Abstract
Host defense consists of two main aspects, namely, immune response to invading pathogens and suppression of tumor development. A family of transcription factors, IFN regulatory factors (IRFs), has recently gained much attention in terms of its critical role in linking these two aspects of host defense, wherein IRF5 was previously shown to play a critical role in the induction of proinflammatory cytokines by activation of Toll-like receptors. In the present study, using IRF5 gene-targeted mice (Irf5−/− mice), we demonstrate another facet of the IRF5 function in the regulation of immune response and tumor suppression. We show that IRF5 is critical for antiviral immunity by showing that Irf5−/− mice are highly vulnerable to viral infections, accompanied by a decrease in type I IFN induction in the sera. Furthermore, we show that Irf5−/− fibroblasts are resistant to apoptosis upon viral infection, resulting in an enhanced viral propagation. Finally, we provide evidence that IRF5 is critical for the induction of apoptosis, but not in cell cycle arrest, in response to DNA damage and that IRF5 functions as a tumor suppressor by acting on a pathway that may be distinct from that for p53. These results, together with the dual regulation of IRF5 gene expression by IFN signaling and p53, may provide a new link in the transcriptional network underlying antiviral immunity and tumor suppression.
Keywords: apoptosis, IRF, p53
An efficient and regulated cellular response is central to host defense, and it is coordinated by a genetic regulatory network in which a given transcription factor controls the expression of a set of diverse target genes depending on the cell type and/or the nature of cellular stimuli. Signaling in the immune system elicited by infection with viruses or bacteria usually leads to the production of type I IFN and inflammatory cytokines, resulting in the elimination of these pathogens (1, 2). Yet another important aspect of immune signal is the induction of apoptosis, which may be termed an altruistic suicide, which inhibits the further spread of infectious pathogens (3). This apoptotic response is highly similar to the response to genotoxic agents, such as ionizing radiation and certain anticancer drugs, which also induce cellular apoptosis, that is, one of the hallmarks of tumor suppression (4, 5). Indeed, recent studies have expanded the concept of a close relationship between the immune signaling system and oncogenesis (6, 7).
IFN regulatory factor (IRF) family members have been implicated as critical transcription factors that sense various environmental stresses and induce various genes required for an adequate cellular response (8–12). The first family member to be characterized was IRF1, which plays a critical role in the induction of both antiviral immunity and apoptosis of cells exposed to DNA-damaging agents (13, 14). Indeed, IRF1 cooperates with the tumor suppressor p53 to induce cell cycle arrest through the induction of p21WAF1/Cip1, and IRF1 induces apoptosis of cells expressing an oncogene. In addition, IRF8 (also known as the IFN consensus sequence binding protein; ICSBP) is critical to the induction of IL-12 in response to microbial infections, and it also contributes to suppression of cell transformation in hematopoietic cells (15).
IRF5 is an essential transcription factor that acts downstream of Toll-like receptors (TLRs), where IRF5 directly interacts with MyD88, an essential adaptor of TLRs, and regulates the TLR-dependent induction of proinflammatory cytokines, such as IL-6 and IL-12 (9). Several studies, employing overexpression or RNA interference assays using cultured cell lines, have suggested that IRF5 plays a role in inducing antiviral responses (16) as well as stress-induced cell cycle arrest and apoptosis (17–19); however, it remains to be determined in detail how these responses are affected in primary cells carrying a null mutation in the Irf5 alleles.
In this study we first examined the role of IRF5 in viral infection using gene-targeting mice. We show that Irf5−/− mice are highly vulnerable to viral infections. Virus-infected Irf5−/− mice exhibited a significant decrease in the induction levels of serum type I IFN and IL-6, and the virus-mediated induction of these cytokines was also partially impaired in Irf5−/− macrophages in vitro. Interestingly, mouse embryonic fibroblasts (MEFs) from Irf5−/− mice showed normal type I IFN production by these viruses, but they did not readily undergo virus-induced apoptosis, allowing a more efficient virus replication than WT MEFs. We also rigorously examined the role of IRF5 in cell cycle arrest and apoptosis of MEFs and adduce evidence that IRF5 is critical to the induction of apoptosis, but not cell cycle arrest, in response to DNA damage. Furthermore, we demonstrate a tumor-suppression activity of IRF5 via a pathway that is presumably distinct from that of p53. We will discuss our results, along with those previously reported by others, in the light of the complexities of the transcriptional network underlying the immune and antioncogenic responses induced by IRF5 and p53.
