Abstract
Phytophthora infestans (Mont.) de Bary caused the 19th century Irish Potato Famine. We assessed the genealogical history of P. infestans using sequences from portions of two nuclear genes (β-tubulin and Ras) and several mitochondrial loci P3, (rpl14, rpl5, tRNA) and P4 (Cox1) from 94 isolates from South, Central, and North America, as well as Ireland. Summary statistics, migration analyses and the genealogy of current populations of P. infestans for both nuclear and mitochondrial loci are consistent with an “out of South America” origin for P. infestans. Mexican populations of P. infestans from the putative center of origin in Toluca Mexico harbored less nucleotide and haplotype diversity than Andean populations. Coalescent-based genealogies of all loci were congruent and demonstrate the existence of two lineages leading to present day haplotypes of P. infestans on potatoes. The oldest lineage associated with isolates from the section Anarrhichomenun including Solanum tetrapetalum from Ecuador was identified as Phytophthora andina and evolved from a common ancestor of P. infestans. Nuclear and mitochondrial haplotypes found in Toluca Mexico were derived from only one of the two lineages, whereas haplotypes from Andean populations in Peru and Ecuador were derived from both lineages. Haplotypes found in populations from the U.S. and Ireland was derived from both ancestral lineages that occur in South America suggesting a common ancestry among these populations. The geographic distribution of mutations on the rooted gene genealogies demonstrate that the oldest mutations in P. infestans originated in South America and are consistent with a South American origin.
Keywords: late blight, oomycetes, phylogeography, Solanum tuberosum, stamenopiles
Phytophthora infestans (Mont.) de Bary causes late blight of potato and tomato and is one of the world's most devastating plant diseases (1). P. infestans caused the 19th century Irish Potato Famine, which led to the starvation and death of more than one million people and precipitated a massive human migration from Ireland to North America. Speculation about the origin of P. infestans and the source of inoculum for the epidemics began soon after the catastrophe and remains the subject of debate (2–6).
Nineteenth-century scientists thought that P. infestans originated in the South American Andes (currently Bolivia, Ecuador, and Peru) (2, 3), the center of origin of the cultivated potatoes and other Solanaceous species (7, 8), and assumed cospeciation of the pathogen with its host. The first three potato varieties to succumb to late blight in Europe in 1845 were named “Lima,” “Peruviennes,” and “Cordilieres” (9). Potatoes were imported from South America, and historical records of the potato disease in the Andes suggest that it was endemic in the region (6). However, only a few clonal lineages of P. infestans have been described from the Andes, so the Andean hypothesis has not been generally accepted (4, 10–15).
Others suggest that the center of origin of P. infestans is in the central highlands of Mexico's Toluca Valley because high nuclear genetic diversity and the presence of sexual reproduction of the pathogen occurs there (4, 5, 11, 12). Sexual reproduction is less common in Andean populations and evidence for host adaptation and reproductive isolation has been reported (16). Sexual recombination generates high variability in P. infestans progeny (17–19). High levels of nuclear genetic variability found in central Mexico could be the result of sexual reproduction and not of ancestry. The introduction of the A2 mating type into Europe resulted in a shift from low to high nuclear genetic diversity in the Netherlands, particularly in places where both mating types were found together, mirroring the diversity found in central Mexico (18, 20–22). Greater diversity in a place may be due to a particular history of founder effects, extinctions, and expansions of local populations. In contrast, there is less mitochondrial diversity in Toluca Mexico and the predominance of one maternal lineage suggests either a single maternal origin for this population or selection (23, 24). The mitochondrial genome is inherited maternally as a unit in P. infestans, without genetic recombination (23).
It was suggested that P. infestans originally migrated from Mexico to the United States in infected wild potato tubers in the 19th century to cause famine-era epidemics (4, 11). In the U.S., the pathogen infected potatoes and then spread to Europe and the rest of the world (4). Spread of a single clonal lineage, the US-1 (Ib mtDNA haplotype) was proposed (4). The US-1 lineage is not found widely in extant Mexican populations of P. infestans (12, 23, 24), whereas this lineage is still found in other populations around the world including the Andes. We sequenced the mtDNA from historic specimens of P. infestans from the Irish famine and found the Ia haplotype was common (25, 26). The US-1 lineage (Ib mtDNA haplotype) did not cause the famine, but was identified in more recent samples from the Andean region in Ecuador and Bolivia (26). This finding suggests either extinction of the US-1 lineage from the Mexican population or a non-Mexican origin of this lineage. We published the mitochondrial genome sequences of extant mtDNA haplotypes of P. infestans (27). Two independent ancestral lineages, the type I (Ia,Ib) and type II (IIa,IIb), are derived from a common ancestor and the type I lineage is more closely related to the common ancestor.
A third theory, known as the Three-Step or Hybrid theory, suggests Mexico as the center of origin of the pathogen, but that the source of inoculum for the 19th century epidemics originated from the South American Andes (5, 13). It was speculated that P. infestans migrated first from Mexico to the South American Andes centuries before the 1840's and was subsequently dispersed from the Andean region to the U.S. and Europe (5).
Here, we describe a population genetic and phylogeographic approach using coalescent analysis and mitochondrial and nuclear gene genealogies to address two questions. First, does mitochondrial and nuclear DNA evidence justify the specific hypothesis that a common ancestor of P. infestans originated in South American populations? Second, what can be inferred about the source of inoculum that caused the 19th century epidemics?
Results
DNA Sequence Variability.
