Abstract
DNA barcoding should provide rapid, accurate and automatable species identifications by using a standardized DNA region as a tag. Based on sequences available in GenBank and sequences produced for this study, we evaluated the resolution power of the whole chloroplast trnL (UAA) intron (254–767 bp) and of a shorter fragment of this intron (the P6 loop, 10–143 bp) amplified with highly conserved primers. The main limitation of the whole trnL intron for DNA barcoding remains its relatively low resolution (67.3% of the species from GenBank unambiguously identified). The resolution of the P6 loop is lower (19.5% identified) but remains higher than those of existing alternative systems. The resolution is much higher in specific contexts such as species originating from a single ecosystem, or commonly eaten plants. Despite the relatively low resolution, the whole trnL intron and its P6 loop have many advantages: the primers are highly conserved, and the amplification system is very robust. The P6 loop can even be amplified when using highly degraded DNA from processed food or from permafrost samples, and has the potential to be extensively used in food industry, in forensic science, in diet analyses based on feces and in ancient DNA studies.
INTRODUCTION
DNA barcoding is a relatively new concept (1,2), aiming to provide rapid, accurate and automatable species identifications by using a standardized DNA region as a tag (3). As recently pointed out by Chase et al. (4), there are two categories of potential DNA barcode users: taxonomists and scientists in other fields (e.g. forensic science, biotechnology and food industry, animal diet).
According to the current technology, the ideal DNA barcoding system should meet the following criteria. First, it should be sufficiently variable to discriminate among all species, but conserved enough to be less variable within than between species. Second, it should be standardized, with the same DNA region as far as possible used for different taxonomic groups. Third, the target DNA region should contain enough phylogenetic information to easily assign species to its taxonomic group (genus, family, etc.). Fourth, it should be extremely robust, with highly conserved priming sites, and highly reliable DNA amplifications and sequencing. This is particularly important when using environmental DNA where each extract contains a mixture of many species to be identified at the same time. Fifth, the target DNA region should be short enough to allow amplification of degraded DNA. Unfortunately, such an ideal DNA marker does not exist. However, for different category of users (i.e. taxonomists versus scientists in other fields), the five criteria listed above will not be equally important. For example, a high level of variation with sufficient phylogenetic information will be most important for taxonomists. In contrast, the levels of standardization and robustness will be most important in forensics or when analyzing processed food.
So far, methodological papers published on DNA barcoding have typically dealt with the most suitable region of the genome according to the taxonomists' point of view [e.g. Ref. (5–7)]. In animals, the 5′ fragment of the mitochondrial gene for the cytochrome oxidase subunit I (COI or COXI) represents a good candidate [e.g. Ref. (5,8,9)]. However, thereis no consensus in the scientific community, and 16S rRNA, another mitochondrial gene, or the nuclear ribosomal DNA have also been proposed as useful barcoding markers (7,10). In plants, the situation is much more difficult, because both the mitochondrial and chloroplast genomes are evolving too slowly to provide enough variation. For taxonomists, the current strategy is to sequence several DNA regions (4), including both nuclear and chloroplast fragments such as the internal transcribed spacer (ITS) region of the 18S–5.8S–26S nuclear ribosomal cistron (11) or the chloroplast trnH–psbA region (6).
In this study, we approach the plant DNA barcoding problem in another way, by emphasizing the point of view of scientists other than taxonomists, looking for standardized and robust methodologies. For this purpose, we must find a genome region as variable as possible, but bearing the possibility of designing highly conserved PCR primers that amplify a very short DNA region, of no more than 100–150 bp. Such a short region should allow reliable amplifications of even highly degraded DNA found in processed food or in fossil remains. Up to now, when working with substrates such as ancient DNA, the strategy has been to use primers based on the chloroplast rbcL gene (12), but this system only allows in most cases the identification of families, not genera or species.
The chloroplast trnL (UAA) intron may represent a good target region for our purpose. Its sequences have been widely used for reconstructing phylogenies between closely related species (13–15) or for identifying plant species (16,17). Nevertheless, it is widely recognized that it does not represent the most variable non-coding region of chloroplast DNA (18), but it bears some unique advantages. Universal primers for this region were designed ∼15 years ago (19), and subsequently extensively used, mainly in phylogenetic studies among closely related genera and species (20). The evolution of the trnL (UAA) intron has been thoroughly analyzed and is well understood (21,22). Furthermore, this region is the only Group I intron in chloroplast DNA (23,24). This means that it has a conserved secondary structure (25,26) with alternation of conserved and variable regions (22). As a consequence, the alignment of diverse trnL intron sequences might allow the design of new versatile primers embedded in conserved regions and amplifying the short variable region in between.
