Abstract
HIV-1 integrase (IN) catalyses integration of a DNA copy of the viral genome into the host genome. Specific interactions between retroviral IN and long terminal repeats (LTR) are required for this insertion. To characterize quantitatively the influence of the determinants of DNA substrate specificity on the oligomerization status of IN, we used the small-angle X-ray scattering (SAXS) technique. Under certain conditions in the absence of ODNs IN existed only as monomers. IN preincubation with specific ODNs led mainly to formation of dimers, the relative amount of which correlated well with the increase in the enzyme activity in the 3′-processing reaction. Under these conditions, tetramers were scarce. Non-specific ODNs stimulated formation of catalytically inactive dimers and tetramers. Complexes of monomeric, dimeric and tetrameric forms of IN with specific and non-specific ODNs had varying radii of gyration (Rg), suggesting that the specific sequence-dependent formation of IN tetramers can probably occur by dimerization of two dimers of different structure. From our data we can conclude that the DNA-induced oligomerization of HIV-1 IN is probably of importance to provide substrate specificity and to increase the enzyme activity.
INTRODUCTION
Replication of retroviruses depends on integration of a double-stranded DNA copy of the retroviral genome into the host cell nuclear genome [reviewed in (1)]. The integration step is catalysed by the retroviral enzyme integrase (IN), whose recognition sequence is located at the ends of the viral long terminal repeats. This LTR sequence is critical for site-specific cleavage and integration (2,3). Human immunodeficiency virus type (HIV-1) IN catalyses two reactions in order to insert both ends of the proviral DNA into the host cell genome: (i) the 3′-processing, in which the two nucleotides (GT) from the 3′-ends of linear viral DNA are removed, leaving CA dinucleotide at each 3′-end; (ii) the strand transfer or joining reaction in which the processed viral DNA ends are inserted into the host DNA.
HIV-1 IN possesses three independent structural and functional domains as determined by structural, complementation and mutational analyses. The amino-terminal domain (residues from 1 to 50) contains a conserved HHCC motif that binds to one atom of zinc (4). This region is involved in protein–protein interactions and may contribute to the specific recognition of viral DNA ends (5). The central catalytic core domain (residues 50–212) contains a D,D(35)E motif conserved among retroviral and retrotransposon INs (6). This motif is essential for the catalysis. The carboxy-terminal domain (residues 213–288) is involved in non-specific DNA binding and IN oligomerization necessary for the integration process (7,8).
While numerous structures of IN catalytic core-containing fragments have been shown to be dimeric (9–11), the structure of neither a full-length IN nor an IN · DNA complex has been determined. Although it is not yet known how the three domains are positioned in the active oligomeric enzyme, a higher-order complexity has to be invoked if the conformation is to be reconciled with an enzymic activity capable of concerted integration. In comparison with the Mu transposase, which is structurally homologous to HIV-1 IN (12), it has been proposed that a tetramer of IN is required for the integration reaction (13). The most recent structures point to the existence of a tetramer (a dimer of dimers) (11). Moreover, tetramers have been isolated from human cells expressing HIV-1 IN (14). Recently it was shown that HIV-1 IN forms stable synaptic complexes in which a tetramer of IN is stably associated with a pair of viral DNA ends. The results define the series of stable nucleoprotein complexes that mediate retroviral DNA integration and show that active dimers are part of tetrameric forms of IN (15).
In solution, HIV-1 IN exists in a dynamic equilibrium of monomers, dimers, tetramers and high-order oligomers (16). We have recently analysed the activity of different purified oligomeric forms of recombinant IN obtained after stabilization by platinum crosslinking and shown that these forms do not share the same in vitro catalytic properties (17). While monomers were inactive for all specific IN activities, dimers were able to catalyse the 3′-processing and the insertion of only one LTR into a short target DNA. In contrast, a tetramer of IN catalysed the full-site integration of the two viral LTR ends into a target DNA.
Time-resolved fluorescence anisotropy studies showed that the most competent form for catalysis corresponds to a dimer bound to one viral DNA end, whereas high-order complexes such as aggregates predominate during the second phase when activity drops off (18). It was supposed that a single dimer is required for 3′-processing, with a dimer of dimers responsible for the subsequent full integration.