A total of 3,265 nucleotides were sequenced corresponding to 2,010 nucleotides in the two regions of the mitochondrial genome, P3 (rpl14, rpl5, tRNAs) and P4 (part of cox 1) and 1,255 nucleotides in two single-copy nuclear genes, RAS (Intron Ras+Ras) and B-tubulin [supporting information (SI) Table 2].
Sequence diversity ranged from 0.20% to 2.23% (SI Table 2). Sequence diversity estimates were 0.65% for mitochondrial gene regions and 1.03% for single-copy nuclear genes. Four nucleotide substitutions leading to amino acid changes were found in the P3 region, but no amino acid changes were observed in the other genes examined. Two synonymous substitutions were detected for Ras and one for the B-tubulin gene (SI Table 2). Two nucleotide changes were observed for each polymorphic site consistent with an infinite-sites model. Isolates of P. infestans sensu lato (now Phytophthora andina) from the section Anarrhichomenum from Ecuador were highly polymorphic across all loci and sequence diversity estimates were higher for both mitochondrial (1.44%) and for Intron Ras (6.69%) regions, than isolates from potato (SI Table 2). Seven nucleotide substitutions leading to amino acid changes were found in the P3 region in isolates from the section Anarrhichomenum (SI Table 2).
Nucleotide diversity (π) estimates for the pooled (total) sample was 2.20 × 10−3 for the mitochondrial loci and 3.25 × 10−3 for the nuclear locus (SI Table 3). The average per-nucleotide expected heterozygosity, θw, for the pooled sample was 1.39 × 10−3 for the mitochondrial loci and 2.57 × 10−3 for the nuclear locus. When section Anarrhichomenum isolates were included in the mitochondrial data set (Pooled Anarr; SI Table 3), π and θw were elevated to 2.82 × 10−3 and 3.51 × 10−3, respectively. Sequence data from these isolates were not included in the nuclear data set.
When samples were partitioned into South American (SA) and non-South American (NSA) populations, both populations had similar estimates of nucleotide diversity in the mitochondrial loci. However, the magnitude of nucleotide diversity in the nuclear locus of SA (4.18 × 10−3) populations was approximately double that of NSA (2.22 × 10−3) populations (SI Table 3). The SA population also showed higher nucleotide diversity in the mitochondrial loci when section Anarrhichomenum isolates was included (SI Table 3). Surprisingly, the Toluca Mexico population showed low levels of nucleotide diversity for both nuclear and mitochondrial loci compared with other populations, and only accounted for 20% and 42% of the total mitochondrial and nuclear nucleotide diversity, respectively (SI Table 3).
Tests of neutrality and population subdivision. Neutrality tests were not significant for the pooled (total) sample across both mitochondrial and nuclear loci (SI Table 3), so the equilibrium model of neutral evolution could not be rejected. Positive values for several neutrality tests are indicative of an excess of intermediate-frequency variants. Two frequently sampled haplotypes in the mitochondrial loci and four repeatedly sampled haplotypes in the nuclear locus (IRRAS) were observed (Table 1). Two mitochondrial haplotypes (H1 and H2) were sampled at the same frequency, whereas in the nuclear IRRAS locus, one haplotype (H2) was sampled at a high frequently and three other haplotypes (H1, H4, H6) were sampled at intermediate frequencies (Table 1). At one extreme, the maintenance of two haplotypes at equal frequencies in the Brazilian and Bolivian populations resulted in significant positive values for all neutrality tests (SI Table 3). Negative values for several test statistics are indicative of an excess of low frequency alleles and it's possible that some populations have not yet reached equilibrium as a result of a recent bottleneck or selective sweep. Two of the three variable sites found in the Mexican sample had a minor allele frequency of 10% compared with the most common allele, which had a frequency of 50%. This distribution of alleles in the Mexican population is consistent with drift and a strong founder effect. Small sample sizes and the presence of population subdivision are known to limit the power of the neutrality tests so we subsequently tested for population subdivision (28, 29).
Table 1.
Distribution of haplotypes, base substitutions, and insertion/deletion events in mitochondrial and nuclear genes of P. infestans
| Mitochondrial (P3 + P4) | Nuclear (IRRAS) | ||
|---|---|---|---|
| Locus | |||
| Position in GenBank database accession | 111111111 | ||
| 23333333333333333999999000000000 | 1111111 | ||
| 90001344445558899567899111111111 | 5566670011233 | ||
| 80777002771693567203934348888999 | 6901602678105 | ||
| 80230642069849364919843804689013 | 1630018487331 | ||
| Position in combined consensus | 11111111111111111 | ||
| 112344555668800234666889999999 | 11123344456 | ||
| 34111446116138901637367771122222 | 6901601467983 | ||
| 02452864281061586242176137912346 | 3852232821775 | ||
| Site number | 11111111112222222222333 | 1111 | |
| 12345678901234567890123456789012 | 1234567890123 | ||
| Site type | tttttvvvtvvvttvttvtvtttttvtttttv | tvvtttvtvtttv | |
| Character type | i-----i---ii-iiii------i-------- | i-iiiiiiiiiii | |
| Substitution type | rsrssrrrsrrsssssss | ss | |
| Consensus | TGTGTATTCATAGTCACGGGTCACATCACGAC | CGAAAGGGGTGTT | |
| Haplotype (frequency) | |||
| H1 (23) | C.....A...GC.CAGT......T........ | H1 (19) | TTCG.A....ACA |
| H2 (24) | ................................ | H2 (93) | ............. |
| H3 (1) | .........C...................... | H3 (2) | ......TA.C... |
| H4 (1) | .................C.............. | H4 (14) | .......A..... |
| H5 (3) | ......................G......... | H5 (1) | .......A..... |
| H6 (2) | CAC.CTAGT...A.A...ATCT..GGTGTAGA | H6 (20) | .......A.C... |
| H7 (4) | C..A..A...GC.CAGT......T........ | H7 (1) | ...........C. |
| H8 (2) | C............................... | H8 (1) | T.CG.A....ACA |
| H9 (1) | C.........G..CAGT......T........ | H9 (1) | ....G........ |
| H10 (1) | C.....A...GC.CA.T......T........ | H10 (1) | ....G..A.C... |
| H11 (1) | ....G..AT.... | ||
| H12 (2) | ....G..AT.... |
t, transitions; v, transversions; i, phylogenetically informative sites; −, uninformative sites; r, nonsynonymous (i.e., replacement) substitutions; s, synonymous substitutions. Numbering in vertical columns is that of GenBank accession no. U17009 for mitochondrial loci and accession no. U30474 for nuclear loci.
