Abstract
The spliceosome is a dynamic macromolecular machine that catalyzes the excision of introns from pre-mRNA. The megadalton-sized spliceosome is composed of four small nuclear RNPs and additional pre-mRNA splicing factors. The formation of an active spliceosome involves a series of regulated steps that requires the assembly and disassembly of large multiprotein/RNA complexes. The dynamic nature of the pre-mRNA splicing reaction has hampered progress in analyzing the structure of spliceosomal complexes. We have used cryo-electron microscopy to produce a 29-Å density map of a stable 37S spliceosomal complex from the genetically tractable fission yeast, Schizosaccharomyces pombe. Containing the U2, U5, and U6 snRNAs, pre-mRNA splicing intermediates, U2 and U5 snRNP proteins, the Nineteen Complex (NTC), and second-step splicing factors, this complex closely resembles in vitro purified mammalian C complex. The density map reveals an asymmetric particle, ≈30 × 20 × 18 nm in size, which is composed of distinct domains that contact each other at the center of the complex.
Keywords: Cdc5, cryo-EM, pre-mRNA splicing, Schizosaccharomyces pombe
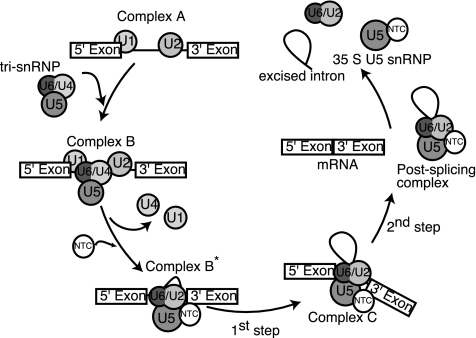
One model of pre-mRNA splicing posits that the spliceosome is assembled in a step-wise fashion (1, 2). Spliceosome assembly begins with the recognition of the 5′ splice site and branch-point sequences of the pre-mRNA by the U1 small nucleotide RNP (snRNP) and the U2 snRNP respectively (Complex A, Fig. 1). After binding of the U4/U6.U5 tri-snRNP, the U4/U6 snRNA duplex is replaced by a U2/U6 snRNA duplex (Complex B, Fig. 1). Furthermore, the U1 snRNA base pairing at the 5′ splice site is disrupted and exchanged for base pairing between the 5′ splice site and the U6 snRNA. The subsequent addition of another complex, the Nineteen Complex (NTC), and the release of the U1 and U4 snRNPs marks the transition from an inactive to an active spliceosome composed of the NTC and the U5 and U2/U6 snRNPs (Complex B* and C, Fig. 1). 5′-Splice site cleavage and lariat formation, followed by 3′-splice site cleavage and exon ligation, occur within the activated spliceosome.
Fig. 1.
Schematic model of pre-mRNA splicing.
Structural information about the organization of spliceosomal complexes is still sparse, mainly because of their large sizes and dynamic natures. Difficulties in isolating sufficient quantities of pure stable spliceosomal complexes have so far limited x-ray crystallographic studies to a few isolated spliceosome components, with much focus centered on the core snRNP Sm and Lsm proteins (3–17). A promising structural approach for obtaining information about spliceosome organization is single-particle cryo-EM, a powerful technique that is ideal for determining the structures of large dynamic complexes at protein concentrations too low for crystallization trials. EM structures of four distinct mammalian spliceosomal complexes have been presented, providing snapshots of the spliceosome at distinct stages of the splicing reaction (18–21). Here we show that the Schizosaccharomyces pombe U5.U2/U6 complex represents a spliceosomal particle involved in the late stages of pre-mRNA splicing and describe the 3D structure of this multi-snRNP particle by cryo-EM. The determination of this structure represents another step toward understanding the organization of a native multi-snRNP spliceosomal complex involved in the late stages of splicing and demonstrates that S. pombe provides a genetically tractable system in which to purify and structurally characterize complexes involved in pre-mRNA splicing.
