Abstract
Arrays of >5,000 Saccharomyces cerevisiae proteins were screened to identify proteins that can preferentially bind a small RNA hairpin that contains a clamped adenine motif (CAM). A CAM is required for the replication of Brome Mosaic Virus (BMV), a plant-infecting RNA virus that can replicate in S. cerevisiae. Several hits were selected for further characterization in Nicotiana benthamiana. Pseudouridine Synthase 4 (Pus4) and the Actin Patch Protein 1 (App1) modestly reduced BMV genomic plus-strand RNA accumulation, but dramatically inhibited BMV systemic spread in plants. Pus4 also prevented the encapsidation of a BMV RNA in plants and the reassembly of BMV virions in vitro. These results demonstrate the feasibility of using proteome arrays to identify specific RNA-binding proteins for antiviral activities. Furthermore, the effects of Pus4 suggest that the CAM-containing RNA motif provides a regulatory link between RNA replication and encapsidation.
Keywords: Brome mosaic virus, protein–RNA interaction, pseudouridine synthase 4, yeast proteome chip, viral RNA replication
Positive-stranded RNA viruses include important pathogens that cause diseases in plants, animals, and humans (1). Cellular proteins, such as nucleic acid modification enzymes and innate immunity receptors, have critical roles in modulating pathogen infection (2–4). A better understanding of how cellular and viral factors interact could lead to the identification of proteins with antiviral activities.
Brome mosaic virus (BMV) is a model positive-stranded RNA virus that infects plants (5–7). A property of BMV that has led to significantly improved understanding of its interactions with host factors is its ability to replicate and transcribe its genome in Saccharomyces cerevisiae (8). Other RNA and DNA viruses have since been found to replicate in yeast (9–12). Another useful feature of BMV is that the cis-acting sequences (promoters) for RNA synthesis by the BMV replication enzyme complex have been identified and characterized (6). A multifunctional tRNA-like structure within the 3′ untranslated portion of BMV RNAs contains the promoter for minus-strand RNA synthesis (13), regulates translation (14), and facilitates viral assembly (15). The structure of the core promoter for BMV minus-strand RNA replication was solved by NMR spectroscopy and consists of a short stem with a three-nucleotide loop called stem-loop C (SLC) (16). The triloop in SLC contains an adenine that is displaced from the central stack and held in place by an intramolecular network of interactions to form a clamped adenine motif (CAM) (16). The structures of RNAs with single nucleotide changes which abolished CAM formation have also been determined (17). However, an unbiased, comprehensive screening for host protein that can recognize this critical CAM has not been conducted because of a lack of proper tool.
The protein microarray technologies can dramatically accelerate functional profiling at the proteome level (18, 19). Using such an unbiased protein chip approach, Hall et al. (20) demonstrated that a metabolic enzyme can specifically regulate the transcription of a mitochondrial gene. Given the wide range of cellular functions in which RNAs play vital roles, we used yeast protein arrays to identify proteins which could bind a CAM-containing RNA derived from BMV and analyzed the effects of select proteins on BMV infection.
Results
The CAM-Binding Proteins Are Identified by Using Protein Arrays.
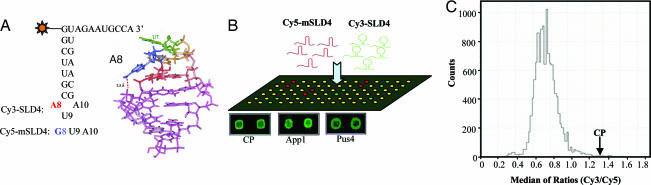
A yeast proteome chip was probed with a 1:1 molar ratio of a Cy3-labeled CAM-containing RNA (SLD4) and a Cy5-labeled RNA (mSLD4), which contains a nucleotide substitution that abolished the formation of the CAM (Fig. 1A). We have observed that the CP could specifically recognize SLC (C.K., unpublished observation). Therefore, purified BMV capsid protein (CP) was printed on these arrays to serve as a control (21). Each protein was printed in duplicate and the results from two independent arrays were averaged. The binding signals from BMV CP and two of the positives are illustrated in Fig. 1B. The ratios of the Cy3/Cy5 signals for the whole yeast proteome array generally followed a normal distribution, with approximately a dozen of the proteins having signals comparable or better than that of the BMV CP (Fig. 1C). Among these hits are heat shock proteins, enzymes for RNA modification, and proteins involved in translation (Table 1).
Fig. 1.