Hudson's tests were performed to quantify population genetic structure within and among populations (28, 30). Pairwise comparisons between SA and NSA populations showed significant genetic structure for both mitochondrial (P = 0.0000, KST = 0.2143, KS = 3.8625) and nuclear loci (P = 0.0265, KST = 0.0175, KS = 2.5082). For the mitochondrial loci P3 and P4, the differentiation was marginally significant within the SA populations with the section Anarrhichomenum isolates included (P = 0.0237, KST = 0.1292, KS = 4.7311) or excluded (P = 0.0190, KST = 0.2303, KS = 2.7602), indicating moderate gene flow within SA. By comparison, the NSA populations were strongly subdivided (P = 0.0013, KST = 0.31364, KS = 2.8229). For the nuclear IRRAS locus, we observed significant genetic differentiation between the SA (P = 0.0001, KST = 0.2347, KS = 2.4530) and NSA (P = 0.0001, KST = 0.1162, KS = 1.6608) populations.
Among the NSA populations, the Toluca population was genetically differentiated from the U.S. and Irish populations for both mitochondrial and nuclear loci (SI Tables 4 and 5). However, the U.S. and Irish populations were not significantly differentiated from each other. For the mitochondrial loci, the U.S. and Irish populations were not genetically differentiated from the Peruvian and Ecuadorian populations (SI Table 4). For the nuclear locus, the Irish population was also not genetically differentiated from the Peruvian population (SI Table 5). The Mexican population was not genetically differentiated from Peruvian and Ecuadorian populations for the nuclear locus (SI Table 5), but was differentiated from all SA populations for the mitochondrial loci (SI Table 4). For the nuclear locus, the Brazilian and Bolivian population were not different from each other but were significantly differentiated from the Peruvian and Ecuadorian populations (SI Table 5). When the analysis was performed with paired localities from four populations, Hudson's tests for population subdivision showed the same trend. For the mitochondrial loci, there was significant population differentiation between Brazil and Bolivia (BRABO) and Mexico and Costa Rica (MECO) (P = 0.0093, KST = 1.0000, KS = 0.0000, KT = 1.8461), Peru and Ecuador (PEECU) and MECO (P = 0.0000, KST = 0.4567, KS = 2.1698, KT = 3.9938), MECO and U.S. and Ireland (USIR) (P = 0.0040, KST = 0.2075, KS = 2.4776, KT = 3.1264). For the nuclear locus, genetic differentiation was marginally significant between BRABO and USIR (P = 0.0438, KST = 0.0800, KS = 3.6543, KT = 3.9720), and highly significant between BRABO and PEECU (P = 0.0000, KST = 0.2649, KS = 2.3557, Kt = 3.2051); and BRABO and MECO (P = 0.0000, KST = 0.3403, KS = 1.8029, KT = 2.7331).
Migration Analysis.
We simultaneously estimated population divergence time, population mean mutation rate and direction of migration, if present, between SA and NSA with the isolation with migration (IM) coalescent model (31). For the nuclear locus, migration was nonzero and significantly higher from SA to NSA (m2 = 14.6) than from NSA to SA (m1 = 4.3) when moving forward in time. For the mitochondrial region, migration appeared to be equilibrating from SA to NSA (m2 = 2.6) and from NSA to SA (m1 = 4.0). For both mitochondrial and nuclear loci, the complete posterior probability distribution for migration parameters, which contains 90% of the probability, could be estimated only for migration from SA to NSA (SI Table 6). The migration likelihood surfaces were too rough for estimating the 90% posterior probability density intervals from NSA to SA; however, three independent runs supported these parameter estimates.
Genealogical Analysis.