More specifically, our objective in this paper is to evaluate the power and the limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding, and to assess the possibility for designing a new system allowing species identification with highly degraded DNA.
MATERIALS AND METHODS
General strategy
The power and the robustness of the trnL intron for DNA barcoding were first evaluated with the data available in GenBank. Then, they were evaluated on two specific datasets by sequencing the whole intron for more than 100 plant species originating from the same environment, and by compiling sequences of the main plants used in the food industry. Finally, we tested the robustness of a new pair of internal primers applied on different substrates supposed to contain highly degraded DNA.
Primer used
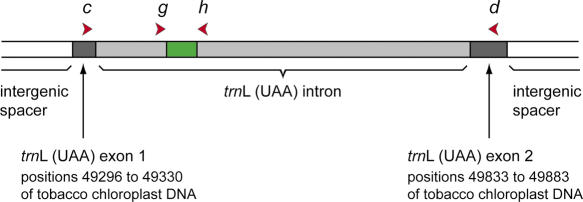
Figure 1 presents the location of the primers in the chloroplast trnL (UAA) gene, and Table 1 gives their sequences. The primers c and d are from Taberlet et al. (19). This fragment encompasses the entire trnL (UAA) intron plus a few base pairs on each side belonging to the trnL (UAA) gene itself. The primers g and h were designed for this study on two highly conserved regions after aligning various sequences, either from GenBank or produced earlier in the Grenoble laboratory.
Figure 1.
Position of the primers c, d, g and h on the chloroplast trnL (UAA) gene. The P6 loop amplified with primer g and h is indicated in green.
Table 1.
Sequences of the two universal primer pairs amplifying the trnL (UAA) intron
| Name | Code | Sequence 5′–3′ |
|---|---|---|
| c | A49325 | CGAAATCGGTAGACGCTACG |
| d | B49863 | GGGGATAGAGGGACTTGAAC |
| g | A49425 | GGGCAATCCTGAGCCAA |
| h | B49466 | CCATTGAGTCTCTGCACCTATC |
Length of the amplified fragment with primers c–d in tobacco: 456 bp. Length of the amplified fragment with primers g–h in tobacco: 40 bp. The code denotes the 3′-most base pairs in the published tobacco cpDNA sequence (23). Primers c and d are from Taberlet et al. (19). Primer g and h were designed for this study (France patent no 2 876 378; April 14, 2006).
The Arctic plant dataset
We analyzed 123 arctic plant samples collected between 1998 and 2003, partly taken from herbarium specimens and partly from field-collected, silica-dried leaf samples deposited at the Natural History Museum in Oslo. Total DNA was extracted from around 10 mg of dried leaf tissue with the DNeasy 96 Plant Kit (Qiagen), following the manufacturer's protocol. Double-stranded DNA amplifications were performed in volumes of 25 μl containing 2.5 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer and 1 U of AmpliTaq Gold® DNA polymerase (Applied Biosystems). The trnL (UAA) intron was amplified with primers c and d (19). Following an activation step of 10 min at 95°C for the enzyme (Applied Biosystems specification), the PCR mixture underwent 35 cycles of 30 s at 95°C, 30 s at 50°C and 2 min at 72°C on a GeneAmp PCR system 2720 (Applied Biosystems). To remove excess primers and deoxynucleotide triphosphates after amplification, PCR products were purified on QIAquick PCR Purification Kit columns (Qiagen), according to the manufacturer's instructions. Sequencing was performed, on both strands, using the BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) in volumes of 20 μl containing 20 ng of purified DNA and 4 pmol of amplification primer, according to the manufacturer's specifications. Sequencing reactions underwent 25 cycles of 30 s at 96°C, 30 s at 50°C and 4 min at 60°C. Excess dye terminators were removed by a spin-column purification. Sequencing reactions were electrophoresed for 45 min on an ABI PRISM®3100 Genetic Analyzer (Applied Biosystems) using 36 cm capillaries and POP-4™ polymer.
The Food dataset
Seventy-two sequences of the main plants used in the food industry were retrieved from GenBank or sequenced following the previous protocol. For this analysis, we restricted our investigations to the short fragment amplified with the g–h primer pair.