Substrate recognition by IN is critical for retroviral integration (reviewed in (19)). In order to catalyse cleavage and strand transfer reactions, IN must recognize the viral DNA ends in a sequence-specific manner and the host target DNA in a sequence-independent manner. The most important sequence feature specifying the viral attachment site is a CA/TG dinucleotide pair, invariably found at the site where it joins to the host DNA. The presence of CA immediately upstream of the cleavage site in the LTR is a highly conserved feature of all retroviruses. These conserved bases are crucial for the recognition of viral DNA ends by IN. We have previously analysed some mechanistic aspects of substrate specificity for HIV-1 IN by studying the effect of oligodeoxynucleotides (ODNs) on the 3′-processing reaction (20,21). The ODNs were single- or double-stranded molecules of different lengths and with sequences, either related (specific) or unrelated (non-specific) to the HIV-1 U5 end of the LTR. All ODNs were able to interact with IN, although the enzyme affinity was ∼10–20-fold higher for specific than for non-specific ODNs of the same length. Non-specific ODNs were found to competitively inhibit the processing reaction. In contrast, preincubation of IN with specific ODNs resulted in an activation of the 3′-processing reaction. There are various possible reasons for this activation: (i) the influence of specific ligands on the oligomeric form of IN; (ii) stimulation of the transition between an inactive IN molecule to an active one due to complex formation with specific ODNs; (iii) a change in the enzyme conformation leading to optimal catalysis.
When IN was used in an ‘activated form’ (obtained by preincubation of IN with Mn2+ before initiating the processing reaction), specific ODNs were also found to be competitive inhibitors towards the DNA substrate. Under these conditions, a significant inhibition was obtained in the presence of nanomolar concentrations of specific ds ODNs (20,21).
Several lines of evidence suggest that the oligomeric state of IN may be important for discrimination between specific and non-specific DNAs (16,17,22,23). DNA-induced formation of HIV-1 IN oligomers have been reported previously using fluorescence fluctuation spectroscopy (24). We have recently shown that preincubation of IN with specific DNA leads to an increase in the relative amount of Pt2+-crosslinked dimeric and tetrameric of the enzyme bound to DNA (17).
In this work we have investigated the mechanism of formation of IN oligomers using small-angle X-ray scattering (SAXS). This technique is a powerful tool that yields information on the overall shape, size and structure of biological macromolecules in solution. SAXS provides not only models of particle shapes, but also answers to important functional questions. Its application is thus particularly useful in studying enzymes where large structural or conformational changes take place in the presence of their substrates. The SAXS approach has allowed us to directly observe the different steps in the IN oligomerization process due to the presence of specific ODNs. Kinetic SAXS experiments allowed us to analyse the structural changes of IN in response to protein–ligand interactions, and to study the kinetics of oligomerization. We showed that once bound to DNA, IN undergoes substantial oligomerization.
MATERIALS AND METHODS
Materials
Reagents were purchased from Sigma and ICN. Electrophoretically homogeneous HIV-1 IN was purified from the JSC 310 protease-deficient yeast strain transformed with the IN expression plasmid pHIV1SF2IN as previously described (20).
Oligonucleotides
Synthesis, purification and characterization of all homo- and hetero-ODNs were performed as described previously (25) and their concentration was determined according to (26). The following ODNs were used: non-specific ss A5 and ds A21 · T21; specific ss 5′-GCAGT and ss 5′-GTG TGG AAA ATC TCT AGC A (19-CA), ss 5′-GTG TGG AAA ATC TCT AGC AGT (21-GT), ss 5′-ACT GCT AGA GAT TTT CCA CAC (21-COM, complementary to 21-GT and to 19-CA), ds 21-GT (21-GT · 21-COM) and ds 19-CA (19-CA · 21-COM).
Enzyme assay
The ds 21-GT substrate used for 3′-end processing was prepared by annealing 21-GT and 21-COM ODNs for 2 min at 90°C, followed by slow cooling. The ds ODN was then labelled at the 3′-end with [α-32P]dGTP and [α-32P]TTP in the presence of the exonuclease-free Klenow fragment of E. coli DNA polymerase I as previously described (21,27). The IN activity was determined by following the 3′-end processing reaction at 30°C. The standard reaction mixture (20–100 µl) contained: 20 mM HEPES/NaOH (pH 7.5), 10 mM DTT, 0.1 mM EDTA, 4 mM NaCl, 7.5 mM MnCl2, 0.05% Nonidet P-40 and 1.5–3 nM 3′-end labelled ds [32P]21-GT as the substrate. The mixture was incubated for 2–60 min in the presence of 5–40 nM IN, then the reaction was stopped by transferring 10–50 µl aliquots to 40 µl of ice-cold 20 mM Tris–HCl containing 100 mM NaCl, and DNA (2 mg/ml), after thorough mixing, 1 ml of cold 8% trichloroacetic acid was added. The solutions were kept on ice for 3–4 h to allow precipitation. The precipitates were pelleted by centrifugation at 2°C for 20 min (15 000 rpm), and 1 ml of the supernatant was used to count the radioactivity. All measurements were taken within the linear regions of the time courses and the enzyme concentration curves.