Results and Discussion
Purification and Characterization of the S. pombe U5.U2/U6 snRNP Complex.
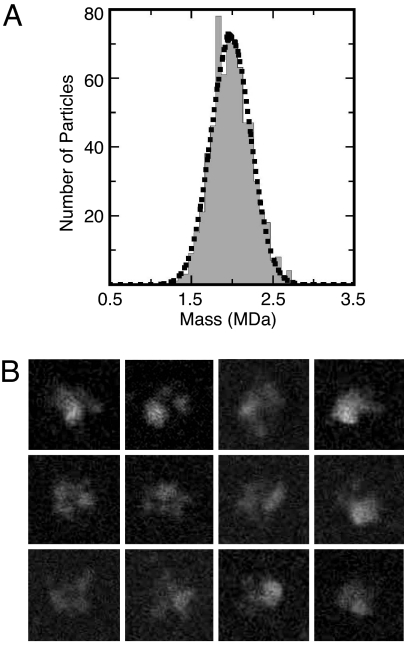
Using a tandem affinity purification strategy (22, 23) targeting S. pombe Cdc5, we isolated a stable complex of splicing factors that includes the U2, U5, and U6 snRNAs (24). The purified U5.U2/U6 particle is homogeneous, as judged by negative/stain EM (data not shown) and sucrose gradient centrifugation (24). To obtain an accurate molecular mass of the S. pombe U5.U2/U6 spliceosomal complex, we analyzed freeze-dried unstained particles by scanning transmission EM (STEM) analysis. The 663 selected particles segregated into one Gaussian-shaped curve, indicating a complex with a mass of 2.0 MDa (±194 kDa) (Fig. 2A). The unstained particles in general had a tri-lobe appearance and looked very similar to U5.U2/U6 particles seen in vitrified ice [Fig. 2B and supporting information (SI) Fig. 6A]. Summing the mass of the proteins identified in the U5.U2/U6 complex by MS (24) yields a molecular mass of ≈3.0 MDa; however, the sum of spliceosome components found just in the U2/U6 and U5 snRNPs and the NTC equals ≈2.2 MDa, a value much closer to the STEM-determined mass of the U5.U2/U6 particles. This suggests that the S. pombe U5.U2/U6 complex, similar to the mammalian C complex (18), is likely composed of a core group of tightly bound proteins that interact with a set of less tightly associated pre-mRNA splicing factors.
Fig. 2.
Mass measurement of the S. pombe U5.U2/U6 complex by STEM. (A) Histogram showing mass distribution of S. pombe U5.U2/U6 complexes. (B) Gallery of representative unstained S. pombe U5.U2/U6 complex included in the STEM analysis. Although the images had relatively low contrast, many particles displayed a characteristic tri-lobe shape that yielded masses in the range of the histogram peak. Side length of individual images is 53 nm.
A majority of S. pombe U2, U5, and U6 snRNPs sediment in a stable U5.U2/U6 spliceosomal complex (24–26). The difficulty in establishing a robust in vitro splicing system using S. pombe extracts may be due to the stability of this interaction and has thwarted efforts to characterize the biochemical activity of the tandem affinity-purified U5.U2/U6 complex. However, both the snRNA and protein compositions of this complex (24) reveal striking similarities to the human 45S B* spliceosomal complex (27) and the human C complex (28). Like these spliceosomal particles, the S. pombe U5.U2/U6 complex contains the U2, U5, and U6 snRNAs and U2 and U5 snRNP components as well as second-step splicing factors and the NTC (reviewed in ref. 29). These similarities suggested that the U5.U2/U6 complex might also contain pre-mRNA splicing intermediates.