Bioinformatic approach
PCR were simulated on the full plant division of GenBank download from NCBI server on the December 14, 2005 (ftp://ftp.ncbi.nlm.nih.gov/genbank). This release corresponds to 731 531 entries. The electronic PCR software (ePCR) was specially developed for this study. It is based on the agrep algorithm (27) that allows identifying occurrences of a small pattern (corresponding to a PCR primer) on a large text (genomic sequence) with a fixed maximum mismatch count. This strategy is more relevant than simple blast queries, which are not suitable to identify similarity on nucleic sequences when the query sequence (here oligonucleotide sequence) is too short. Our ePCR software allows specifying maximum mismatch count, minimum and maximum length of the amplified region and takes care to also retrieve taxonomic data from analyzed entries. It works on Genbank, EMBL or fasta formatted sequence files (in the latter case, taxonomic data must be encoded in a special format on the title line). The ePCR software is available for academic users upon e-mail request to Eric Coissac (eric.coissac@inrialpes.fr).
ePCR was realized on GenBank data, first with the c and d primers, second with the g and h primers, third on a short rbcL fragment with the h1aF and h2aR primers (12), and finally with eight primer pairs found in Shaw et al. (18). ePCR was also realized on the arctic plant dataset with the c and d primers (after adding the c and d sequences on each side of the sequenced PCR product), and with the g and h primers.
Next, amplicon databases constructed by the ePCR software were analyzed to extract taxonomic specificities of the amplified sequences. This analysis used the taxonomic classification provided by NCBI to assess taxonomic relationships between sequences. The main goal of this analysis was to determine the proportion of the species, genera and families unambiguously identified by the sequences amplified via ePCR. A taxon (species, genus or family) was defined as ‘unambiguously identified’ if all the sequences associated with this taxon are not found in any other taxa. To limit the influence of the taxonomic coverage of the GenBank database, we discarded genera represented by only one species and families represented by only one genus. The same measure of specificity was applied to the arctic plant dataset described above. We also assessed the intraspecific variation of the whole trnL intron and of the short P6 loop fragment by extracting, from the GenBank amplicon database constructed by the ePCR software, all the species represented by more than one entry.
Primer ‘universality’
The universality of the four primers c, d, g and h was examined by comparing their sequences with homologous sequences, either from GenBank (for primers c, d, g and h) or produced in this study (for primers g and h).
Robustness of the system for biotechnological applications
To illustrate the possibility of using the g–h primer pair in biotechnology, we retrieved from GenBank some sequences corresponding to common plant species frequently used in food industry. To demonstrate the robustness of the system using the g and h primers, we tried to amplify this fragment in several highly degraded templates, such as processed food (four samples: brown sugar from sugar cane, cooked potatoes, cooked pasta and lyophilized potage), human feces (two samples) and permafrost samples (four samples). Appropriate criteria for the retrieval of highly degraded DNA were followed (28). This included DNA extraction and PCR setup in dedicated and isolated ancient DNA facilities in Grenoble and Copenhagen, and the use of multiple extraction and PCR blank controls. Importantly, the permafrost sample had been drilled spiking the drilling apparatus with a recognizable bacterial vector (pCR4-TOPO; Stratagene) to test for contamination during drilling and handling. After arrival (frozen) in the laboratory, ∼2–3 cm of the core surfaces was removed. The outer scrape and the interior core material were subjected to DNA extractions followed by 40 cycles of PCR using vector-specific primers T3/T7. No vector contaminants were detected in the inner core extracts used for the plant DNA studies. For processed food, total DNA was extracted from 50 mg of dried material using the DNeasy Tissue Kit (Qiagen) following the manufacturer's instructions. The DNA extract was recovered in a volume of 200 μl. Total DNA was extracted according to Godon et al. (29) and to Willerslev et al. (30) for the human feces and the permafrost sample, respectively. DNA amplifications were carried out using the primers g and h in final volume of 25 μl, using 2.5 μl of DNA extract as template. The amplification mixture contained 1 U of AmpliTaq® Gold DNA Polymerase (Applied Biosystems), 10 mM Tris–HCl, 50 mM KCl, 2 mM of MgCl2, 0.2 mM of each dNTPs, 1 μM of each primer (for some experiments, the g primer was labeled with the HEX fluorochrome, or the h primer was labeled with the FAM fluorochrome), and 200 mg/ml of BSA (Roche). After 10 min at 95°C (Taq activation), the PCR cycles were as follows: 35 cycles of 30 s at 95°C, 30 s at 55°C and 30 s at 72°C, except for the sugar extract for which we performed 50 cycles, and for the amplifications with the fluorescent g primer for which we removed the elongation time in order to reduce the +A artefact (31,32). PCR products obtained with the fluorescent g or h primers were electrophoresed for 35 min on an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems) using 36 cm capillaries and POP-4™ polymer. PCR products obtained with non-fluorescent primers were either directly sequenced, or cloned (except for the permafrost samples) if the sequences obtained with direct sequencing were not readable (i.e. a mixture of different sequences).