Results
IRF5 Is Essential for the Host Defense Against Viral Infection.
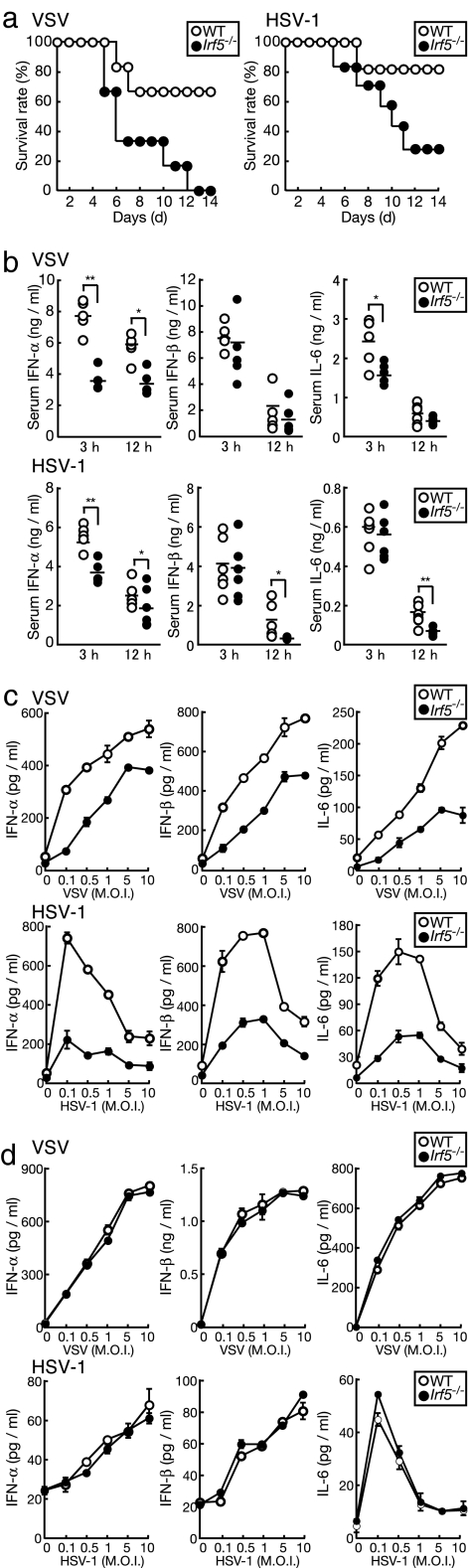
We first examined whether IRF5 is involved in host defense against infections by DNA and RNA viruses. WT and Irf5−/− mice were infected with vesicular stomatitis virus (VSV) or herpes simplex virus type 1 (HSV-1), and the survival of these mice was monitored daily. As shown in Fig. 1a, Irf5−/− mice showed a much higher vulnerability than WT mice to VSV and HSV-1 infections: 0% and 33% of Irf5−/− mice survived, whereas 67% and 83% of WT mice survived 14 days after VSV and HSV-1 infections, respectively. We also analyzed the induction levels of IFN-α, IFN-β, and IL-6 by ELISA. As shown in Fig. 1b, Irf5−/− mice showed a significant decrease in IFN-α induction level compared with WT mice and, although not as pronounced as IFN-α, significant decreases in IFN-β and IL-6 induction in the sera at 3 h or 12 h after infection.
Fig. 1.
Role of IRF5 in the host defense against viral infections. (a) WT and Irf5−/− mice (six mice per group) were infected with VSV (Left) or HSV-1 (Right). The survival of these mice was monitored daily for 14 days. (b) Sera from each mouse in a were collected at 3 and 12 h after infection, and IFN-α, IFN-β, and IL-6 levels were measured by ELISA. Each point represents an individual mouse. ∗, P < 0.05; ∗∗, P < 0.005. (c) CD11b+ splenic macrophages from WT and Irf5−/− mice were infected with VSV or HSV-1 at the indicated multiplicity of infection (M.O.I.) for 24 h. Cytokine concentrations in the supernatants were measured by ELISA. (d) WT and Irf5−/− MEFs were infected with VSV or HSV-1 for 24 h. Cytokine concentrations were measured by ELISA. Results shown are the means ± SD of triplicate determinations for c and d.