Screening the proteome arrays with an RNA containing a CAM. (A) The sequence of the RNA SLD4 containing the CAM motif is shown along with a structure of the CAM (12). The clamped adenine is residue eight in SLD4 and a substitution to a guanine that abolishes CAM formation in RNA mSLD4 is shown below SLD4. Other nucleotides in mSLD4 are identical to those in SLD4. (B) A scheme illustrating the probing of the protein array. An equal mixture of SLD4 with a 5′ Cy3, and mSLD4 with a 5′ Cy5 was used as the probe. Images of the protein spots of two of the positive hits, Pus4 and App1, and the internal positive control, the BMV CP, are shown below the array. (C) The distribution of the results (the median of Cy3/Cy5 ratios) from the array screen. The signal of BMV CP protein is identified within the graph.
Table 1.
A summary of the 12 proteins that were the best hits from the arrays and their putative functions
| Name | Cy3/Cy5 ratio* | Function |
|---|---|---|
| Hsp104 | 2.38 ± 0.01 | Heat shock protein |
| Orl1 | 1.72 ± 0.05 | UDP-N-acetylglucosamine pyrophosphorylase |
| App1 | 1.49 ± 0.04 | Actin patch binding protein |
| YLR257W | 1.46 ± 0.04 | Unknown |
| Mdg1 | 1.42 ± 0.05 | Signal transduction in the mating response |
| Zeo1 | 1.41 ± 0.03 | Regulator of the Pkc1p XMpk10 pathway |
| Met16 | 1.37 ± 0.04 | Homocysteine methyltransferase |
| Sse1 | 1.34 ± 0.03 | Heat shock protein |
| Pus4 | 1.29 ± 0.03 | Pseudouridine synthase |
| Etc1 | 1.28 ± 0.01 | 2-Encyl thioester reductase |
| Met1 | 1.26 ± 0.09 | Mitochondrial translation elongation factor G |
| Tim10 | 1.25 ± 0.01 | tRNA guanine methyltransferase |
| BMV capsid | 1.39 ± 0.09 | BMV RNA encapsidation |
The Cy3/Cy5 ratios (±SEM) were averaged from the results at two independent chips, each of which contained two samples of each protein.
The top 12 hits were individually purified for additional characterization. To confirm their ability to bind RNA, each candidate protein was cross-linked by UV irradiation to radiolabeled SLD4, followed by electrophoresis on a denaturing protein gel. BSA at four times the concentration of the candidate proteins was used as a negative control, and did not cross-link SLD4 (Fig. 2A). Eleven of the 12 candidate proteins were confirmed to bind SLD4, although the signals were weak for two (Zeo1 and Sse1). Mdg1 was the only one that did not cross-link SLD4. However, the input template was rapidly degraded in reactions with Mdg1, and we suspect the presence of contaminating ribonuclease activity and did not pursue this result further.
Fig. 2.
Characterization of the candidate proteins identified in the protein array experiments. (A) Confirmation of SLD4-protein binding by the 12 candidate proteins using a cross-linking assay. Each cross-linking reaction was performed with 20 μg/ml of each purified candidate protein and 1 nM of SLD4 radiolabled with 32P at its 5′ end. (B) Expression of the S. cerevisiae TyA Gag protein did not significantly affect BMV plus-strand RNA accumulation. The identities of the BMV RNAs are noted to the right of the image of the Northern blot analysis. The images of gel slices below the Northern blots show a cellular rRNA stained with ethidium bromide that serves as loading controls for the RNAs. (C) Effects of expressing the empty vector Qri1, Pus4, or App1 on BMV RNA accumulation. The images are of the Northern blots detecting minus- and plus-strand BMV RNAs, as identified by the symbols to the left of the images. The blots were probed first for minus-strand RNAs to generate the top images, then stripped by boiling in a low salt buffer at 95°C and then probed for plus-strand RNA accumulation. The agarose gel strips contain a cellular rRNA that serves as a loading control. Quantification of RNA accumulations were performed with a phosphorimager.
Pus4, App1, and Qri1 were the three strongest binders to SLD4 (Fig. 2A). Pus4 is a pseudouridine synthase that isomerizes uridine to pseudouridines in tRNAs (22). This finding is interesting because the tRNA-like structure at the 3′ untranslated region of BMV RNAs contains pseudouridines (23). App1 is an actin patch protein that could function in intracellular trafficking (24, 25). Qri1 is an UDP-N-acetylglucosamine pyrophosphorylase that is involved in cell wall polysaccharide formation (26).
