 K with the exception of EXSY spectra that were recorded at 313 K for faster exchange. Standard one-, two-, and three-dimensional spectra were recorded and processed as described elsewhere (39).
Isolation and Mapping of Motile dgrA suppressors.
A plasmid carrying dgrA (pBBR::dgrA) was conjugated into a C. crescentus recA mutant strain, and 150 individual transconjugants were patched onto PYE swarmer plates. Motile dms mutants were isolated and analyzed by immunoblot with an α-DgrA antibody. Mutants with reduced DgrA levels were discarded. The rest was analyzed by retransforming plasmids into the recA mutant strain to distinguish between intra- and extragenic suppressors. Intragenic mutations were identified by sequencing. The extragenic suppressor (dms0541) was mapped by Tn5 linkage (40) and cotransduction with phage ΦCR30, and it was identified by sequencing.
Supplementary Material
Acknowledgments
We thank Suzette Moes for handling of MS samples, Martha Gerber and Flora Mauch for help with the suppressor screen, Jacob Malone (University of Basel) for a gift of P. aeruginosa DNA, and Colin Manoil (University of Washington, Seattle, WA) for providing plasmid pIT2. This work was supported by Swiss National Science Foundation Fellowships 3100A0-108186 (to U.J.) and 31-109712 (to S.G.).
Abbreviations
- c-di-GMP
cyclic diguanosine monophosphate
- DgrA
diguanylate receptor A
- MS/MS
tandem mass spectrometry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607738104/DC1.
Residue numbering for PA4608 as in Biological Magnetic Resonance Bank ID code 6514, which differs from that in Protein Data Bank structure 1YWU by +22, is used throughout this work.
References
- 1.Kolter R, Greenberg EP. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt AJ, Ryjenkov DA, Gomelsky M. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 7.Bobrov AG, Kirillina O, Perry RD. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 8.Tamayo R, Tischler AD, Camilli A. J Biol Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin MY, Nikolskaya AN, Koonin EV. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 10.Ulrich LE, Koonin EV, Zhulin IB. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Aldridge P, Jenal U. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- 13.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 14.Levi A, Jenal U. J Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. J Biol Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 16.Choy WK, Zhou L, Syn CK, Zhang LH, Swarup S. J Bacteriol. 2004;186:7221–7228. doi: 10.1128/JB.186.21.7221-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boles BR, McCarter LL. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazmierczak BI, Lebron MB, Murray TS. Mol Microbiol. 2006;60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kader A, Simm R, Gerstel U, Morr M, Romling U. Mol Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, Benziman M., de Vroom E, Fidder A, de Paus P, Sliedregt LA, et al. J Biol Chem. 1990;265:18933–18943. [PubMed] [Google Scholar]
- 21.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 22.Amikam D, Galperin MY. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 23.Ryjenkov DA, Simm R, Romling U, Gomelsky M. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 24.Ko M, Park C. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 25.Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. Proteins. 2006;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 26.Spera S, Bax A. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 27.Dosset P, Hus J-C, Blackledge M, Marion D. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 28.Gober JW, E JC. In: Prokaryotic Development. Shimkets LJ, Brun YVB, editors. Washington, DC: Am Soc Microbiol; 2000. pp. 319–339. [Google Scholar]
- 29.Jenal U, White J, Shapiro L. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Shapiro L. J Bacteriol. 1992;174:3327–3338. doi: 10.1128/jb.174.10.3327-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta J, Agrawal RK, Frank J. Proc Natl Acad Sci USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komarova AV, Tchufistova LS, Supina EV, Boni IV. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorensen MA, Fricke J, Pedersen S. J Mol Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 34.Belas R, Suvanasuthi R. J Bacteriol. 2005;187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim K, Oh J, Han D, Kim EE, Lee B, Kim Y. Biochem Biophys Res Commun. 2006;340:1028–1038. doi: 10.1016/j.bbrc.2005.12.108. [DOI] [PubMed] [Google Scholar]
- 38.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 39.Grzesiek S, Bax A, Hu JS, Kaufman J, Palmer I, Stahl SJ, Tjandra N, Wingfield PT. Protein Sci. 1997;6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.