RESULTS
The three datasets
Via the ePCR with primers c and d we retrieved 1308 sequences from GenBank, corresponding to 706 species, 366 genera and 119 families (excluding all sequences with at least one ambiguous nucleotide, and excluding genera with a single species and families with a single genera). With primers g and h, we retrieved 18 200 sequences, corresponding to 11 404 species, 4215 genera and 410 families. These 18 200 sequences give a good evaluation of the number of chloroplast trnL (UAA) intron sequences in GenBank. The much lower number obtained for the c–d ePCR is simply due to the fact that the recorded sequences do not contain the primer sequences, and thus are not ‘amplified’ via our ePCR approach. The arctic plant dataset produced for this study consists of 132 species, 58 genera and 28 families (GenBank accession nos DQ860511–DQ860642). The food dataset analyzed for primers g and h, consists of 72 species, 64 genera and 37 families retrieved from GenBank, or produced for this study (GenBank accession numbers of species sequenced for this study: EF010967–EF010973).
For all datasets, the length of the sequences amplified with c and d varies from 254 to 767 bp, and the length of the P6 loop amplified with g and h varies from 10 bp in Cuscuta indecora to 143 bp in Schoenoplectus littoralis.
Universality of primer sites
Table 1 presents the sequences of the two primer pairs c–d, and g–h. Figure 2 shows the exact positions of the four primers used in the secondary structure RNAs produced by both the trnL (UAA) exon and the trnL (UAA) intron. Primers g and h are located on highly conserved catalytic parts of the intron, leading to the amplification of the short P6 loop.
Figure 2.
Positions of the primers c and d on the secondary structure of the trnL (UAA) exon (A) and of the primers g and h on the secondary structure of the trnL (UAA) intron (B) for Nymphaea odorata [modified from Ref. (33)]. Highly conserved elements of the catalytic core (P, Q, R1, R2 and S) are located in grey boxes. The P6 loop, amplified with primers g and h, is identified by green letters. The 3′ ends of each of the four primers c, d, g and h are marked out by an arrow and their positions are identified by red letters.
Table 2 shows the variation at the priming sites. Only sequence variants with a frequency of more than 0.005 were listed. Primers c and d are highly conserved among land plants, from Angiosperms to Bryophytes. Even in some algae, this primer pair has the potential to produce PCR products. The very large number of trnL (UAA) intron sequence retrieved as well as those produced for this study allowed an extensive evaluation of the universality of primers g and h. These new primers are highly conserved in Angiosperms and Gymnosperms.
Table 2.
Sequence variation of priming site for primer c, d, g and h
Only variants at a frequency higher than 0.005 are indicated. A total of 1014 and 14 145 GenBank entries were used for the primer pairs c–d and g–h, respectively. %: percentage of sequence variants found in GenBank. Species: Example of species corresponding to the sequence variant. Acc. no.: accession number in GenBank.
Proportions of species, genera and families identified
Table 3 shows the percentage of species, genera and families properly identified using the primer pairs c–d and g–h in both the GenBank and arctic plant datasets, and the primer pair h1aF–h2aR (12). Globally, on the GenBank dataset, the entire trnL (UAA) intron and the P6 loop amplified with primers g and h allow the identification of 67.3 and 19.5% of the species without taking into account single species within a genus, respectively. However, these values are probably underestimates, because of the possibility of misidentification in GenBank (i.e. a wrong species assignment, either by misidentification of the specimen, by problems of synonymy or by PCR contamination). The ePCR using other primer pairs found in Shaw et al. (18), which amplify psbB-psbH, rpoB-trnC (GCA), rpS16 intron, trnD (GUC)-trnT (GGU), trnH (GUG)-psbA and trnS (UGA)-trnfM (CAU), never retrieved more than 100 sequences, and were not taken into account. Table 4 illustrates the sequence variation of g-h amplicons for commonly eaten plant species.
Table 3.
Percentages of species, genera and families identified using the chloroplast trnL (UAA) intron, the P6 loop of this intron and comparison with another primer pairs
| cpDNA gene and dataset | Length variation (bp)a | No. of species/genera/ families analyzedb | Species (%) | Genus (%) | Family (%) |
|---|---|---|---|---|---|
| Chloroplast trnL (UAA) intron amplified with primers c and d. GenBank dataset | 254–767 | 706/366/119 | 67.28 | 86.34 | 100.00 |
| Chloroplast trnL (UAA) intron amplified with primers c and d. Arctic plant dataset | 355–653 | 103/47/24 | 85.44 | 100.00 | 100.00 |
| P6 loop of trnL intron amplified with primers g and h. GenBank dataset | 10–143 | 11 404/4225/310 | 19.48 | 41.40 | 79.35 |
| P6 loop of trnL intron amplified with primers g and h. Arctic plant dataset | 22–83 | 106/48/25 | 47.17 | 89.58 | 100.00 |
| P6 loop of trnL intron amplified with primers g and h. Food dataset | 22–65 | 72/64/37 | 77.78 | 87.50 | 100.00 |
| P6 loop of trnL intron amplified with primers g and h. Subset of the GenBank datasetc | 10–127 | 1524/1525/244 | 24.02 | 59.48 | 90.57 |
| rbcL amplified with primers h1aF and h2aR (12). Subset of the GenBank datasetc | 91–98 | 1524/1525/244 | 15.09 | 37.51 | 68.03 |
Note that these estimates were made by taking into account genera with more than two species for the species identification, families with more than two genera for genus identification, and orders with more than two families for family identification.