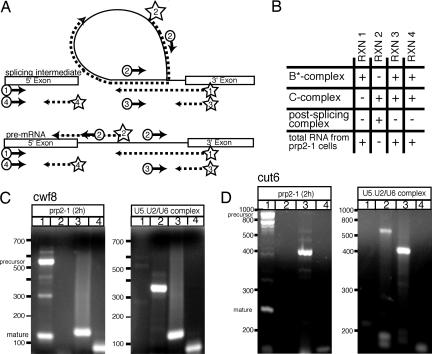
To test this possibility, we used RT-PCR to probe RNA extracted from U5.U2/U6 particles for the presence of pre-mRNA, mRNA, lariat, 5′ exon, and 3′ exon. Reverse transcriptase can read through the 2′–5′ linkage in templates such as those found in lariat structures of the pre-mRNA splicing reaction (30) (Fig. 3A). RNA extracted from arrested S. pombe prp2–1 cells was used as a control for detecting both pre- and mature mRNA. S. pombe prp2 encodes the large subunit of the splicing factor U2AF (31). Fig. 3B summarizes the amplification products expected from different spliceosomal complexes and from total RNA extracted from arrested prp2–1 cells. Because we expected the U5.U2/U6 complex to contain a mixture of different transcripts, we probed for cwf8 and cut6, because these contain introns of lengths easily detected by RT-PCR (385 and 621 nucleotides, respectively). RNA extracted from the U5.U2/U6 complex contained both cwf8 and cut6 lariat intermediates (Fig. 3 C and D, lanes 2) but did not contain pre-mRNA and spliced RNA (Fig. 3 C and D, lanes 1). Products were also obtained in reaction 3, designed to detect the connection of the intron with the 3′ exon (Fig. 3 C and D, lanes 3), indicating that some particles contain lariats attached to the 3′ exons. Last, products corresponding to the 5′ exons of cut6 and cwf8 (85 and 120 nucleotides, respectively) were also detected (Fig. 3 C and D, lanes 4). PCR products were sequenced to confirm their identity, and no products were obtained in the absence of reverse transcriptase (data not shown). From these results, we conclude that at least some U5.U2/U6 particles contain transcripts that have undergone the first but not the second step of pre-mRNA splicing, indicating that the S. pombe U5.U2/U6 complex closely resembles human C complex (Figs. 1 and 3B). These results do not rule out the possibility that other U5.U2/U6 complexes are free of pre-mRNA splicing intermediates or contain free lariat, representing a population of postsplicing particles formed after the second step of pre-mRNA splicing (Fig. 1). Nevertheless, the presence of RNA splicing intermediates within the U5.U2/U6 particles demonstrates that this complex represents a stable multi-snRNP complex in the late stages of the splicing reaction. Further, the preponderance of this stable, C like complex at steady state suggests that the slowest, or perhaps most regulated, step of pre-mRNA splicing in vivo is the second step of the splicing reaction, at least in fission yeast.
Fig. 3.
Identifying RNAs present in the S. pombe U5.U2/U6 complex. (A) RT-PCR was used to characterize RNA extracted from U5.U2/U6 particles. Exons are represented by boxes; introns are represented by solid lines. Positions of primers used in reactions 1–4 are shown in the diagram. Primers used for first-strand synthesis are marked by a star. These primers and/or circled primers were used for subsequent amplifications. The arrows depict the direction of DNA synthesis. The dotted lines represent first-strand synthesis. (B) Summary of PCR amplification products expected in each reaction from different spliceosomal complexes and from RNA extracted from arrested prp2–1 cells. Reaction 1 detects pre- or mature mRNA, reaction 2 detects lariat RNA, reaction 3 detects the connection of the intron with its 3′ exon, and reaction 4 detects the 5′ exon. “+” denotes product expected and “-” denotes no product expected. (C and D) RT-PCR products from reactions probing for the cwf8 (C) and cut6 (D) transcripts in RNA extracted from arrested prp2–1 cells or RNA extracted from U5.U2/U6 particles. Reactions are the same as in A and B. PCR products separated on 3% Nusieve gels and detected with ethidium-bromide.