Two SLD4-Binding Proteins Can Inhibit BMV Infection.
Qri1, App1, Pus4, and several other cellular and viral proteins were examined for effects on BMV RNA levels in plants. Each protein was expressed transiently using Agrobacterium vectors in Nicotiana benthamiana along with the BMV RNAs (27). A feature of Agrobacterium expression system is that the several hundred copies of the recombinant T-DNA could be integrated per cell; therefore, recombinant protein expression could be adjusted by varying the density and composition of the inoculum infiltrated into leaves (28). The expression of the empty vector, unrelated RNA binding proteins from S. cerevisiae, namely TyA Gag and Dis3, and two RNA-binding proteins from hepatitis C virus (29–32) had no discernable effects on BMV RNA levels in the agroinfiltrated leaves (ref. 33 and Fig. 2B). Thus, the effects of Qri1, App1, and Pus4 are not simply due to the expression of an RNA-binding protein. Further indication for specificity is that Qri1, App1, and Pus4 preferentially inhibited plus-strand, but had less of an effect on minus-strand BMV RNA levels. In fact, Pus4 reproducibly increased minus-strand RNA levels, up to 130%. For the plus-strand BMV RNAs, Qri1 decreased RNA accumulation only at the higher inoculum concentrations, whereas App1 and Pus4 decreased the viral RNA levels in a concentration-dependent manner, reducing RNA levels by up to a third relative to the controls (Fig. 2C). Pus4 also decreased translation from BMV RNA3 by ≈2-fold [see supporting information (SI) Fig. 6A]. Thus, Pus4 and App1 will be the focus of the reminder of the study.
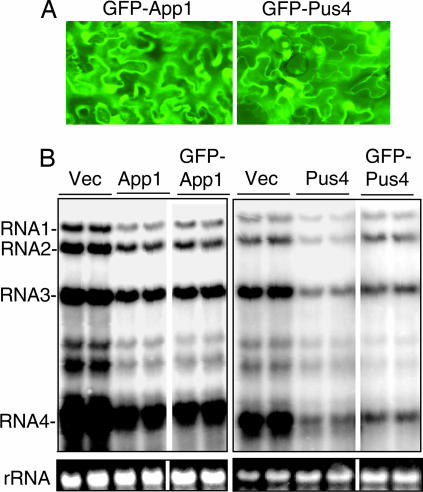
To ensure that the effects of Pus4 and App1 are the direct result from expression of the recombinant proteins in plant cells, we made and expressed App1 or Pus4 with either N- or C-terminal fusions to the GFP. All four constructs resulted in the detection of GFP signal. In N. benthamiana cells the cytoplasm appears to as a thin band around the central vacuole. GFP signal from the fusions were detected in this band as well as in the nuclei, which appear as bright oblong circles (Fig. 3A and data not shown). In addition, when coinfiltrated with A. tumafaciens that expresses the BMV RNAs and N-terminal fusions, GFP–App1 and GFP–Pus4 were able to suppress plus-strand BMV RNA levels the 2- to 3-fold that we had previously observed for the untagged App1 and Pus4, although GFP–Pus4 was slightly affected for this activity (Fig. 3B). The C-terminal GFP fusion of App1 also suppressed BMV RNA levels, but Pus4-GFP lost the inhibitory activity (data not shown), likely due to perturbation of required structure(s). Nonetheless, these results demonstrate that the inhibitory activities of App1 and Pus4 are correlated with expression of the recombinant proteins in N. benthamiana.
Fig. 3.
Expression of Pus4 and App1 in plants. (A) Fluorescent microscopic images of plant cells expressing GFP-App1 and GFP-Pus4 chimeric proteins. GFP signals were visualized and photographed using a Zeiss Axioplan 2 microscope using GFP optimised FITC filter (at 543 nm excitation and 505- to 530-nm emission). (B) Images of autoradiograms demonstrating that GFP-App1 and GFP-Pus4 are capable of inhibiting BMV plus-strand RNA accumulation. The layout of the gel images is identical to those in Fig. 2B. Two independently derived samples were tested for each manipulation and all of the samples were originally from one blot.
Pus4 and App1 Affect BMV Systemic Spread in Planta.