Because IRF5 is expressed at high levels in splenocytes (9, 20), we next purified CD11b+ splenic macrophages from WT and Irf5−/− mice and examined IFN-α, IFN-β, and IL-6 production in response to viral infections by ELISA. The production of these cytokines by macrophages infected with VSV or HSV-1 was markedly decreased in Irf5−/− cells compared with WT cells (Fig. 1c). Interestingly, when the same experiment was carried out by using MEFs, we observed an essentially normal production of these cytokines in the Irf5−/− background, despite the efficient nuclear translocation of IRF5 in virus-infected MEFs [Fig. 1d and supporting information (SI) Fig. 6 a and b]. Therefore, it appears that the contribution of IRF5 to type I IFN and IL-6 production is cell-type-dependent, where IRF5 participates in cytokine induction in cells such as macrophages that express IRF5 at higher levels than MEFs. Taking together the above and previous results, the serum type I IFN level in virus-infected mice is largely if not entirely dependent on hematopoietic cells (see Discussion).
IRF5-Dependent Apoptosis of Virus-Infected Cells.
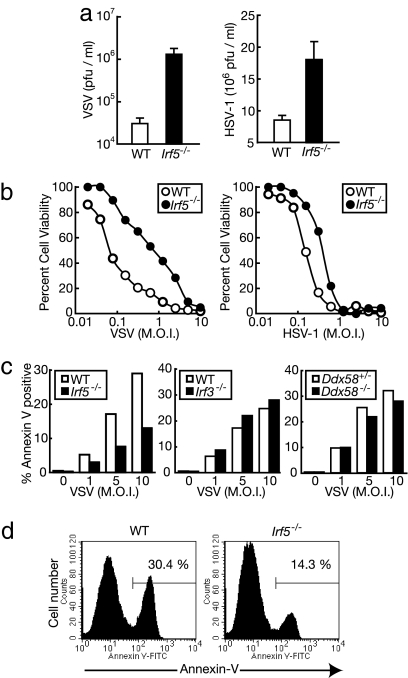
Does the decrease in serum type I IFN induction level in the virus-infected Irf5−/− mice totally account for their vulnerability? To further assess the role of IRF5 in antiviral immunity, we infected Irf5−/− MEFs with VSV or HSV-1, and virus titer was monitored in the supernatant of infected cells. We observed that the virus titer was markedly higher in the Irf5−/− MEFs than in WT MEFs (Fig. 2a). Interestingly, when we determined the number of viable cells by crystal violet staining, we found that Irf5−/− MEFs were more resistant to cell death upon infection with VSV or HSV-1 than WT MEFs (Fig. 2b). WT MEFs infected with VSV eventually underwent apoptosis as revealed by annexin V staining, and this apoptotic response was markedly suppressed in Irf5−/− MEFs (Fig. 2 c and d). On the other hand, the virus-induced apoptosis of Irf3−/− MEFs was similar to that of MEFs from a littermate WT mouse (Fig. 2c). These results indicate that IRF5, not IRF3, plays a critical role in the induction of apoptosis of MEFs in response to viral infections and suggest that this function of IRF5, at least in part, accounts for the vulnerability of Irf5−/− mice to viruses, as shown in Fig. 1a.
Fig. 2.
IRF5 is required for apoptotic response against viral infections. (a) A total of 1 × 105 WT (open bars) and Irf5−/− (filled bars) MEFs were infected with VSV or HSV-1 (multiplicity of infection of 1) for 24 h, and virus titers of the supernatant were determined. Results shown are the means ± SD of triplicate determinations. (b) WT (open circles) and Irf5−/− (filled circles) MEFs were infected with serially diluted VSV or HSV for 36 h. After infection, cells were fixed and their viability was evaluated by crystal violet staining. (c and d) WT and mutant MEFs were left untreated or were infected with serially diluted VSV for 24 h, and apoptotic responses were monitored by annexin V and propidium iodide staining. The percentages of propidium iodide-negative and annexin V-positive cells are shown in c. One representative example of three for each group is shown. A representative histogram of annexin V binding of VSV-infected WT or Irf5−/− MEFs (gated on PI-negative cells) is shown in d.
The induction of type I IFN and IL-6 in MEFs against VSV infection is mediated by retinoic acid-inducible gene I (RIG-I), an essential molecule that senses intracellular virus-derived RNA and evokes antiviral responses (21, 22). Thus, it was of interest to examine the role of RIG-I in virus-induced apoptosis, that is, whether the activation of IRF5 by VSV for the induction of apoptosis depends on the RIG-I pathway. Interestingly, Ddx58 (RIG-I gene)-deficient MEFs exhibited the apoptotic response similar to those from a heterozygous littermate mouse on VSV infection (Fig. 2c). Thus, IRF5 may be activated by a RIG-I-independent pathway for the induction of apoptosis.