A Reliable Low-Resolution Model of the U5.U2/U6 Complex Obtained by Random Conical Tilt Reconstruction of Cryo-Negatively Stained Samples.
Even if a sample is structurally homogenous, it can be difficult to obtain a reliable 3D reconstruction from images of vitrified particles with the angular reconstitution approach (32). This is particularly true when particles lack any inherent symmetry, such as the U5.U2/U6 complex. In these cases, it is safer to use the random conical tilt approach (33) to calculate a 3D reconstruction, in which the orientation parameters of the imaged particles are unambiguously defined. However, random conical tilt reconstructions calculated from specimens prepared by conventional negative staining often exhibit severe structural deformations because of flattening upon drying and incomplete stain embedding (34, 35). To minimize these preparation artifacts, we prepared the U5.U2/U6 complex by cryo-negative staining (34, 36). Image pairs of the same specimen areas were recorded at tilt angles of 55° and 0°. The images of the untilted specimens were used to classify the particles into 20 classes (SI Fig. 6). Many of the 20 classes showed particles with very similar structural features that clearly resembled each other. The images from the tilted specimens corresponding to six particular classes were chosen to calculate separate 3D reconstructions (Fig. 4A and SI Fig. 6, marked with *). Structures calculated from particles found in four of the six representative classes looked very similar and could easily be aligned (Fig. 4A Lower, first four structures). The differences between the four structures may be explained by the presence of some structural flexibility in the upper lobes of the complex, by the small number of particles used for each reconstruction, and/or by variations in the staining of the imaged particles. The additional two structures were obviously smaller than the other four structures and most likely represent the lower region of the larger structures (Fig. 4A Lower, last two structures). These classes (≈10% of the total particles) likely represent U5.U2/U6 complexes that are in different stages of disintegration and were excluded from further analysis. To further improve the reconstruction the tilted images corresponding to the four representative classes (Fig. 4A Upper, first four classes) and particles from additional classes that clearly resembled these averages (SI Fig. 6, marked with a “+”) were combined and used to calculate a 3D reconstruction by using FREALIGN (37).
Fig. 4.
Random conical tilt reconstruction of the S. pombe U5.U2/U6 complex in cryo-negative stain. (A) Six representative class averages of U5.U2/U6 particles selected from images of untilted specimens in cryo-negative stain (Upper) and the random conical tilt reconstructions calculated from the corresponding particles selected from the images of the tilted specimens (Lower). The number of particles in each projection average is shown in the lower right corner of each average. The last two classes result in 3D structures that are substantially smaller than the first four structures. Side length of individual panels is 54 nm. (Scale bar, 5 nm.) (B) 3D reconstruction of the S. pombe U5.U2/U6 complex filtered to 40-Å resolution combining particles from the first four classes in A and particles associated with additional class averages that clearly resemble these first four classes (see SI Fig. 7; merged classes are marked with a “+”). The complex is rotated about the vertical axis in 60° steps or about the horizontal axis by 90° or 180° steps (in reference to the top left structure), as indicated by arrows. (Scale bar, 5 nm.)
The 3D structure of the U5.U2/U6 complex in cryo-negative stain is presented in Fig. 4B. The structure, ≈23 × 18 × 15 nm in dimension, consists of three distinct lobes. It shares a striking similarity to the density map of the human C complex, as determined by the cryo-negative staining approach (18), providing direct evidence that spliceosomal complexes purified from different organisms are structurally conserved. Both structures are composed of three globular lobes of similar sizes that join together at the center of the complex. Small differences evident between the structures are most likely because of the different species the complexes originate from.
Cryo-EM and 3D Reconstruction.