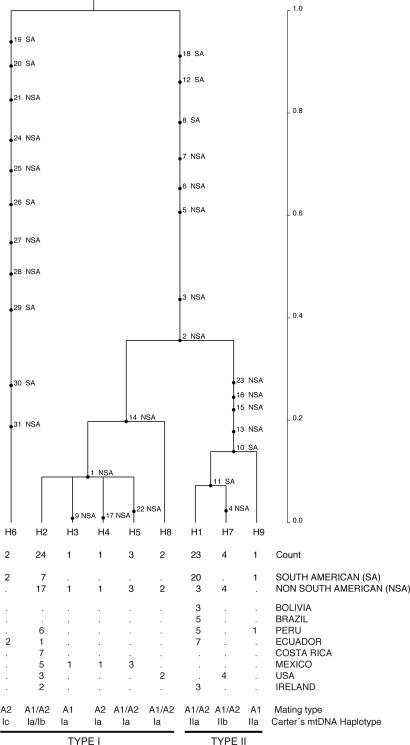
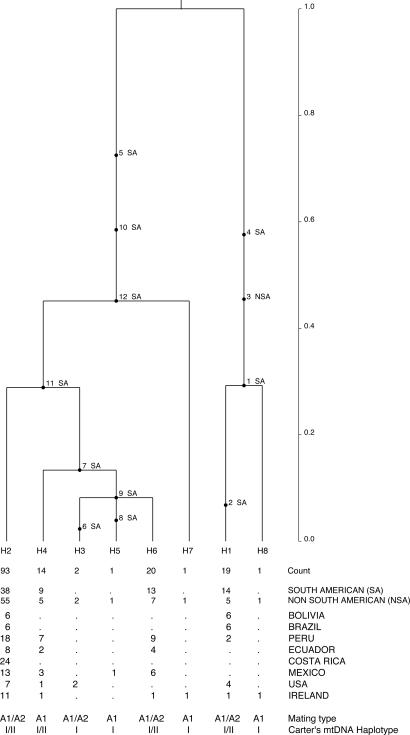
Two ancestral lineages were present in the mitochondrial (Fig. 1) and nuclear (Fig. 2) gene genealogies (32). The oldest lineage in the mitochondrial genealogy was haplotype H6 and included only isolates from the section Anarrhichomenum from Ecuador (Fig. 1). The other distinct lineage gave rise to the extant haplotypes of P. infestans from potato, the wild Solanum species examined, and tomato. Haplotype H6 was highly differentiated from all other isolates. Haplotypes of P. infestans from potato diverged more recently in the mitochondrial (T < 0.35) and nuclear (T < 0.5) genealogies compared with the time to the most recent common ancestor of all lineages (T = 1.0). The distribution of haplotypes among populations and the isolates associated with each haplotype are shown in SI Table 7.
Fig. 1.
The rooted coalescent-based gene genealogy showing the distribution of mutations for South American (SA: Peru, Ecuador, Bolivia, Brazil) and non-South American (NSA: Costa Rica, Mexico, U.S., Ireland) populations for the mitochondrial (P3+P4) loci of Phytophthora infestans generated using GENETREE (32). Time scale is in coalescent units of effective population size. The direction of divergence is from the top (past–oldest) to the bottom (present–youngest). The numbers below the tree from top to bottom designate each distinct haplotype and its count (i.e., the number of occurrences of the haplotype in the sample), the count of each haplotype in each population, the mating type of the isolates, and the mtDNA haplotype (33). Maximum likelihood estimates of the tree with the highest root probability, standard deviation (SD) and Watterson's theta (55) were as follows: Likelihood = 3.5848 ×10−22, SD = 4.3213 ×10−19, theta = 5.0. Estimates were based on 10 million coalescent simulations and five independent runs to ensure convergence. Simulations were performed assuming constant population size.
Fig. 2.
The rooted coalescent-based gene genealogy showing the distribution of mutations for South American (SA: Peru, Ecuador, Bolivia, Brazil) and non-South American (NSA: Costa Rica, Mexico, U.S., Ireland) populations for the IRRAS locus of Phytophthora infestans generated using GENETREE (32). Time scale is in coalescent units of effective population size. The direction of divergence is from the top (past–oldest) to the bottom (present–youngest). The numbers below the tree from top to bottom designate each distinct haplotype and its count (i.e., the number of occurrences of the haplotype in the sample), the count of each haplotype in each population, the mating type of the isolates, and the mtDNA haplotype (33). Maximum likelihood estimates of the tree with the highest root probability, standard deviation (SD) and Watterson's theta (55) were as follows: Likelihood = 3.5320 × 10−23, SD = 4.8689 × 10−20, theta = 2.0. Estimates were based on 10 million coalescent simulations and five independent runs to ensure convergence. Simulations were performed assuming constant population size.
In the mitochondrial gene genealogy (Fig. 1), mutations 23 and 16, found in P4 and P3, respectively, resulted in the EcoRI restriction sites identified to differentiate between type I and II lineages (33, 34, 35). These mutations separate the type II lineage from the common ancestral lineage containing both type I and II at T = 0.35 (Fig. 1). Haplotype H6 contains isolates of P. andina and is also type I and ancestral to type II. Within the type II lineage, haplotype IIb (H7) was the first to diverge from the type II common ancestor that includes haplotype IIa (H1 and H9). The two most frequently sampled haplotypes, H1 and H2, which correspond to types II and I, respectively, contain isolates of both A1 and A2 mating types (Fig. 1). The type I haplotype (H2) was globally distributed, with the exception of the Brazilian and Bolivian populations. The type II haplotype (H1) was also widely distributed, although it was not found in the Toluca Valley and Costa Rican populations (SI Table 7). Haplotype H7 (type IIb) includes the US6, US11, US12, and US13 genotypes, and haplotype H8 includes the US-14 and US-17 genotypes (SI Table 7). These genotypes previously had been considered either tomato specific isolates or sexual recombinants (36).
The same mtDNA haplotypes were associated with different nuclear backgrounds (Fig. 2), as has been observed by others using different molecular markers (23, 24). The two most common nuclear haplotypes (H1 and H2) of each ancient lineage contained isolates of both mtDNA lineages (types I and II) and of both mating types (A1 and A2). Two other common nuclear haplotypes (H4 and H6) also contained isolates from the two mtDNA lineages. Haplotype H1 contained primarily isolates of the type II mitochondrial lineage and A2 mating type. The Peruvian isolates PCZ 007 and PCZ 050 were the only isolates of the H1 haplotype that were of the type I lineage and A1 mating type. In general, the type I mtDNA lineage showed more nuclear diversity than the type II mtDNA lineage. All eight nuclear haplotypes contained isolates of the type I mtDNA lineage, but only four of the nuclear haplotypes contained isolates of the type II mtDNA lineage (Fig. 2).