aLength in base pairs excluding primers.
bExcluding families with a single genera, genera with a single species and species alone in a genus except for food dataset.
cBased on species in common between the g–h and the h1aF–h2aR datasets.
Table 4.
Example of P6 loop [trnL (UAA)] sequences of commonly eaten plant species amplified with primers g and h
| Common name | Scientific name | P6 loop sequence amplified with primers g and h | Acc. no. |
|---|---|---|---|
| Cacao | Theobroma cacao | ATCCTATTATTTTATTATTTTACGAAACTAAACAAAGGTTCAGCAAG CGAGAATAATAAAAAAAG | EF010969 |
| Beet | Beta vulgaris | CTCCTTTTTTCAAAAGAAAAAAAATAAGGATTCCGAAAACAAGAATAAAAAAAAAG | EF010967 |
| Sugarcane | Saccharum officinarum | ATCCCCTTTTTTGAAAAAACAAGTGGTTCTCAAACTAGAACCCAAAGGAAAAG | AY116253 |
| Wheat | Triticum aestivum | ATCCGTGTTTTGAGAAAACAAGGGGTTCTCGAACTAGAATACAAAGGAAAAG | AB042240 |
| Rye | Secale cereale | ATCCGTGTTTTGAGAAAACAAGGGGTTCTCGAACTAGAATACAAAGGAAAAG | AF519162 |
| Rice | Oryza sativa | ATCCATGTTTTGAGAAAACAAGCGGTTCTCGAACTAGAACCCAAAGGAAAAG | X15901 |
| Millet | Panicum miliaceum | ATCCCTTTTTTGAAAAAACAAGTGGTTCTCAAACTAGAACCCAAAGGAAAAG | AY142738 |
| Strawberry | Fragaria vesca | ATCCCGTTTTATGAAAACAAACAAGGGTTTCAGAAAGCGAGAATAAATAAAG | EF010971 |
| Apricot | Prunus armeniaca | ATCCTGTTTTATTAAAACAAACAAGGGTTTCATAAACCGAGAATAAAAAAG | EF010968 |
| Sour cherry | Prunus cerasus | ATCCTGTTTTATTAAAACAAACAAGGGTTTCATAAACCGAGAATAAAAAAG | EF010970 |
| Maize | Zea mais | ATCCCTTTTTTGAAAAACAAGTGGTTCTCAAACTAGAACCCAAAGGAAAAG | NC_001666 |
| Garden pea | Pisum sativum | ATCCTTCTTTCTGAAAACAAATAAAAGTTCAGAAAGTGAAAATCAAAAAAG | EF010972 |
| Common bean | Phaseolus vulgaris | ATCCCGTTTTCTGAAAAAAAGAAAAATTCAGAAAGTGATAATAAAAAAGG | AY077945 |
| Johnson grass | Sorghum halepense | ATCCACTTTTTTCAAAAAAGTGGTTCTCAAACTAGAACCCAAAGGAAAAG | AY116244 |
| Lettuce | Lactuca sativa | ATCACGTTTTCCGAAAACAAACAACGGTTCAGAAAGCGAAAATCAAAAAG | U82042 |
| Sunflower | Helianthus annuus | ATCACGTTTTCCGAAAACAAACAAAGGTTCAGAAAGCGAAAATAAAAAAG | U82038 |
| Wild oat | Avena sativa | ATCCGTGTTTTGAGAGGGGGGTTCTCGAACTAGAATACAAAGGAAAAG | X75695 |
| Barley | Hordeum vulgare | ATCCGTGTTTTGAGAAGGGATTCTCGAACTAGAATACAAAGGAAAAG | X74574 |
| Potato | Solanum tuberosum | ATCCTGTTTTCTGAAAACAAACAAAGGTTCAGAAAAAAAG | EF010973 |
| Tomato | Solanum lycopersicum | ATCCTGTTTTCTGAAAACAAACCAAGGTTCAGAAAAAAAG | AY098703 |
| Egg plant | Solanum melongena | ATCCTGTTTTCTCAAAACAAACAAAGGTTCAGAAAAAAAG | AY266240 |
| Radish | Raphanus sativus | ATCCTGAGTTACGCGAACAAACCAGAGTTTAGAAAGCGG | AF451576 |
| Cabbage | Brassica oleracea | ATCCTGGGTTACGCGAACAAAACAGAGTTTAGAAAGCGG | AF451574 |
Among all the amplicons retrieved from GenBank by using the ePCR software, the percentage of species represented by more than a single entry was 11% for the whole trnL intron and 14% for the P6 loop. This subset of sequences allowed to estimate the lower and upper limits of the intraspecific variability. The lower limit was estimated assuming no variation in species represented by a single entry in GenBank, and the upper limit by taking into account only species represented by more than one entry in GenBank. The intraspecific variability lies between 5.9 and 55.0% for the whole intron, and 3.4 and 24.1% for the P6 loop. However, the upper values certainly represent a large overestimation of the real values, because a single entry in GenBank might correspond to many analyzed individuals from the same species. Furthermore, for the P6 loop, the intraspecific polymorphism does not compromise the species identification in 85 cases out of 481.