Role of IRF5 in Cell Cycle Arrest and Apoptosis in Response to Genotoxic Stresses.
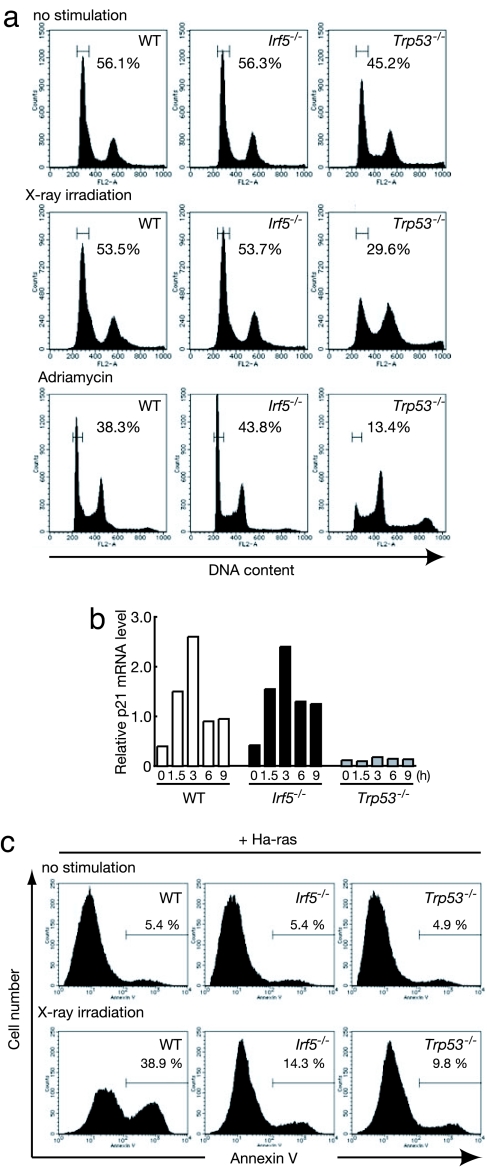
The balance between cell cycle arrest and apoptosis is critical to the host's ability to eliminate cancerous cells, while saving the normal cells. IRF5 has been implicated in the induction of cell cycle arrest and apoptosis in response to genotoxic stresses (17–19). Indeed, IRF5 undergoes nuclear translocation upon DNA damage (ref. 17 and our unpublished data). This finding, together with the above results, prompted us to examine in detail whether IRF5 also contributes to DNA-damage-induced cell cycle arrest and apoptosis. Irf5−/− MEFs were subjected to ionizing irradiation or treated with adriamycin and subjected to cell cycle analysis; WT MEFs and MEFs deficient in the p53 gene (Trp53−/− MEFs) were treated similarly. In contrast to Trp53−/− MEFs, which showed a low G0/G1 rate (29% upon ionizing irradiation; 13% upon adriamycin treatment), WT and Irf5−/− MEFs were substantially arrested in the G0/G1 phase of the cell cycle after irradiation or adriamycin treatment (Fig. 3a). These results indicate that IRF5 is dispensable for DNA-damage-induced cell cycle arrest at least in these experimental settings. Consistently, a normal induction of p21WAF1/Cip1 mRNA was observed in Irf5−/− MEFs upon ionizing irradiation, collectively indicating a dissociation of the IRF5 function from that of p53 (Fig. 3b).
Fig. 3.
Role of IRF5 in DNA damage-induced apoptosis. (a) WT, Irf5−/−, and Trp53−/− MEFs were left untreated (Top), γ-irradiated (5 Gy; Middle), or treated with 0.5 μg/ml adriamycin (Bottom) for 24 h, and then the cells were stained with PI and analyzed by flow cytometry to measure DNA content. (b) The steady-state mRNA levels of p21WAF1/Cip1 were measured by RNA blotting in WT and mutant MEFs after irradiation or adriamycin treatment. (c) WT and mutant MEFs expressing activated Ha-ras were γ-irradiated (20 Gy). After 48 h, the percentage of annexin V-FITC-positive cells was determined by flow cytometry. One representative example of three is shown.