Several unique haplotypes were found in the Mexican population (Toluca Valley), but these haplotypes were descendents from only one of the ancient lineages in both mitochondrial (Fig. 1) and nuclear (Fig. 2) gene genealogies, whereas Peruvian haplotypes descended from both ancestral lineages. In the mitochondrial genealogy one isolate from Peru (PHU006) represented a unique haplotype (H9) and an Ecuadorian isolate (3252) was inferred as recombinant haplotype (H10) (SI Table 7). The U.S. and Irish haplotypes were also derived from both ancestral lineages and unique haplotypes were observed for these populations (Fig. 1). The rooted nuclear gene genealogy showed a predominantly SA origin of mutations throughout its entire coalescent history (Fig. 2). The rooted mitochondrial genealogy also indicated a SA origin for mutations close to the common ancestor of P. infestans and P. andina (Fig. 1). In the mitochondrial genealogy, the older mutations originated in SA populations, and divergence of the two major haplotype lineages as well as the lineage from the section Anarrhichomenum, occurred in SA, suggesting that these populations are older than NSA populations.
Discussion
Geographical Origin of P. infestans.
Population genetic theory predicts that ancestral populations will have increased polymorphisms compared with more recent, nonancestral populations (37). Our data do not support the Mexican center of origin of P. infestans (4, 12). First, the three divergent ancestral lineages were found in the South American Andes. If we consider only the two lineages associated mainly with potatoes, both were found in the Andes and only one in Toluca Mexico. Alternatively, it is possible that the ancestral lineages originated in the Toluca Valley but the second lineage became very low in frequency or disappeared via chance (drift) or selection from there. Given that the ancestral lineage associated with potatoes and not found in the Toluca has a high frequency among populations of P. infestans in many different countries around the world (23), it is unlikely that this lineage would have disappeared by chance from the Toluca. Selection has been proposed to explain the presence of only one mtDNA haplotype (type Ia) of P. infestans in Toluca Mexico (18). One possible mechanism might be that domestic potatoes provide a different environment and therefore selected different mtDNA haplotypes than wild Solanum species (23, 24). However, so far the same mtDNA haplotype (type Ia) has been found associated with both wild and cultivated potatoes in Toluca (24). No indication of selection in either mitochondrial or nuclear loci for the Mexican population or the pooled sample was found (SI Table 2). The neutrality tests were consistent with a model of neutral evolution. However, it is important to note that nonsignificant results do not completely rule out the action of natural selection (29). It is more likely that the ancestral lineages have an Andean origin. Our data support a single genetic origin or founder effect for the Toluca population.
Extant haplotypes found in the Andes were derived from both ancient lineages, whereas haplotypes found in the Toluca Valley of central Mexico, even rare haplotypes, were always derived from only one of the ancient lineages for the nuclear (Fig. 2) and mitochondrial (Fig. 1) loci. Only one maternal lineage gave rise to the haplotypes of the mtDNA lineages found in Toluca Valley and supports other reports of the monomorphic condition for the mitochondrial haplotypes for the Toluca Valley population (23, 24).
The pattern of genetic variability in the Toluca Mexico population for the mitochondrial loci is consistent with a strong founder effect not seen in other populations (SI Table 3). In addition, Hudson's tests for population subdivision (SI Tables 4 and 5) showed that Toluca populations are genetically differentiated from Peruvian and Ecuadorian populations for the mitochondrial loci, but not the nuclear locus. Levels of differentiation estimated from mitochondrial and autosomal nuclear loci are expected to differ at equilibrium because of their effective population size differences. The mitochondrial locus with a lower effective size often has its diversity more strongly affected by historical events such as founder effects or bottlenecks than do autosomal nuclear genes (37, 38).
Nucleotide diversity in South American populations was higher for both mitochondrial and nuclear loci compared with the Toluca Mexico population (SI Table 3). Summary statistics, particularly diversity estimates (θw and π) are inflated when the sample includes deeply divergent lineages, as is evident for the U.S. and Irish populations (SI Table 3). Sampling bias or population admixture and not ancestry might be a possible explanation for the high diversity observed in South American populations compared with the Toluca Valley population. Sampling bias is unlikely to explain the results because each isolate from the Toluca Valley is a unique genotype (12, 24) and the isolates used in this study included different allozymes genotypes and mating type (SI Table 8). In addition, a similar sampling scheme was applied to each population. If isolates of P. infestans immigrated from differentiated populations into South America, rather than the reverse scenario, a potential bias due to population admixture could occur.
Nuclear and mitochondrial data showed evidence of gene flow between South American and non South American populations in both directions. However, both loci indicated different patterns in the direction of gene flow. Data from the nuclear IRRAS locus suggest that gene flow was very high in the past from South America to non South America (Fig. 2 and SI Table 6), whereas on a more recent time scale, the mitochondrial loci data suggest that migration from South America to non South America is equilibrating (Fig. 1 and SI Table 6). Our interpretation is that gene flow was originally from South America and more recently equilibration has occurred between the regions. Although evidence suggests that Mexico is the source of recent migrations of P. infestans into Europe and to other areas of the world (4, 39, 40), earlier migrations of P. infestans are more likely from Peru, because potato, tomato, and other Solanaceous crops originated there and were used and spread by ancient cultures (7). Modern populations of P. infestans in other countries resemble those in South America (13, 41)
The lineage associated with the section Anarrhichomenum was found only in the South American Andes (Ecuador). This lineage fits the morphological description for P. infestans (15), but recent molecular analysis indicates that these isolates are a new species called P. andina (42). This shared morphology also supports the Andean Highlands of South America as ancient center of diversity of P. infestans. Recently, other lineages morphologically similar to P. infestans have been associated with wild and cultivated Solanaceae in Ecuador (15) and Peru (43), suggesting that the Andes are a “hot spot” for diversification in the genus Phytophthora.