Using cryo-EM, we collected images of U5.U2/U6 particles in vitrified ice adsorbed to a thin carbon layer. The individual U5.U2/U6 particles were clearly visible in the raw images (SI Fig. 7 A). The 30,671 selected particles were band-pass-filtered and subjected to multireference alignment, multivariate statistical analysis, and classification to obtain averages of the characteristic views of the molecule with improved signal-to-noise ratio (SI Fig. 7B Bottom). Using the program FREALIGN (37), the class averages were aligned directly to the low-resolution cryo-negative stain model (Fig. 4B), and the structure was refined until stable at a resolution of 40 Å. The averages, rather than the individual particle images, were initially used to ensure that propagation of any residual preparation artifacts found in the cryo-negative stain model were minimized in the final 3D reconstruction calculated from the artifact-free molecules embedded in vitrified ice. Five refinement cycles were done by using the class averages. From cycle six onward, the individual particle images were aligned to the model. The density map was refined and corrected for the contrast transfer function by using FREALIGN (37). The Fourier shell correlation (FSC) curve calculated from our final density map suggests a resolution of either 29 or 23 Å based on the FSC = 0.5 and FSC = 0.143, respectively (38) (SI Fig. 7C). The distribution of the Euler angles of the particles shows that the complex assumes randomly distributed orientations within the vitrified ice layer (SI Fig. 7D). Reprojections from the density map (SI Fig. 7B Middle) are very similar to the corresponding raw particle images (SI Fig. 7B Top) and class averages (SI Fig. 7B Bottom), demonstrating the consistency of the 3D reconstruction with the projection data. The refined density map was contoured so that the molecular mass of the complex is 2.0 MDa, matching the molecular mass determined by STEM analysis (Fig. 2A).
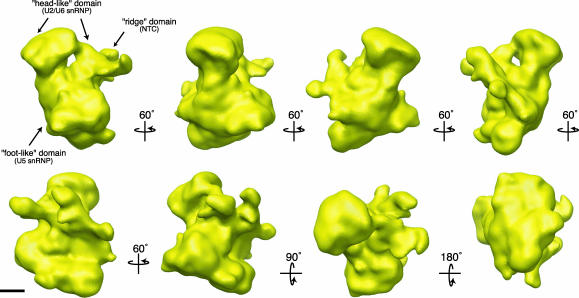
The S. pombe U5.U2/U6 complex, ≈30 × 20 × 18 nm in size, consists of three distinct lobes (Fig. 5). The top portion of the complex, which we term the “head-like” domain, is bilobal in shape with a diameter of ≈20 nm and a height of ≈8 nm. The head is connected on one side to the large bottom domain and, on the other side, to a ridge-like extension. These two connections are separated by a small cavity. The most massive part of the structure is the bottom “foot-like” domain that is generally triangular in shape and has dimensions of ≈18 × 18 × 22 nm. Running along the side of this large base is a ridge-like structure that contains a number of spiny protrusions. As with our cryo-negative stain model, this structure also shares a striking similarity to the density map of the human C complex as determined by cryo-negative staining (18).
Fig. 5.
Surface representation of the S. pombe U5.U2/U6 complex at 29 Å. Views of the U5.U2/U6 complex tilted stepwise about the vertical axis by 60° steps or about the horizontal axis by 90° or 180° steps (in reference to the top left structure), as indicated by arrows. The complex is composed of three lobes that likely represent the U5 and the U2/U6 snRNPs, and the NTC. The surface representation of the complex is contoured to a mass of 2.0 MDa. (Scale bar, 5 nm.)
Implications for the Arrangement of the Spliceosome.
Based on their relative size and available structural and biochemical data, we have tentatively assigned the three lobes as the U5 snRNP, the U2/U6 snRNP and the NTC (Fig. 5). The protein and snRNA composition of spliceosome complexes purified from human, Saccharomyces cerevisiae and S. pombe cells are highly conserved (reviewed in ref. 29). Therefore, in analogy to human spliceosomal complexes, we assigned the largest, “foot-like” lobe of the S. pombe U5.U2/U6 complex to the U5 snRNP (Fig. 5). This interpretation is consistent with the cryo-negative stain structure of the mammalian U5 snRNP (21), which has similar dimensions as the bottom base of our U5.U2/U6 complex and is also roughly triangular in shape. The U5 snRNP initially enters the splicing cycle as part of the tri-snRNP (Fig. 1). Concomitant with dissociation of U1 and U4, additional proteins, such as members of the NTC, are recruited to the mammalian spliceosome, and the U5 snRNP is reorganized into a larger particle (27). Many of these additional proteins are present in our U5.U2/U6 complex (24) and are reported to remain associated with the U5 snRNA even after the completion of the splicing reaction and the disassembly of the spliceosome (27).