Robustness of the system using the g and h primers
We obtained PCR products with 35 cycles for all the samples analyzed, except for the sugar sample, for which 50 cycles were necessary. After electrophoresis of the fluorescent PCR products, some samples gave a single peak (data not shown; sugar, cooked potatoes, cooked pasta) while all the other samples gave a multi-peak profile. The sequences obtained after direct sequencing for the three samples that gave a single peak correspond to sugarcane (Saccharum officinarum), potato (Solanum tuberosum) and wheat (Triticum vulgare). Figure 3 illustrates the multi-peak profiles obtained after electrophoresis of the fluorescent PCR products for more than 20 000 years old permafrost sample, and for a human fecal sample. The PCR products of the lyophilized potage and of the human feces were cloned and sequenced. Table 5 shows the sequences obtained after cloning the PCRproduct obtained from the lyophilized potage. Twenty-three clones were sequenced, and three species were unambiguously identified: leek (Allium porum), potato (S.tuberosum) and onion (Allium cepa). The same approach was used for the human feces, and the plant species identified are banana (Musa acuminata), lettuce (Lactuca sativa) and cacao (Theobroma cacao).
Figure 3.
Example of multi-peak profiles obtained after capillary electrophoresis of the fluorescent PCR products obtained using the g and h primers. (A) Permafrost sample drilled from Main River Ice Bluff (N.E. Siberia, 64.06N, 171.11E), between 21 050 and 25 440 years old (uncalibrated 14C years, based on AMS dating of plant macrofossils from the section); g fluorescent primer; each peak represents at least one arctic plant species. (B) Human feces sample; h fluorescent primer; three of the four main peaks have been identified after cloning and sequencing: peak 1, nonidentified; peak 2, banana (Musa acuminata); peak 3, lettuce (Lactuca sativa); and peak 4, cacao (Theobroma cacao).
Table 5.
Sequences obtained after cloning the PCR product from the lyophilized potage
| Sequence obtained 5′–3′ | Species | Number of clones |
|---|---|---|
| ATCTTTATTTTTTGAAAAACAAGGGTTTAAAAAAGAGAATAAAAAAG | Leek (Allium porum) | 19 |
| ATCCTGTTTTCTGAAAACAAACAAAGGTTCAGAAAAAAAG | Potato (Solanum tuberosum) | 3 |
| ATCTTTCTTTTTTGAAAAACAAGGGTTTAAAAAAGAGAATAAAAAAG | Onion (Allium cepa) | 1 |
Note that onion and leek belong to the same genus Allium, and that their sequences differ by a single substitution.
DISCUSSION
DNA barcoding concerns two categories of scientists: taxonomists and scientists in fields other than taxonomy (4). The goal of this paper was to evaluate the potential use of the chloroplast DNA trnL (UAA) intron for plant DNA barcoding in areas other than taxonomy. We will first discuss the drawbacks of this molecular marker, and then its advantages.
The main, and maybe the only but extremely important drawback is the relatively low resolution of the trnL (UAA) intron compared with several other noncoding chloroplast regions. This has already been pointed out in several studies (6,18). It is clear that the trnL intron does not represent the best choice for characterizing plant species and for phylogenetic studies among closely related species. Obviously, this drawback is even more dramatic when using the very short P6 loop (amplified with primers g and h), but on the same subset of species, the short P6 loop performs significantly better than the alternative system used to date when analyzing highly degraded DNA [rbcL fragment amplified with h1aF and h2aR (12)]. Finally, even if the proportion of species unambiguously identified with the P6 loop seems low (around 20%), usually only closely related species are not resolved.