To examine the role of IRF5 in the regulation of oncogenesis, we next induced DNA damage in WT or Irf5−/− MEFs expressing the activated oncogene Ha-ras by x-ray irradiation: it has been reported that the expression of an activated oncogene can sensitize cells to undergo apoptosis in response to DNA damage, one of the hallmarks of tumor suppression (23, 24). As shown in Fig. 3c, Ha-ras-expressing Irf5−/− MEFs did not readily undergo apoptosis (14% annexin V+), whereas a substantial number of WT MEFs expressing the same oncogene undergo apoptosis (38% annexin V+). The resistance of Irf5−/− MEFs to apoptosis is similar to that of Trp53−/− MEFs (9% annexin V+) (Fig. 3c). Thus, on the basis of these findings, both IRF5 and p53 are required to induce apoptosis in oncogene-expressing cells in response to DNA damage.
IRF5 Deficiency Predisposes Cells to Tumorigenic Transformation.
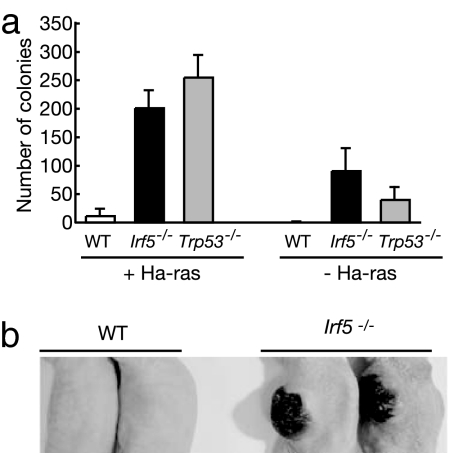
The above findings prompted us to examine the transforming and tumorigenic properties of Ha-ras-expressing Irf5−/− MEFs and Trp53−/− MEFs. As shown in Fig. 4a, Irf5−/− MEFs formed many colonies in methylcellulose gel almost as efficiently as Trp53−/− MEFs, whereas WT MEFs formed few colonies (Fig. 4a Left). It may be worth noting that, under the same culture conditions, both mutant MEFs formed a notable number of colonies without Ha-ras expression (Fig. 4a Right). Furthermore, when Ha-ras-expressing Irf5−/− MEFs were injected s.c. into nude mice, tumors developed within 2 weeks (Fig. 4b and Table 1). In contrast, WT MEFs showed no such property, even though the Ha-ras gene was expressed at similar levels (data not shown). Thus, the loss of IRF5 function predisposes primary fibroblasts to an oncogene-induced transformation.
Fig. 4.
Tumorigenic transformation caused by IRF5 deficiency. (a) WT, Irf5−/−, and Trp53−/− MEFs expressing activated Ha-ras were seeded on soft agar and incubated for 3 weeks. The number of colonies was counted. Results shown are the means ± SD of triplicate determinations. (b) WT and mutant MEFs expressing activated Ha-ras were injected s.c. into BALB/c nude mice. Tumor sizes were evaluated 4 weeks after injection. Two representative mice from each sample are shown.
Table 1.
Tumorigenicity in nude mice
| Genotype | Tumor size (length × width × height), mm |
|---|---|
| WT | ND |
| ND | |
| ND | |
| ND | |
| ND | |
| ND | |
| Irf5−/− | 8 × 8 × 1 |
| 12 × 9 × 1 | |
| 12 × 5 × 1 | |
| 10 × 7 × 1 | |
| 9 × 9 × 4 | |
| 8 × 8 × 4 | |
| Trp53−/− | 23 × 18 × 16 |
| 28 × 24 × 15 | |
| 25 × 14 × 13 |
WT, IRF5−/−, and Trp53−/− MEFs were infected with pGDV12ras retrovirus. Two days after infection, 1 × 106 MEFs were injected s.c. to nude mice to test the tumorigenicity of the cells. Tumor size was determined by caliper measurements after 4 weeks of injection. ND, not detected.
Induction of IRF5 and Proapoptotic Genes.