In our density map, the foot-like domain makes extensive contact with the upper two head-like lobes that we interpret as the U2/U6 snRNP (Fig. 5). This interpretation is consistent with the proposed organization of the mammalian C complex (18). In addition, the structure of the mammalian tri-snRNP also contains a bilobal upper domain, composed of the U4/U6 snRNP, which interacts closely with U5 snRNP (21). This suggests that the overall position of U5 snRNP may stay globally stable as the U4/U6 snRNA duplex is replaced by a U2/U6 snRNA duplex (Fig. 1, transition from complex B to complex B*).
Making extensive contacts with both the upper “head-like” lobes and the “foot-like” base is a “ridge” domain that wraps around one side of the S. pombe U5.U2/U6 complex (Fig. 5). As suggested by Jurica et al. (18) for the mammalian C complex, this domain could represent the NTC. The NTC binds the spliceosome concomitant with U1 and U4 snRNP release and is essential for spliceosome activation and stabilization (Fig. 1) (39, 40). The location of this subcomplex between the upper and lower domains of the U5.U2/U6 complex would be advantageous for its important role in stabilizing the spliceosome during the major U6 snRNA binding transitions that must occur during the transition from an inactive to active spliceosome.
Our interpretation of the density map of the S. pombe U5.U2/U6 complex is consistent with many biochemical and structural details known about individual snRNP particles and provides a picture of the macromolecular arrangement of a splicing complex involved in the late stages of pre-mRNA splicing. The complexity and dynamic nature of the splicing reaction have made it difficult to isolate homogenous populations of spliceosomes suitable for structural analysis. We have shown that the unusually stable S. pombe U5.U2/U6 complex purified by targeting Cdc5 represents a spliceosomal particle involved in the late stages of pre-mRNA splicing that it is amenable to structural analysis. The 3D structure of this complex constitutes an important step toward understanding the detailed architecture of the spliceosome and demonstrates that S. pombe provides a powerful system for purifying and structurally characterizing spliceosomal complexes.
Materials and Methods
Purification of the U5.U2/U6 Complex.
The U5.U2/U6 complex was purified by using the tandem affinity purification strategy as described (24).
STEM.
STEM was carried out in the Brookhaven National Laboratory STEM facility with tobacco mosaic virus included as an internal control. For mass measurements, freeze-dried S. pombe U5.U2/U6 particles were prepared by the wet film technique.
The microscope operation and data acquisition are both computer controlled. The scattered electrons are collected in two annular detectors. The number of scattered electrons from a pixel is directly proportional to the mass thickness in that pixel. The digital data from the large angle detector are used for mass analysis. By summing over the pixels containing the particle of interest and subtracting the thin carbon substrate, the mass of that particle can be determined. Each micrograph consists of 512 × 512 pixels of 1 nm. Software programs developed at the STEM facility (PCMass) were used for these analyses.
RNA and RT-PCR.
Total RNA was prepared from cells by extraction with hot acidic phenol as described (41). First-strand synthesis was performed with the SuperScript First-strand Synthesis System (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. One hundred nanograms of RNA was used for each reaction. cwf8 and cut6 products were resolved on 3% and 2% (wt/vol) Nusieve agarose gels (BMA, Rockland, ME), respectively.
Specimen Preparation and EM.