It is interesting to note that the relatively low resolution of the trnL (UAA) intron is logically linked to a lower intraspecific variation, compared with other noncoding regions of chloroplast DNA (18). Nevertheless, even the short P6 loop can present some intraspecific variation, due in 21.2% of the cases to the presence of a T (or A) stretch of >10 bp long.
However, the strong drawback posed by the relatively low resolution is compensated by several advantages. First, the primers used to amplify both the entire region (c and d) and the P6 loop (g and h) are extremely well conserved (Table 2), from Bryophytes to Angiosperms for the c–d primer pair, from Gymnosperms to Angiosperms for the g–h pair. The primers g and h are much more conserved than the primers h1aF and h2aR (12) targeting a protein sequence, and thus having much more variable positions. This advantage is particularly important when amplifying multiple species within the same PCR. Second, the number of trnL (UAA) intron sequences available in databases is already very high, by far the most numerous among noncoding chloroplast DNA sequences, allowing in many cases the identification of the species or the genus. Finally, the robustness of both systems (the entire intron and the P6 loop) also represents an important advantage. This last advantage might be linked to the two previous ones, because a robust system will incite scientists to use this region, increasing the number of sequences in databases, and the robustness mainly comes from the primer universality.
Actually, in some situations, the relatively low resolution of the trnL intron can be largely compensated by the possibilities of standardization. In many situations, the number of possible plant species is restricted, reducing the impact of the relatively low resolution. In our arctic plant dataset, the number of species unambiguously identified among 123 is close to 50% for the P6 loop, and close to 85% for the entire intron. In the same way, the eaten plant species are few and taxonomically diverse, and can be identified in most cases. Even the short P6 loop allows the identification of the three commonly eaten species of the genus Solanum (potato, tomato and eggplant), which differ by a single mutation (see Table 4). However, the P6 loop does not allow the identification of the different cultivars of the same species [specifically, of Brassica oleracea (Brussels sprouts, Kohl rabi, Broccoli, etc.) or of Phaseolus vulgaris (different cultivated varieties)]. In addition, the P6 loop cannot distinguish most of the species of the genus Prunus (apricot, peach, cherry, etc.).
To conclude, the trnL (UAA) intron, despite its relatively low resolution, provide a unique opportunity for plant DNA barcoding in the biotechnology area, because of the universality of the c–d and g–h primers, of the robustness of the amplification process, and of the possibility of developing highly standardized procedures. Furthermore, the low-intraspecific variation represents an important advantage if the amplicons are detected by hybridization. Even the short P6 loop allows to gather valuable information about plant identification and will undoubtedly become the marker of choice for highly degraded template DNA. This P6 loop has the potential to be extensively used in food industry, in forensic science, in diet studies based on feces, and in permafrost analyses for reconstructing past plant communities.
Acknowledgments
This study has been financially supported by an ECLIPSE II grant (CNRS). We thank Dietmar Quandt for help with Figure 2, and Jean-Pierre Furet for extracting the DNA from human fecal samples. E.W. wants to thank Andrei Sher and James Haile for helping with sample collection, Tina Brand for assisting the lab work and The Wellcome Trust, UK and the National Science Foundation, DK for financial support. F.P. is supported by the French ‘Institut National de la Recherche Agronomique’. C.B. thanks Reidar Elven and Hanne H. Grundt for help with the arctic plant sample collection and the Research Council of Norway (grant 146 515/420) for funding. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Floyd R., Abebe E., Papert A., Blaxter M. Molecular barcodes for soil nematode identification. Mol. Ecol. 2002;11:839–850. doi: 10.1046/j.1365-294x.2002.01485.x. [DOI] [PubMed] [Google Scholar]
- 2.Hebert P.D.N., Cywinska A., Ball S.L., de Waard J.R. Biological identification through DNA barcodes. Proc. R. Soc. Lond., B. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebert P.D.N., Gregory T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005;54:852–859. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- 4.Chase M.W., Salamin N., Wilkinson M., Dunwell J.M., Kesanakurthi R.P., Haidar N., Savolainen V. Land plants and DNA barcodes: short-term and long-term goals. Philos. Trans. R. Soc. B Biol. Sci. 2005;360:1889–1895. doi: 10.1098/rstb.2005.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert P.D.N., Ratnasingham S., de Waard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kress W.J., Wurdack K.J., Zimmer E.A., Weigt L.A., Janzen D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl Acad. Sci. USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vences M., Thomas M., van der Meijden A., Chiari Y., Vieites D. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front. Zool. 2005;2:5. doi: 10.1186/1742-9994-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert P.D.N., Penton E.H., Burns J.M., Janzen D.H., Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert P.D.N., Stoeckle M.Y., Zemlak T.S., Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautz D., Arctander P., Minelli A., Thomas R.H., Vogler A.P. A plea for DNA taxonomy. Trends Ecol. Evol. 2003;18:70–74. [Google Scholar]