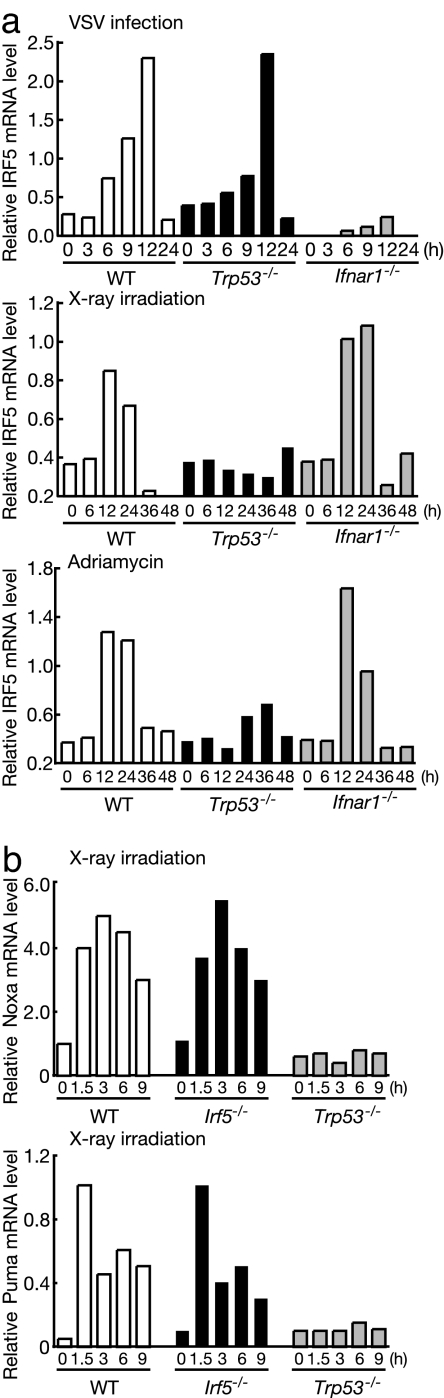
It has been reported that IRF5 is induced by viral infections or p53 activation (19, 25). As shown in Fig. 5a, the IRF5 mRNA induction by VSV is normal in Trp53−/− MEFs, but is completely abrogated in MEFs from mice deficient in IFNAR1 (Ifnar1−/− MEFs), a component of the type I IFN receptor complex. These data indicate that the virus-dependent IRF5 gene induction depends on the type I IFN signal but not on p53. Conversely, IRF5 mRNA induction by DNA damage induced by x-ray or adriamycin is p53-dependent but IFN-independent (Fig. 5a). Thus, IRF5 expression is under dual, mutually independent, control mechanisms, so as to evoke immune and antitumor responses of a cell.
Fig. 5.
Normal induction of Noxa and Puma in Irf5−/− cells. (a) WT, Ifnar1−/−, and Trp53−/− MEFs were left untreated, infected with VSV (multiplicity of infection of 1), γ-irradiated (20 Gy), or treated with 0.5 μg/ml adriamycin for the indicated time periods. The induction of IRF5 mRNA was analyzed by quantitative real-time RT-PCR. (b) WT and mutant MEFs were γ-irradiated for the indicated periods. Noxa and Puma mRNA levels were determined by RNA blotting.
In view of the fact that the p53-dependent apoptosis is mediated, at least in part, by the induction of proapoptotic genes such as Noxa and Puma (26–28) and that the IRF5 gene is up-regulated by p53, we examined whether these proapoptotic genes are also regulated by IRF5. As shown in Fig. 5b, RNA blotting revealed that all these genes were normally induced in response to x-ray irradiation. These findings suggest that IRF5 regulates proapoptotic gene(s) other than the above-mentioned genes to induce apoptotic response to DNA damage.
Discussion
The IRF family of transcription factors has gained much attention for its contribution to the regulation of the immune system and oncogenesis (8, 9, 11–15). Thus far, little is known about the function of IRF5 of this family, although it has gained much attention in the context of the TLR-mediated gene induction program (9, 29). Our present study using gene-targeted mice clearly revealed the function of IRF5 beyond the context of TLR signaling, namely, its cell-type-dependent contribution to type I IFN induction and apoptosis in response to viral infection or DNA damage. On the basis of the present findings, one may infer the following events: IRF5 is transcriptionally induced by viral infection through type I IFN signaling or by DNA damage through activated p53; and then, induced IRF5 is activated by an as yet unknown TLR-independent mechanism presumably in the cytoplasm and translocates to the nucleus to induce type I IFN and proapoptotic gene(s).