Because Pus4 or App1 did not inhibit minus-strand BMV RNA accumulation, it is less likely that they affected the recognition of SLC by the BMV replicase. To seek an explanation for the effect on plus-strand RNA accumulation, we examined whether Pus4 and App1 would affect BMV systemic spread, which occurs through encapsidated plus-strand RNAs (34). The third leaf of N. benthamiana was agroinfiltrated to express the BMV genomic RNAs and either the empty vector, App1, or Pus4 (Fig. 4A). The BMV CP was readily detected in the infiltrated leaves, although its level is slightly less in the presence of Pus4 (Fig. 4B, leaf 3). Detection of BMV CP in the higher leaves is expected in a systemic infection, which preferentially spread from the lower toward the upper leaves in a source to sink direction (35, 36). In the higher leaves, BMV was detected when expressed in the presence of the vector control (Fig. 4B, leaves 4 and 5). However, coexpression with Pus4 and App1 resulted in no detectable CP in these leaves (Fig. 3B), indicating that systemic infection was affected.
Fig. 4.
App1 and Pus4 specifically affect BMV RNA infection in plants. (A) A schematic representation of a N. benthamiana plant used for Agro inoculation and examination of virion formation. (B) App1 and Pus4 inoculated at 0.2 OD595 can decrease spread of BMV in plants. The N. benthamiana leaves from which the samples were prepared are noted at the bottom of the gel images. The larger gel images are of Coomassie blue-stained SDS/PAGE. The samples were plant lysates that had the cell debris removed by a 10,000 × g centrifugation. The bands corresponding to the CP and Rubisco are identified to the right of the gel images. Below the larger gel images are images of Coomassie blue-stained CP from virions that were purified through a sucrose density cushion. (C) Effects of a 1.0 OD595 inoculum of Agrobacterium expressing empty vector, Pus4, or App1. (D) Replication-independent encapsidation of BMV RNA. RNA R3CP, which contains the CP coding sequence and the 3′ UTR of RNA3, is capable of being encapsidated but incapable of replication. R3CP was infiltrated into N. benthamiana along with the CP and either the empty vector, Pus4, or App1. The Western blot result shows signals for the CP in the plant lysates or sucrose density gradient-enriched virions probed with antisera to the BMV CP. Rubisco from the total lysates is shown below the image of the autoradiogram used to purify the virions.
To analyze BMV virion formation, lysates from the third, fourth, and fifth leaves were centrifuged through a sucrose cushion, which prevents the sedimentation of the free CPs. The virions were then detected in denaturing protein gels. Again, no CP was detected in the fourth or fifth leaves in the presence of App1 or Pus4 (Fig. 4B). In fact, we noted that a more severe decrease in the level of the virions in comparison to the CP level, suggesting that the CP produced did not successfully form virions. To examine this further, a plant was infiltrated to express a 5× higher level of Pus4 or App1. Although the overall CP level had only a modest reduction in comparison to the control, a more severe reduction in the virion formation was observed in the presence of Pus4 (compare Fig. 4 C to B), consistent with the idea that Pus4 affected virion formation.
Because Pus4 reduced BMV RNA levels as well as virion production, we seek to distinguish whether its effect on RNA encapsidation requires RNA replication. To do this, we used a truncated version of BMV RNA3, R3CP, which was replication-defective, but could serve as template for translation of the CP and encapsidation (27). The amount of CP produced from R3CP is much lower than that from the replication-competent BMV RNA3, thus necessitating detection of the CP by Western blots. Western blots of the virions that sedimented through a sucrose cushion showed that the empty vector and App1 did not reduce virion formation, whereas Pus4 did so by >90% (Fig. 4D). These results indicate that Pus4 could affect BMV encapsidation independent of RNA replication. Furthermore, App1's effect on systemic spread occurs by a different mechanism than through encapsidation.
Pus4 Affects Virion Formation by Competition Against the CP.
We hypothesize that Pus4 may compete with the BMV CP to affect plus-strand RNA encapsidation. To test this, BMV CP was printed onto glass slides and its binding to Cy3-SLD4 in solution was determined in the presence of competitor proteins. BSA and Qri1 had minimal effects on SLD4 binding to the CP, whereas App1 also had a modest effect (Fig. 5A). In contrast, Pus4 at a molar ratio of 1:1 or 2:1 to Cy3-SLD4 reduced CP binding to SLD4 to 45% and 28%, respectively (Fig. 5A). These results suggest that Pus4 and the CP bind to the same or overlapping features within SLD4.
Fig. 5.