Uranyl formate (0.7% wt/vol) was used for cryo-negative staining as described (34, 36). To prepare samples in vitrified ice, a thin layer of carbon film supported by a holey carbon grid (Quantifoil Micro Tools, Jena, Germany) was glow-discharged and used to adsorb purified U5.U2/U6 particles. Grids were blotted and frozen in liquid ethane by using a Vitrobot (FEI, Hillsboro, OR).
For specimens prepared by cryo-negative staining or vitrification, electron micrographs were taken under low-dose conditions (total electron dose of ≈25 e−/Å2) at a nominal magnification of ×50,000 at defocus values ranging from −3.5 to −6 μm by using an Oxford (Oxford, U.K.) cryo-transfer holder in a FEI Tecnai 200 kV electron microscope equipped with a field emission electron source.
All images were recorded on Kodak (Melville, NY) SO-163 film. Film was developed at 20°C for 10 min by using full-strength Kodak D-19 developer. Micrographs were digitized with a Zeiss (Oberkochen, Germany) SCAI scanner by using a pixel size of 7 μm, and 3 × 3 pixels were averaged to yield a pixel size of 4.2 Å at the specimen level. The defocus values of all images were determined by using the program CTF-FIND3 (42).
Random Conical Tilt Reconstruction.
Pairs (5, 821) of cryo-negatively stained U5.U2/U6 particles were selected interactively from both the images of the untilted and 55° tilted sample (100 image pairs) by using WEB, the display program associated with the SPIDER software package (43), and windowed into 128 × 128-pixel images. The untilted images were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment and K-means classification specifying 20 output classes (SI Fig. 7). Many of the 20 classes had very similar structural features. 3D structures were individually calculated for six representative classes by using the random conical tilt approach (44) implemented in the SPIDER package (SI Fig. 6, marked with *). To generate the best model possible, 4,605 particles associated with 12 classes were combined and used to generate a final structure in FREALIGN (37) (SI Fig. 6, merged classes marked with a “+”). One of the structures calculated by the random conical tilt approach was used as an initial model to align the individual images. The final 3D reconstruction was low-pass-filtered to 40-Å resolution for use as the initial model to align the images of the vitrified particles. The surface rendering of the structure was performed with the program Chimera (45).
Image Processing of Particles in Vitrified Ice.
A total of 30,671 particles were selected interactively from images of vitrified U5.U2/U6 taken at defocus values ranging from −3.5 to −6.0 μm by using the program Ximdisp associated with the MRC program suite (46). The particles were windowed into 128 × 128-pixel images. To obtain averages of the characteristic views of the molecule with improved signal-to-noise ratio, the images from the untilted samples were band-pass filtered (100- to 30-Å resolution) and subjected to three rounds of multireference alignment, multivariate statistical analysis, and classification into 200 classes by using the IMAGIC-V package (47).
To minimize propagating any residual artifacts in the low-resolution cryo-negative stain 3D model, the above 200 class averages were aligned to the cryo-negative stain 3D model by using the program FREALIGN (37). After initial Euler angle assignment, a 3D reconstruction was calculated. The model was refined by matching the class averages to projections from the refined model at a resolution of 45 Å. Refinement cycles were iterated until no changes could be detected in the 3D model (five cycles total). The resulting low-resolution structure was used as the initial input model for aligning all of the 30,671 particle images. FREALIGN was used to refine the orientation parameters of the particles and to correct the images for the contrast transfer function (CTF) of the electron microscope (37), assuming a 7% contribution of the amplitude contrast to the total CTF (42). 10% of the particles with the highest phase residuals were omitted from the three-dimensional reconstruction of the U5.U2/U6 complex. The contouring threshold was calculated by using a protein density of 0.8 Da/Å3 and a complex mass of 2.0 MDa. The final reconstruction was filtered to a resolution of 29 Å. The surface rendering of the structure was performed with the program Chimera (45).