IRF5 plays a critical role in the induction of type I IFN and IL-6 in response to viral infections, particularly in hematopoietic cells, in which IRF5 is expressed at high levels. Because virus-mediated type I IFN induction is entirely dependent on IRF7 (11, 30), we infer that activated IRF5 might form a heterodimer with IRF7 to act on type I IFN promoters. However, we cannot rule out an indirect effect of IRF5, and this issue needs to be clarified. Considering the finding that RIG-I and a serine threonine kinase, Tank-binding kinase 1 (TBK1), are essential for the type I IFN gene induction against VSV infection (22), IRF5 may be activated by RIG-I and TBK1. Indeed, our preliminary data showed that IRF5 may be modified and detected as a shifted band by immunoblot analysis using lysate from cells cotransfected with TBK1 and IRF5. Interestingly, however, VSV-mediated apoptosis is induced independent of RIG-I (Fig. 2c), indicating that IRF5 might also be activated by a RIG-I-independent signaling pathway. Although further careful analysis is required to gain further insight into IRF5-dependent apoptosis, we infer that this RIG-I-independent pathway, possibly via ataxia-telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR) pathways, might involve an as yet unidentified sensing system activated by severe cellular stresses caused by viral infections. This notion may corroborate with the reports that genotoxic stresses induce the activation of p53 via ATM/ATR and that the virus-activated p53 is also critical to antiviral host defense (3).
It is well documented that when cells are exposed to genotoxic stress, a choice between two major fates, life or death, is made (4, 5). As this decision is crucial, many regulatory mechanisms are in place to ensure an appropriate choice. The survival response is coupled to the induction of cell cycle arrest at checkpoints in different phases of the cell cycle (4, 5). This permits DNA repair, which may be associated with the induction of genes promoting cell survival. In contrast, the induction of distinct molecular events that culminate in the activation of common cell death pathways mediates cell death (4, 5). The final outcome is probably determined by the type and amount of damage to the genome. The tumor suppressor p53, activated in response to DNA damage, induces both of these cellular responses. Therefore, a key question is how do cells regulate the balance of p53 activities to ensure that healthy cells survive and genetically damaged cells that might lead to tumor formation are eliminated. IRF5 is transcriptionally induced through the activation of p53, and indeed Irf5−/− cells showed a similar phenotype to Trp53−/− cells, indicating that IRF5 is essential to the apoptotic response. In contrast, unlike previous reports (17, 19), our findings indicate that the induction of the p21WAF1/Cip1 gene and the induction of cell cycle arrest normally occur in Irf5−/− cells. Therefore, although IRF5 is one of the target genes of p53, it seems to be selectively involved in apoptosis but not in cell cycle arrest. We therefore infer that IRF5 is a key member of the molecular switch that evokes either cell cycle arrest or apoptosis. Furthermore, the induction of the known p53-dependent proapoptotic genes is normally observed in the absence of IRF5 (Fig. 5b), suggesting the presence of an as yet unknown gene activation program for apoptosis mediated by IRF5. Finally, in the light of our present and previous findings, it will be of great interest to examine the status of the IRF5 gene and its expression in human cancers as well as immunodeficiencies.
Materials and Methods
Mice and Cell Culture.
The generation of Irf3−/−, Irf5−/−, and Trp53−/− mice was described previously (3, 9, 30). Ifnar1−/− mice were purchased from B & K Universal Group, and BALB/c nude mice were from CLEA Japan (Osaka, Japan). MEFs were prepared following a standard procedure. Ddx58-deficient MEFs were kindly provided by O. Takeuchi and S. Akira (Osaka University, Osaka, Japan). Splenic CD11b+ cells were collected by using CD11b MicroBeads and a MACS column (Miltenyi Biotec, Bergisch Gladbach, Germany).
Viral Infections.
VSV and HSV-1 were prepared as described previously (30). Mice were intravenously infected with 1 × 108 pfu of VSV or 1 × 107 pfu of HSV-1, and blood was collected at 3 and 12 h after infection. Levels of IFN-α, IFN-β, and IL-6 were monitored by commercial ELISA kits according to the manufacturer's instructions (ELISA kits for IFN-α and IFN-β were from PBL Biomedical Laboratories, Piscataway, NJ, and a kit for IL-6 was from R&D Systems, Minneapolis, MN). The t test was used for statistical analysis. For in vitro infection, cultured cells were infected with serially diluted VSV or HSV-1 for the indicated time periods. Virus titers in the supernatants were determined by standard plaque assay, and cell viability was assessed by crystal violet staining as described previously (3).
RNA Analysis.
RNA extraction, RT-PCR, and RNA blot analysis were performed as described previously (3). Quantitative real-time RT-PCR analysis was performed by using a LightCycler and SYBRGreen system (Roche, Indianapolis, IN). Data were normalized by the level of G3PDH expression in each sample. The following oligonucleotide primers were used: G3PDH, 5′-CTCATGACCACAGTCCATGC-3′ and 5′-CACATTGGG-GGTAGGAACAC-3′; IRF5, 5′-TAGAGGCTACCCAGGAGCAA-3′ and 5′-GCCCACTCCAGAACACCTTA-3′. For RNA blot analysis, equal amounts of total RNA (5 μg) were loaded in each lane. Probes for Noxa, Puma, and p21WAF1/Cip1 were previously described (3). Signal quantification was done by using NIH image software.