Source of Origin of the Isolates of P. infestans That Caused Early Epidemics.
The second question of interest was the source of origin of the isolates of P. infestans that caused the potato famine. The geographic origin of isolates in Ireland and the U.S. can be estimated using as a criterion the number of shared haplotypes between the areas considered nonancestral and the putative ancestral areas (in our case, central Mexico or South America) (40). Populations of P. infestans in the South American Andes are derived from the two ancestral lineages found in both mitochondrial (Fig. 1) and nuclear (Fig. 2) genealogies. Populations of P. infestans in the U.S. and Ireland also contained members derived from both ancestral lineages and were more similar to SA populations than to the central Mexico population, indicating that these populations shared a common ancestor with the South American populations as has been previously suggested by others based on race composition, allozyme markers and nuclear DNA content (13). Populations from the U.S. and Ireland were not genetically different from the Peruvian population for both mitochondrial and nuclear loci. The most common haplotypes corresponding to each ancient lineage in each genealogy were always found in SA populations within the present day haplotypes. The U.S. and Irish populations were not genetically differentiated from the Peruvian populations, strongly suggesting that Peru was the source of origin of the population that spread to these two continents. These data fit the historical records as well, because it is clear that Peruvian potatoes were being shipped to the U.S. and Europe during the famine era, whereas Mexico had no domesticated potato production at the time (1, 6, 9).
Evolution of mtDNA Haplotypes of P. infestans.
The evolution of mitochondrial diversity in P. infestans has been the subject of much research (23, 24, 27, 34). Four mitochondrial haplotypes have been described in P. infestans: Ia, Ib, IIa, and IIb (33, 34). Gavino and Fry (23) proposed that haplotype Ib was ancestral to the other known haplotypes, although they did not completely rule out the hypothesis that haplotype Ia might be the ancestral haplotype with limited sequence data in their study. Type II haplotypes (IIa and IIb) were considered derived from type I and closely related (23, 34, 35). Recently, Flier et al. (24) suggested that the Ia rather than Ib represents the ancestral type of mtDNA haplotype in P. infestans. Our mitochondrial genealogy shows that type I and II lineages split from a common ancestor, and thus neither lineage can be considered ancestral to the other (Fig. 1). However, the type I haplotypes are more closely related to the ancestral type because fewer mutations occurred in this lineage than the type II after splitting from the common ancestor (Fig. 1). In addition, type I haplotypes share the same nucleotide states (C, sites 17 and 24, SI Table 3) with the closely related South American species Phytophthora andina, associated with the section Anarrhichomenum. Nucleotide T present in type II haplotypes is considered derived and is not present in the common ancestor. The same nucleotide state found in the P4 region of type I haplotypes of P. infestans was recently found in three other species closely related to P. infestans, Phytophthora mirabilis, Phytophthora ipomoeae, and Phytophthora phaseoli (42), suggesting the ancestral condition of the nucleotide state present in type I haplotypes and a potential South American origin of this clade. We sequenced the whole mitochondrial genomes of the other three haplotypes of P. infestans and clarified their evolutionary relationships (27).
Conclusion
Analysis of the mitochondrial and nuclear loci of P. infestans strongly supports a South American center of origin of this pathogen. The evolutionary history of P. infestans is proposed as follows: an ancestral population of Phytophthora diverged into different lineages in the South American Andes in association with wild Solanum species. Two of the divergent lineages gave rise to the extant haplotypes of P. infestans capable of infecting potato, tomato, and some wild Solanum species. Other lineages evolved into distinct species, closely related to P. infestans and morphologically identical to it (section Anarrhichomenum isolates). Host specificity became the driving force for maintaining the divergent lineages in Ecuador and Peru. An Andean source of inoculum initiated epidemics first in the U.S. and then Ireland that led to the famine. Our data provide strong evidence for an “out of South America” origin of this destructive plant pathogen and clearly demonstrate that the oldest mutations in the ancestral strains occurred in South America.
Materials and Methods
Sampling and DNA Extraction.
Nonrandom sampling was conducted to obtain isolates (n = 94) with a range of genotypic and phenotypic diversity from eight countries associated with the origin and migration hypotheses of this plant pathogen (SI Table 8). All isolates were from potato (Solanum tuberosum), except isolates from Mexico (Toluca Valley), Ecuador, and Costa Rica. Isolates of P. infestans sensu lato (now P. andina) from Ecuador were from the Solanum section Anarrhichomenum (15). Total genomic DNA was extracted from mycelium using a standard cetyltrimethylammonium bromide (CTAB) protocol (25). DNA was diluted 1:10 or 1:100 (3–10 ng/μl) for further use.
DNA Amplification and Sequencing.