Mechanism of inhibition by Pus4. (A) The competitive binding of Pus4 and the BMV CP to SLD4. The purified BMV CP was printed on the glass slides as 12 arrays with quadruplicate per array. For each binding assay, duplicate arrays were incubated with Cy3-SLD4 and competitor proteins. The Cy3-SLD4 was premixed with BSA (0.2 mg/ml), Qri1, App1, or Pus4 at 1:1 or 2:1 (protein to SLD4) molar ratios. The median of fluorescence density (F532) was used as binding signal. Each data point was averaged from eight binding signals (±SEM). (B) The binding of Pus4 to SLD4 versus mSLD4. The purified Pus4 were printed on the glass slides as 12 arrays with quadruplicate per array. Each binding assay was performed in duplicate with Cy3-SLD4 or Cy3-mSLD4 at concentrations of 0.2, 0.5, 2.0, and 10 μM. The median of fluorescence density (F532) was taken as the binding signal. Each data point was averaged from binding signals of eight spots (±SEM). (C) The active site of Pus4 is not required for inhibition of the plus-strand BMV RNA3 and RNA4 accumulation. The Agrobacterium cultures expressing Pus4, mPus4, and App1 were infiltrated at an OD595 of 0.5. (D) Electron micrographs of the uranyl acetate-stained images from BMV virion reassembly reactions performed in the presence of BSA or Pus4 at equal concentration to the CP. (Scale bar, 50 nm.) (Insets) Averaged images of 50 individual particles from the reassembly reactions. Numbers at the bottom of the micrographs are the means and one standard error from four independent micrographs of each reassembly reaction.
To better define the binding of Pus4 and the CP, we determined their binding affinities for SLD4 and mSLD4. Pus4 and CP printed on glass slides were incubated with increasing concentrations of SLD4 or mSLD4. Pus4 bound SLD4 with a Kd of 0.4 μM, whereas the CP has a Kd of 0.7 μM (SI Fig. 6B). Furthermore, an independently performed fluorescent anisotropy experiments Pus4 was able to bind SLD4 with a Kd of 0.9 μM. A Kd for mSLD4 could not be determined because the binding isotherm would not reach saturation even at the highest concentration of mSLD4 tested (2 μM). These results are consistent with the hypothesis that the Pus4 could compete with the CP to bind to plus-strand BMV RNAs and prevent virion formation.
To examine whether the RNA modification activity of Pus4 is required to inhibit BMV RNA levels, we mutated aspartate 75 of Pus4 to an alanine. This residue is required for Pus4 to recognize uridine (22). The mutant construct, named mPus4, reduced the BMV RNA3 and RNA4 to a level comparable to that seen with wild-type Pus4, suggesting that enzymatic activity of Pus4 is not critical for its inhibition of BMV infection (Fig. 5C). In this experiment, the BMV replication enzymes 1a and 2a were expressed transiently from mRNAs, and the more severe effect on RNA3 level may reflect the absence of RNA1 and RNA2 that could titrate out some of the Pus4 molecules.
Lastly, to confirm that Pus4 could affect virion formation, we examined whether Pus4 would affect the in vitro assembly of BMV virions. BMV RNA3 was mixed with a 200 molar excess of dissociated CP and either an equivalent amount of BSA or Pus4. The mixture was then dialyzed for 20 h according to the protocols of Dixit et al. (37) to allow the reassembly of the virions before adding different concentrations of the mixtures onto grids for electron microscopy. The virions formed in the presence of BSA were indistinguishable from those formed in the presence of Pus4 (Fig. 5D). However, the number of virions formed in the presence of Pus4 was 25-fold greater in the sample without Pus4, indicating that Pus4 inhibited BMV virion reassembly in vitro. Similar results were obtained in an independent experiment.
Discussion
In a screen of >5,000 purified proteins on a yeast proteome chip, we identified several that preferentially bind SLC, an RNA containing a CAM. This RNA is the founding member of an RNA triloop in which two of the loop nucleotides are in the central stack (38, 39) and is the core promoter for BMV minus-strand RNA synthesis (13, 16). Eleven of the top 12 proteins were confirmed to bind RNA by UV cross-linking assays (Fig. 2A) and Pus4 preferentially recognized the CAM-containing SLD4 in solution (Fig. 5B). Finally, when expressed in plants, two of the proteins, Pus4 and App1, had mild inhibitory effects on BMV plus-strand RNA levels (Fig. 2), but a dramatic effect on BMV systemic infection in N. benthamiana plants (Fig. 4). The identification of proteins from arrays that bind to viral regulatory RNA elements could, therefore, greatly help in identifying proteins with antiviral activities.