Apoptosis Assay and Cell Cycle Analysis.
Cells were subjected to γ-radiation (20 Gy) or treated with adriamycin (0.5 μg/ml; Sigma, St. Louis, MO). After a 24-h incubation, cells were stained with annexin V coupled to fluorescein isothiocyanate (MBL, Nagoya, Japan) and propidium iodide (PI) (2 μg/ml) in accordance with the manufacturer's instructions. Analysis was performed with a FACSCalibur (BD Biosciences, San Jose, CA). Cell cycle analysis was performed by using the CycleTEST PLUS DNA Reagent Kit according to the manufacturer's instructions (BD Biosciences).
Transformation Assay.
The expression of activated Ha-ras was mediated by retroviral gene transfer, wherein the pGDV12ras was used as described previously (3). MEFs expressing activated Ha-ras were seeded into 60-mm dishes at 1 × 105 cells per dish in a suspension of 1.3% methylcellulose gel dissolved in DMEM supplemented with 10% FCS on top of a bed composed of 0.53% agarose in the same culture medium. The number of colonies formed was determined 3 weeks after seeding. For the tumorigenicity assay, 1 × 106 MEFs expressing activated Ha-ras were injected s.c. in the flanks of 6-week-old BALB/c nude mice.
Supplementary Material
Acknowledgments
We thank Osamu Takeuchi and Shizuo Akira for Ddx58−/− MEFs, Massashi Shishido for technical assistance, and Tsukasa Shibue and David Savitsky for kind advice and critical reading of the manuscript. This work was supported by Kakenhi Grants-in-Aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer” and “Dynamics of Extracellular Environments” from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Uehara Memorial Foundation.
Abbreviations
- TLR
Toll-like receptor
- IRF
IFN regulatory factor
- RIG-I
retinoic acid-inducible gene I
- MEF
mouse embryonic fibroblast
- VSV
vesicular stomatitis virus
- HSV-1
herpes simplex virus type 1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611559104/DC1.
References
- 1.Vilcek J, Sen GS. In: Virology. 3rd Ed. Fields DM, Knipe PM, Howley PM, editors. Philadelphia: Lippincott–Raven; 1996. pp. 375–399. [Google Scholar]
- 2.De Maeyer E, De Maeyer-Guignard J. Interferons and Other Regulatory Cytokines. New York: Wiley; 1988. [Google Scholar]
- 3.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lu X. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 5.Oren M. Semin Cancer Biol. 1994;5:221–227. [PubMed] [Google Scholar]
- 6.Dunn GP, Koebel CM, Schreiber RD. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 7.Belardelli F, Ferrantini M, Proietti E, Kirkwood JM. Cytokine Growth Factor Rev. 2002;13:119–134. doi: 10.1016/s1359-6101(01)00022-3. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 9.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 10.Barnes B, Lubyova B, Pitha PM. J Interferon Cytokine Res. 2002;22:59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- 11.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 12.Honda K, Taniguchi T. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. Nature. 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier MS, Aizawa S, Mak TW, Taniguchi T. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 15.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O'Shea JJ, Singh H, Ozato K. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 16.Barnes BJ, Field AE, Pitha-Rowe PM. J Biol Chem. 2003;278:16630–16641. doi: 10.1074/jbc.M212609200. [DOI] [PubMed] [Google Scholar]
- 17.Hu G, Mancl ME, Barnes BJ. Cancer Res. 2005;65:7403–7412. doi: 10.1158/0008-5472.CAN-05-0583. [DOI] [PubMed] [Google Scholar]
- 18.Cheng TF, Brzostek S, Ando O, Van Scoy S, Kumar KP, Reich NC. J Immunol. 2006;176:7462–7470. doi: 10.4049/jimmunol.176.12.7462. [DOI] [PubMed] [Google Scholar]
- 19.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Cancer Res. 2003;63:6424–6431. [PubMed] [Google Scholar]
- 20.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uze G. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 23.Oren M. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 24.Lowe SW, Ruley HE, Jacks T, Housman DE. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 25.Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H. Oncogene. 2002;21:2914–2918. doi: 10.1038/sj.onc.1205459. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 27.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 28.Nakano K, Vousden KH. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 29.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. J Biol Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 30.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.