Two regions of the mitochondrial genome, P3 (rp114, rp15, tRNAs) and P4 (cox 1) and two single-copy nuclear genes, Ras and β-tubulin, were amplified by PCR (33, 44) (SI Table 9). For the Ras gene, two regions were amplified independently, a 223-bp intron (IntronRas) located in the 5′ untranslated region of the gene and a 600-bp portion (Ras) covering part of exons 3–6 and introns 3–5 (44). Two independent PCRs were done with appropriate controls. PCR products were pooled, purified (QIAquick PCR Purification kits, Qiagen, Valencia, CA), and sequenced directly in the forward and reverse direction. Sequencing reactions were prepared by using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit and analyzed on an ABI PRISM 377 automated sequencer (Applied Biosystem, Foster City, CA).
Sequence Analysis.
Sequences were aligned manually and edited with BioEdit (45). Multiple sequence alignment was also performed in Clustal X (46). All polymorphisms were rechecked from the chromatograms. Isolates of P. infestans are diploid and show disomic inheritance (47), such that individuals can be either homozygous or heterozygous at some loci. Sites showing the presence of two coincident peaks in the forward and reverse sequence chromatograms were observed for Ras and β-tubulin genes indicating heterozygous positions, as a result of the coamplification and simultaneous sequencing of two complementary loci (SI Fig. 3) (48). The two haplotypes within the heterozygote were inferred by using the “haplotype subtraction” method (40). Heterozygous sites were confirmed by restriction digest (SI Fig. 3).
Statistical Analysis.
Sequence data were evaluated for assumptions relevant to estimating evolutionary histories including: no selection, no recombination, random mating within a single population, and random sampling (49). Data analysis was performed using the Java program SNAP Workbench (50) of Carbone et al. (51) (SI Fig. 4). Neutrality and population subdivision tests, migration analysis and genealogical analyses were done (52–60). Complete details of each statistical analysis are provided in SI Text.
Supplementary Material
Acknowledgments
We thank the many colleagues listed in SI Table 8 for providing isolates for this research and Drs. Trudy Mackay, Jeff Thorne, Marc Cubeta, and Julia Hu for useful comments. This work was supported by USDA National Research Initiative Competitive Grants Program Grant 2001-0922 and The Fulbright Scholars LASPAU Academic and Professional Programs for the Americas graduate assistantship (to L.G.-A.).
Abbreviations
- SA
South American
- NSA
non-SA.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EF366671–366732 (P3), EF366733–366794 (P4), EF366795–EF366950 (IR), and EF366951–367106 (RAS)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0611479104/DC1.
References
- 1.Ristaino JB. Microbes Infection. 2002;4:1369–1377. doi: 10.1016/s1286-4579(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 2.Berkeley MJ. J Hort Soc London. 1846;1:9–34. [Google Scholar]
- 3.de Bary A. J Roy Agr Soc. 1876;12:239–268. [Google Scholar]
- 4.Goodwin SB, Cohen BA, Fry WE. Proc Natl Acad Sci USA. 1994;91:11519–11595. doi: 10.1073/pnas.91.24.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrivon D. Plant Pathol. 1996;45:1027–1035. [Google Scholar]
- 6.Abad ZG, Abad JA. Plant Dis. 1997;81:682–688. doi: 10.1094/PDIS.1997.81.6.682. [DOI] [PubMed] [Google Scholar]
- 7.Spooner DM, Mclean K, Ramsay G, Waugh R, Bryan GJ. Proc Natl Acad Sci USA. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volkov RA, Komarova NY, Panchuck I, Hemleben V. Mol Phylogenet Evol. 2003;29:187–202. doi: 10.1016/s1055-7903(03)00092-7. [DOI] [PubMed] [Google Scholar]
- 9.Bourke PM. Nature. 1964;203:805–808. [Google Scholar]
- 10.Reddick D. Chronica Botanica. 1939;5:410–412. [Google Scholar]
- 11.Niederhauser JS. In: Phytophthora. Lucas JA, Shattock RC, Shaw DS, Cooke LR, editors. Cambridge, UK: Cambridge Univ Press; 1991. pp. 25–45. [Google Scholar]
- 12.Grüdnwald NJ, Flier WG. Annu Rev Phytopathol. 2005;43:171–190. doi: 10.1146/annurev.phyto.43.040204.135906. [DOI] [PubMed] [Google Scholar]
- 13.Tooley PW, Therrien CD, Ritch DL. Phytopathology. 1989;79:478–481. [Google Scholar]
- 14.Perez WG, Gamboa JS, Falcon YV, Coca M, Raymundo RM, Nelson RJ. Phytopathology. 2001;91:956–965. doi: 10.1094/PHYTO.2001.91.10.956. [DOI] [PubMed] [Google Scholar]
- 15.Adler NE, Erselius LJ, Chacon MG, Flier WG, Ordoñez ME, Kroon LPNM, Forbes GA. Phytopathology. 2004;94:154–162. doi: 10.1094/PHYTO.2004.94.2.154. [DOI] [PubMed] [Google Scholar]
- 16.Oliva RF, Erselius LJ, Adler NE, Forbes GA. Plant Pathol. 2002;51:710–719. [Google Scholar]