An unexpected result was that less than half of the top hits were known RNA-binding proteins. Of the 12, Pus4, Trm1 (tRNA guanine methyltransferase), and MefI (mitochondria translation elongation factor G) are RNA-binding proteins, and Etr1 (2-Enoyl thioester reductase) can bind single-stranded DNA (40). Of the remaining eight proteins, Qri1, Hsp104, and Sse1 can bind nucleotides, perhaps accounting for the recognition of the CAM (41–43). The biological relevance of RNA binding by the remaining five proteins, including App1, requires additional characterizations. The use of the yeast proteome arrays has led to the identification of novel DNA-binding proteins that activated transcription (20). Our results suggest that the same would be true for the identification of previously unassigned RNA-binding proteins.
Pus4 and App1 inhibit BMV infection in plants by different mechanisms. Based on App1's function in yeast (24, 25), we speculate that it affects intracellular trafficking rather than virus assembly. Pus4 may inhibit systemic infection by BMV through a combination of effects, including decreasing the amount of plus-strand BMV RNAs (Fig. 3), competition with the BMV CP for binding to BMV RNA (Fig. 5A), and reduction in BMV virion formation in plants (Fig. 4). Furthermore, the effect of Pus4 on BMV infection may be due to RNA binding because a mutation in a conserved residue in the Pus4 catalytic pocket retained the ability to inhibit BMV RNA replication (Fig. 5C).
Because SLC is the promoter for BMV minus-strand RNA synthesis, we need to comment on the lack of an effect on minus-strand RNA levels in the presence of App1 and Pus4. BMV replication takes place within a membrane-associated, cave-like chamber that is lined with the replication-associate proteins (5, 7, 44–46). The shuttling of the BMV RNA to the replication factory requires a B-box motif at the 5′ UTR of BMV RNA1 and RNA2, and in the intercistronic region of RNA3 (47, 48). It is possible that Pus4 and/or App1 cannot gain access to BMV RNAs until the RNA exits from the cave in preparation for additional translation and/or encapsidation by the CP.
In fact, the effects of Pus4 and App1 on plus-strand RNA levels may, at least in part, be due to an improper association between the plus-strand BMV RNAs and the capsid that lead to decreased stability of the RNAs. A similar decrease in plus-strand BMV RNAs was observed with a mutation to prevent the translation of the capsid (27). Another evidence consistent with SLC having some role in encapsidation, is that the BMV 3′ UTR was reported to stimulate encapsidation (15), although the capsid binding motif(s) were not mapped in that study. All of these results together point to a testable model where the competition for binding of a small RNA motif by the BMV replicase and the CP will provide a switch to regulate the timing for BMV RNA synthesis and encapsidation.
Materials and Methods
RNA Probe Preparation and Protein Array Assay.
The RNAs were chemically synthesized (Dharmacon, Lafayette, CO) with covalently attached fluorophores Cy3 or Cy5. The RNAs were further purified by gel filtration chromatography to remove unbound dyes before determining their concentrations by spectrometry and their intactness by denaturing gel electrophoresis and staining with Toluidine blue. The yeast proteome microarrays were fabricated by printing ≈5,000 purified yeast proteins in duplicate onto the FAST slides (Whatman, Fordham Park, NJ) (49). The recombinant BMV capsid proteins were purified and added on the chips during the printing of yeast proteome microarrays. The protein chips for RNA probing were preblocked with single-strand DNA mixtures in Superblock buffer (Pierce, Rockford, IL). Cy3-SLD4 and Cy5-mSLD4 were mixed with equal molar concentrations (0.5 μM) and used to probe the protein chips. After washing with TBS-T (Tris buffered saline with 0.05% Tween 20), the slides were dried and scanned with both the Cy3 and Cy5 channels by GenePix 4000B (Molecular Devices, Sunnyvale, CA). The median of ratio (Cy3/Cy5) was used as readout for selecting positive hits. BMV virion assembly in vitro was performed with BMV RNA3 and purified capsid at a ratio of 1:200 according to the protocol in Dixit et al. (37). Pus4 or BSA were added at the level of CP. Virions were applied to glow discharged carbon-coated copper grids and stained with 1% uranyl acetate for 20 s and visualized at ×39,000 with a JEOL 1200EX electron microscope at 100 kV.
Yeast Protein Purification.