Supplementary Material
Acknowledgments
We thank Martha Simon for assistance with STEM imaging. This work was supported by the National Institutes of Health (NIH-GM68900) (K.L.G. and T.W.), and the Giovanni Armenise–Harvard Foundation (T.W.). M.D.O. has been supported by the Jane Coffin-Childs Foundation and the Alfred A. King Trust. K.L.G. is an Investigator of the Howard Hughes Medical Institute. The Brookhaven National Laboratory STEM is a NIH Supported Resource Center (NIH Grant 5 P41 EB2181), with additional support provided by the U.S. Department of Energy Office of Biological and Environmental Research. Molecular graphics images were produced by using the University of California San Francisco (UCSF) Chimera package from the Resource for Biocomputing, Visualization and Informatics at UCSF Francisco (supported by NIH Grant P41 RR-01081).
Abbreviations
- snRNP
small nucleotide RNP
- STEM
scanning transmission EM.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611591104/DC1.
References
- 1.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Tardiff DF, Rosbash M. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins BM, Harrop SJ, Kornfeld GD, Dawes IW, Curmi PM, Mabbutt BC. J Mol Biol. 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- 5.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 6.Mura C, Cascio D, Sawaya MR, Eisenberg DS. Proc Natl Acad Sci USA. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 8.Reuter K, Nottrott S, Fabrizio P, Luhrmann R, Ficner R. J Mol Biol. 1999;294:515–525. doi: 10.1006/jmbi.1999.3258. [DOI] [PubMed] [Google Scholar]
- 9.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 10.Price SR, Evans PR, Nagai K. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 11.Reidt U, Reuter K, Achsel T, Ingelfinger D, Luhrmann R, Ficner R. J Biol Chem. 2000;275:7439–7442. doi: 10.1074/jbc.275.11.7439. [DOI] [PubMed] [Google Scholar]
- 12.Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thore S, Mayer C, Sauter C, Weeks S, Suck D. J Biol Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- 14.Muto Y, Pomeranz Krummel D, Oubridge C, Hernandez H, Robinson CV, Neuhaus D, Nagai K. J Mol Biol. 2004;341:185–198. doi: 10.1016/j.jmb.2004.04.078. [DOI] [PubMed] [Google Scholar]
- 15.Cass DM, Berglund JA. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- 16.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM. Proc Natl Acad Sci USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Luhrmann R, Sattler M. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Nat Struct Mol Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- 19.Boehringer D, Makarov EM, Sander B, Makarova OV, Kastner B, Luhrmann R, Stark H. Nat Struct Mol Biol. 2004;11:463–468. doi: 10.1038/nsmb761. [DOI] [PubMed] [Google Scholar]
- 20.Azubel M, Wolf SG, Sperling J, Sperling R. Mol Cell. 2004;15:833–839. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Luhrmann R, Stark H. Mol Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 24.Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Mol Cell Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald WH, Ohi R, Smelkova N, Frendewey D, Gould KL. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang T, Vilardell J, Query CC. EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 28.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurica MS, Moore MJ. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 30.Vogel J, Hess WR, Borner T. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potashkin J, Naik K, Wentz-Hunter K. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- 32.Van Heel M. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 33.Radermacher M, Wagenknecht T, Verschoor A, Frank J. J Microsc. 1987;146(Pt 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 34.Ohi M, Li Y, Cheng Y, Walz T. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 37.Stewart A, Grigorieff N. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Rosenthal PB, Henderson R. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Ohi MD, Gould KL. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan SP, Kao DI, Tsai WY, Cheng SC. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 41.Collart MA, Oliviero S. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kinston RE, Moore DD, Sledman JG, Smith JA, Sruhy K, editors. New York: Wiley; 1993. pp. 13.12.11–13.12.50. [Google Scholar]
- 42.Mindell JA, Grigorieff N. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 43.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 44.Radermacher M. Ultramicroscopy. 1994;53:121–136. doi: 10.1016/0304-3991(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Crowther RA, Henderson R, Smith JM. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 47.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.