- 17.Goodwin SB, Drenth A, Fry WE. Curr Genet. 1992;22:107–115. doi: 10.1007/BF00351469. [DOI] [PubMed] [Google Scholar]
- 18.Drenth A, Tas ICQ, Govers F. Eur J Plant Pathol. 1993;100:97–107. [Google Scholar]
- 19.Mayton H, Smart CD, Moravec BC, Mizubuti ESG, Muldoon AE, Fry WE. Plant Dis. 2000;84:1190–1196. doi: 10.1094/PDIS.2000.84.11.1190. [DOI] [PubMed] [Google Scholar]
- 20.Sujkowski LS, Goodwin SB, Dyer AT, Fry WE. Phytopathology. 1994;84:201–207. [Google Scholar]
- 21.Bruberg MB, Hannukkala A, Hermansen A. Mycol Res. 1999;103:1609–1615. [Google Scholar]
- 22.Zwankhuizen MJ, Govers F, Zadoks JC. Eur J Plant Pathol. 2000;106:667–680. [Google Scholar]
- 23.Gavino PD, Fry WE. Mycologia. 2002;94:781–793. [PubMed] [Google Scholar]
- 24.Flier WG, Gründwald NK, Kroon LPNM, Sturbaum AK, van den Bosch TBM, Garay-Serrano E, Lozoya-Saldana H, Fry WE, Turkensteen LJ. Phytopathology. 2003;93:382–390. doi: 10.1094/PHYTO.2003.93.4.382. [DOI] [PubMed] [Google Scholar]
- 25.Ristaino JB, Groves CT, Parra GR. Nature. 2001;411:695–697. doi: 10.1038/35079606. [DOI] [PubMed] [Google Scholar]
- 26.May K, Ristaino JB. Mycol Res. 2004;108:471–479. doi: 10.1017/s0953756204009876. [DOI] [PubMed] [Google Scholar]
- 27.Avila-Adame C, Gómez-Alpizar L, Zismann V, Jones KM, Buell CR, Ristaino JB. Curr Gen. 2006;49:39–46. doi: 10.1007/s00294-005-0016-3. [DOI] [PubMed] [Google Scholar]
- 28.Hudson RR, Boos DD, Kaplan NL. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 29.Simonsen KL, Churchill GA, Aquadro CF. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson RR, Slatkin M, Maddison WP. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hey J, Nielsen R. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths RC, Tavaré S. Stat Sci. 1994;9:307–309. [Google Scholar]
- 33.Griffith GW, Shaw DS. Appl Environ Microbiol. 1998;64:4007–4014. doi: 10.1128/aem.64.10.4007-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter DA, Archer SA, Buck KW, Shaw DS, Shattock RC. Mycol Res. 1990;94:1123–1128. [Google Scholar]
- 35.Carter DA, Archer SA, Buck KW, Shaw DS, Shattock RC. In: Phytophthora. Lucas JA, Shattock RC, Shaw DS, Cooke LR, editors. Cambridge, UK: Cambridge Univ Press; 1990. pp. 272–294. [Google Scholar]
- 36.Goodwin SB, Smart CD, Sandrock RW, Deahl KL, Punja ZK, Fry WE. Phytopathology. 1998;88:939–949. doi: 10.1094/PHYTO.1998.88.9.939. [DOI] [PubMed] [Google Scholar]
- 37.Dean MD, Ballard JWO. Mol Phylogen Evol. 2004;32:998–1009. doi: 10.1016/j.ympev.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Arnaud-Haond S, Bonhomme F, Blanc F. J Evol Biol. 2003;16:388–398. doi: 10.1046/j.1420-9101.2003.00549.x. [DOI] [PubMed] [Google Scholar]
- 39.Fry WE, Goodwin SB, Matuszak JM, Spielman LJ, Milgroon MG, Drenth A. Annu Rev Phytopathol. 1992;30:107–129. [Google Scholar]
- 40.Goodwin SB, Cohen BA, Deahl KL, Fry WE. Phytopathology. 1998;84:553–558. [Google Scholar]
- 41.Ghimire SR, Hyde KD, Hodgkiss IJ, Shaw DS, Liew ECY. Phytopathology. 2003;93:236–243. doi: 10.1094/PHYTO.2003.93.2.236. [DOI] [PubMed] [Google Scholar]
- 42.Kroon LPNM, Bakker FT, van den Bosch GBM, Bonants PJM, Flier WG. Fungal Gen Biol. 2004;41:766–782. doi: 10.1016/j.fgb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Garry G, Forbes GA, Salas A, Santa Cruz M, Perez WG, Nelson RJ. Plant Pathol. 2005;54:740–748. [Google Scholar]
- 44.Chen Y, Roxby R. Gene. 1996;181:89–94. doi: 10.1016/s0378-1119(96)00469-6. [DOI] [PubMed] [Google Scholar]
- 45.Hall TA. Nucl Ac Symp. 1999;41:95–98. [Google Scholar]
- 46.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tooley PW, Therrien CD. In: Phytophthora. Lucas JA, Shattock RC, Shaw DS, Cooke LR, editors. Cambridge, UK: Cambridge Univ Press; 1991. pp. 204–217. [Google Scholar]
- 48.Clark AG. Mol Biol Evol. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- 49.Emerson BC, Paradis E, Thébaud C. Trends Ecol Evol. 2001;16:707–716. [Google Scholar]
- 50.Price EW, Carbone I. Bioinfomatics. 2005;21:402–404. doi: 10.1093/bioinformatics/bti003. [DOI] [PubMed] [Google Scholar]
- 51.Carbone I, Liu YC, Hillman BI, Milgroom MG. Genetics. 2004;166:1611–1629. doi: 10.1534/genetics.166.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aylor DL, Price EW, Carbone I. Bioinformatics. 2006;22:1399–1401. doi: 10.1093/bioinformatics/btl136. [DOI] [PubMed] [Google Scholar]
- 53.Myers SR, Griffiths RC. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozas JS, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 55.Watterson GA. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 56.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu Y-X, Li WH. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Y-X. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Templeton AR. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.