To confirm the protein–RNA binding, the clones for potential RNA motif binding proteins were picked up from the yeast library (GST-6XHis tagged) and the proteins were purified from 100-ml yeast cultures in synthetic medium (URA/raffinose) following the typical protocols for GST-protein purification (50). The yeast proteins were eluted from the beads by using the free glutathione. The purity of each protein was examined by gel electrophoresis and Coomassie blue staining and determined to be >95%. The concentrations of the proteins were determined by comparison to a known concentrations of BSA electrophoresed on the same gel.
RNA–Protein Cross-Linking Studies.
A total of 0.2 μg of each purified yeast protein was incubated with 1 nmol of a 5′-labeled SLD4 with in a buffer containing 50 mM Tris·Cl, 4 mM MgCl2, and 25 mM NaCl (pH 8.1) and irradiated with UV at 1200 mJ for 2 min. The cross-linking reactions were subjected to denaturing protein gel electrophoresis and the bands were visualized by using a phosphorimager.
In Planta Analyses.
The expression of App1 and Pus4 was driven by the CaMV 35S promoter coupled with the 5′ nontranslated leader sequence from Tobacco etch virus on a plant transformation vector, pCB-302, in N. benthamiana (51). Briefly, the ORFs of App1 and Pus4 were amplified by PCR using pairs of primers containing the restriction fragments NcoI and XbaI and cloned into the comparable sites in pCB-302. The sequences were then checked by DNA sequencing and transformed into A. tumafaciens strain C58C1 by electroporation (52). The BMV genome and other recombinant constructs were introduced into N. benthamiana as described previously (27). BMV virions were purified from agroinfiltrated N. benthamiana leaves with minor modifications from the protocol of Rao et al. (53). Briefly, leaf samples were homogenized in 1:3 ratio of extraction buffer (250 mM sodium acetate and 10 mM MgCl2, pH 4.5) and vortexed with 10% chloroform for 10 min, centrifuged briefly at 16,000 × g for 15 min, and the supernatant is layered on 10% sucrose cushion prepared in suspension buffer (50 mM sodium acetate and 10 mM MgCl2 at pH 4.5) and subjected to ultracentrifugation for 3 h at 100,000 × g. The viral pellets were dissolved in a minimal volume of suspension buffer and analyzed for PAGE and Western analysis using Amersham (Piscataway, NJ) chemiluminscence ECL kit.
Northern Analysis.
Total RNAs were extracted from 50 mg of leaf tissues as described in (27), and 4-μg equivalent total RNAs were glyoxylated and northern analyzed by using strand specific ribroprobes as described in ref. 54.
Competition Assay and Kd Measurement on Chip.
Purified BMV CP and Pus4 were printed on the glass slides as 12 arrays fit with the 16-well incubation chamber (Whatman). Each array contains quadruplicate dots of the proteins. For the competitive assay, six binding assays were conducted per slide and each slide contains two sets of the arrays. The protein arrays were incubated with Cy3-SLD4 mixed with BSA (0.2 mg/ml), Qri, App1, or Pus4 at 1:1 or 2:1 molar mass ratio to Cy3-SLD4. For the Kd measurement, four binding assays were conducted at the same time on one slide and each assay also has duplicate arrays. The protein arrays were incubated with Cy3-SLD4 or Cy3-mSLD4 at 0.2, 0.5, 2, and 10 μM. After incubation, the slides were washed, dried and scanned with the Cy3 channel by GenePix 4000B (Molecular Devices). Fluorescence density was measured at 532 nm.
Supplementary Material
Acknowledgments
C.K. acknowledges the helpful discussions from the Texas A&M Cereal Killers and National Science Foundation Grant MCB0211996. H.Z. is partially supported by the National Institutes of Health Grants U54RR020839-01, 1R21AI070740-01, and 1R01GM076102-01A1.
Abbreviations
- BMV
Brome mosaic virus
- SLC
stem loop C
- CAM
clamped adenine motif
- CP
capsid protein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611617104/DC1.