- 11.Álvarez I., Wendel J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 12.Poinar H.N., Hofreiter M., Spaulding W.G., Martin P.S., Stankiewicz B.A., Bland H., Evershed R.P., Possnert G., Pääbo S. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 13.Scharaschklin T., Doyle J.A. Phylogeny and historical biogeography of Anaxagorea (Annonaceae) using morphology and noncoding chloroplast sequence data. Syst. Bot. 2005;30:712–735. [Google Scholar]
- 14.McDade L.A., Daniel T.F., Kiel C.A., Vollesen K. Phylogenetic relationships among Acantheae (Acanthaceae): major lineages present contrasting patterns of molecular evolution and morphological differentiation. Syst. Bot. 2005;30:834–862. [Google Scholar]
- 15.Chen S.Y., Xia T., Wang Y.J., Liu J.Q., Chen S.L. Molecular systematics and biogeography of Crawfurdia, Metagentiana and Tripterospermum (Gentianaceae) based on nuclear ribosomal and plastid DNA sequences. Ann. Bot. 2005;96:413–424. doi: 10.1093/aob/mci188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronning S.B., Rudi K., Berdal K.G., Holst-Jensen A. Differentiation of important and closely related cereal plant species (Poaceae) in food by hybridization to an oligonucleotide array. J. Agric. Food Chem. 2005;53:8874–8880. doi: 10.1021/jf0514569. [DOI] [PubMed] [Google Scholar]
- 17.Ward J., Peakall R., Gilmore S.R., Robertson J. A molecular identification system for grasses: a novel technology for forensic botany. Forensic Sci. Int. 2005;152:121–131. doi: 10.1016/j.forsciint.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Shaw J., Lickey E.B., Beck J.T., Farmer S.B., Liu W., Miller J., Siripun K.C., Winder C.T., Schilling E.E., Small R.L. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 19.Taberlet P., Gielly L., Pautou G., Bouvet J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 20.Gielly L., Taberlet P. A phylogeny of the European gentians inferred from chloroplast trnL (UAA) intron sequences. Bot. J. Linn. Soc. 1996;120:57–75. [Google Scholar]
- 21.Quandt D., Stech M. Molecular evolution of the trnL (UAA) intron in bryophytes. Mol. Phylogenet. Evol. 2005;36:429–443. doi: 10.1016/j.ympev.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Quandt D., Müller K., Stech M., Frahm J.P., Frey W., Hilu K.W., Borsch T. Molecular evolution of the chloroplast trnL-F region in land plants. Monogr. Syst. Bot. Missouri Botanic Garden. 2004;98:13–37. [Google Scholar]
- 23.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K., et al. The complete nucleotide sequence of tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer J.D. Plastid chromosomes: structure and evolution. Cell Cult. Som. Cell Genet. Plants. 1991;7A:5–53. [Google Scholar]
- 25.Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982;64:867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- 26.Davies R.W., Waring R.B., Ray J.A., Brown T.A., Scazzocchio C. Making ends meet—a model for RNA splicing in fungal mitochondria. Nature. 1982;300:719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- 27.Wu S., Manber U. Agrep-a fast approximate pattern-matching tool. Proceedings of the USENIX Winter 1992 Technical Conference; Berkeley, CA: USENIX Association; 1992. pp. 153–162. [Google Scholar]
- 28.Willerslev E., Cooper A. Ancient DNA. Proc. R. Soc. Lond. B. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godon J.J., Zumstein E., Dabert P., Habouzit F., Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willerslev E., Hansen A.J., Binladen J., Brand T.B., Gilbert M.T.P., Shapiro B., Bunce M., Wiuf C., Gilichinsky D.A., Cooper A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science. 2003;300:791–795. doi: 10.1126/science.1084114. [DOI] [PubMed] [Google Scholar]
- 31.Brownstein M.J., Carpten J.D., Smith J.R. Modulation of non-templated nucleotide addition by Taq polymerase: primer modification that facilitate genotyping. BioTechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 32.Magnuson V.L., Ally D.S., Nylund S.J., Karanjawala Z.E., Rayman J.B., Knapp J.I., Lowe A.L., Ghosh S., Collins F.S. Substrate nucleotide-determinated non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. BioTechniques. 1996;21:700–709. doi: 10.2144/96214rr03. [DOI] [PubMed] [Google Scholar]
- 33.Borsch T., Hilu K.W., Quandt D., Wilde V., Neinhuis C., Barthlott W. Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J. Evol. Biol. 2003;16:558–576. doi: 10.1046/j.1420-9101.2003.00577.x. [DOI] [PubMed] [Google Scholar]