References
- 1.Knipe DM. Fields Virology. Baltimore: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 2.Samuel CE. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris RS, Liddament MT. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Ahlquist P. Nat Rev Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao C, Sivakumaran K. Mol Plant Pathol. 2000;1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 7.Noueiry AO, Ahlquist P. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 8.Janda M, Ahlquist P. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 9.Alves-Rodrigues I, Galao RP, Meyerhans A, Diez J. Virus Res. 2006;120:49–56. doi: 10.1016/j.virusres.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Proc Natl Acad Sci USA. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price BD, Rueckert RR, Ahlquist P. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Proc Natl Acad Sci USA. 2005;102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman MR, Kao CC. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 14.Barends S, Rudinger-Thirion J, Florentz C, Giege R, Pleij CW, Kraal B. J Virol. 2004;78:4003–4010. doi: 10.1128/JVI.78.8.4003-4010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi YG, Dreher TW, Rao AL. Proc Natl Acad Sci USA. 2002;99:655–660. doi: 10.1073/pnas.022618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim CH, Kao CC, Tinoco I., Jr Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 17.Kim CH, Tinoco I., Jr J Mol Biol. 2001;307:827–839. doi: 10.1006/jmbi.2001.4497. [DOI] [PubMed] [Google Scholar]
- 18.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Bilgin M, Snyder M. Annu Rev Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- 20.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 21.Dragnea B, Chen C, Kwak ES, Stein B, Kao CC. J Am Chem Soc. 2003;125:6374–6375. doi: 10.1021/ja0343609. [DOI] [PubMed] [Google Scholar]
- 22.Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y. Cold Spring Harbor Symp Quant Biol. 2001;66:147–159. doi: 10.1101/sqb.2001.66.147. [DOI] [PubMed] [Google Scholar]
- 23.Baumstark T, Ahlquist P. RNA. 2001;7:1652–1670. [PMC free article] [PubMed] [Google Scholar]
- 24.Samanta MP, Liang S. Proc Natl Acad Sci USA. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friesen H, Colwill K, Robertson K, Schub O, Andrews B. Genetics. 2005;170:555–568. doi: 10.1534/genetics.104.040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynaud A, Facca C, Sor F, Faye G. Yeast. 2001;18:273–282. doi: 10.1002/1097-0061(200102)18:3<273::AID-YEA665>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 27.Gopinath K, Dragnea B, Kao C. J Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. Proc Natl Acad Sci USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesage P, Todeschini AL. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 30.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 31.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 33.Yi G, Gopinath K, Kao C. J Virol. 2006;81:1601–1609. doi: 10.1128/JVI.01991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki N, Arimoto M, Nagano H, Mori M, Kaido M, Mise K, Okuno T. Arch Virol. 2003;148:803–812. doi: 10.1007/s00705-002-0952-x. [DOI] [PubMed] [Google Scholar]
- 35.Biemelt S, Sonnewald U. J Plant Physiol. 2005;163:307–318. doi: 10.1016/j.jplph.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Cronin S, Verchot J, Haldeman-Cahill R, Schaad MC, Carrington JC. Plant Cell. 1995;7:549–559. doi: 10.1105/tpc.7.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixit S, Goicochea N, Daniel M, Murali A, Bronstein L, De M, Stein B, Rotello V, Kao C, Dragnea B. Nano Lett. 2006;6:1993–1999. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 38.Klosterman PS, Hendrix DK, Tamura M, Holbrook SR, Brenner SE. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazoe M, Shirahige K, Rashid MB, Kaneko Y, Nakayama T, Ogasawara N, Yoshikawa H. J Biol Chem. 1994;269:15244–15252. [PubMed] [Google Scholar]
- 41.Gehring AM, Lees WJ, Mindiola DJ, Walsh CT, Brown ED. Biochemistry. 1996;35:579–585. doi: 10.1021/bi952275a. [DOI] [PubMed] [Google Scholar]
- 42.Bosl B, Grimminger V, Walter S. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 43.Yam AY, Albanese V, Lin HT, Frydman J. J Biol Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz M, Chen J, Janda M, Sullivan MJ, Ahlquist P. Mol Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. Proc Natl Acad Sci USA. 2004;101:11263–11268. doi: 10.1073/pnas.0404157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan ML, Ahlquist P. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Noueiry A, Ahlquist P. J Virol. 2003;77:2568–2577. doi: 10.1128/JVI.77.4.2568-2577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Noueiry A, Ahlquist P. J Virol. 2001;75:3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell DA, Marshall TK, Deschenes RJ. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 51.Peng CW, Dolja VV. J Virol. 2000;74:9766–9770. doi: 10.1128/jvi.74.20.9766-9770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Llave C, Kasschau KD, Carrington JC. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao AL, Duggal R, Lahser F, Hall TC. Methods Mol Genes. 1994;4:216–236. [Google Scholar]
- 54.Hema M, Kao C. J Virol. 2004;78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.