Effect of ODNs on the rate of the 3′-processing reaction
To analyse the effect of ODNs on IN activity, the enzyme was preincubated (20–100 µl) at 30°C for 10–60 min under two different conditions. Conditions 1: 20 mM HEPES/NaOH pH 7.5, 10 mM DTT, 0.1 mM EDTA, 50–100 mM NaCl, 2 mM CHAPS, 3% glycerol, 0.05% Nonidet P-40 and 10 nM to 30 µM IN. Conditions 2: 50 mM HEPES/NaOH pH 7.5, 2 mM DTT, 0.1 mM EDTA, 130 mM NaCl, 7 mM CHAPS, 5% glycerol, 0.5% Nonidet P-40 and 40 nM to 70 µM IN. ODNs were used at different concentrations. At various time intervals, aliquots (5–10 µl) of the preincubated mixtures were diluted with a solution containing 20 mM HEPES/NaOH, pH 7.5, 10 mM DTT, 0.1 mM EDTA, 4 mM NaCl, and added to the 3′-processing reaction mixture (the final concentration of IN was 5–10 nM), and the reaction was performed as described previously.
Effect of ODNs on the oligomerization of IN
The effect of specific or non-specific ODNs was analysed by preincubating IN for 30–60 min at 30°C in 40 µl of the solution described earlier (see condition 2) and different concentrations of ODNs. Then the SAXS patterns were obtained.
Small-angle X-ray scattering
SAXS patterns were obtained with a Siemens diffractometer (Germany) by step-by-step scanning using a goniometer and an X-ray scintillation detector (28–31). Small-angle roentgenograms were measured in the angular range h = 0.013–0.22 Å−1, where h = 4π sinθ/λ, 2θ is the scattering angle, and λ is the X-ray wavelength. A special thermostated (20°C) quartz capillary cuvette (0.6 mm in diameter) with a wall thickness of 0.01 mm was used. The radiation wavelength was 1.54 Å. SAXS results were corrected taking into account background scattering, adsorption and collimation, which smoothed the X-ray data. The first step in mathematical processing of the SAXS data and computational checks of functions for size distribution of spherical particles were performed using the computer program and algorithms described earlier (31) as well as optimization programs (32). Results are reported as mean ± SD of at least three different experiments for each sample analysed. Scattering from the solution without protein (background noise) in all experiments was ≤5% of the signal from the solutions containing IN and/or ODNs. The convergence of the experimental and fitted values of I(0) at different concentrations of ODNs and IN for most experiments was within 2–3% except for several experiments with a 4–5% difference in these values.
RESULTS
Conditions of IN study by SAXS technique
In general, the concentration of the ds GT-21 substrate (1–2 nM) in the reaction mixture for 3′-processing reaction is usually ∼10–500 times lower than that of IN (10–500 nM) (1,20,21). An increase in the IN concentration to ∼1–50 µM at a fixed concentration of GT-21 (1–2 nM) leads to a linear increase in the rate of the 3′-processing reaction (21). This means that only <1–5% of IN molecules are active in the reaction and indicates low affinity of IN subunits for each other. We have shown previously that preincubation of IN with different specific ss and ds d(pN)n (n ≥ 3) containing the 3′-terminal sequences of 21-GT or 19-CA significantly increased the enzyme activity in the 3′-processing reaction, while preincubation with the GT dinucleotide and all non-specific d(pN)n (n ≥ 2) led to a significant decrease in the activity (20,21). The increase in the enzyme activity after preincubation with specific ODNs at high concentration of the enzyme was supposed to be the result of a specific shift of the dynamic equilibrium between monomeric and oligomeric forms of IN to formation of the catalytically active enzyme associates (21).
It is known that, the relative amounts of protein particles of different sizes in solution can be quantitatively estimated using the SAXS technique (28–31), but studies of different enzymes by this method demand high concentrations of proteins (2–5 mg/ml). IN is very hydrophobic, poorly soluble and in the absence of salts and non-ionic detergents it usually aggregates at high concentrations forming various oligomeric forms and precipitates (1,20,21). It was shown that even in the presence of non-ionic detergents IN forms dimers, tetramers, octamers and higher-order aggregates (16,22). Therefore, to study the ODN effect on the IN oligomeric state we need to find special conditions providing good solubility of the enzyme at high concentrations in which the spontaneous DNA-uncontrolled oligomerization of the enzyme is excluded. At the same time, under these conditions the enzyme should interact efficient with specific ODNs and their complexation should induce IN oligomerization similar to that for standard reaction mixtures.
In order to increase IN solubility we have used non-ionic detergents (Nonidet P-40, Triton X-100, CHAPS), which stimulate dissociation of non-specific associates and are usually used in the studies of IN (1), and have varied concentrations of all other components of the standard reaction mixture for the 3′-processing reaction. It was found that in a mixture containing 130 mM NaCl, 7 mM CHAPS, 5% glycerol and 0.5% Nonidet P-40 (condition 2, see ‘Materials and Methods’ section), IN is soluble at concentrations ∼1.5–2.5 mg/ml and according to SAXS data (see later) exists only in monomeric form. A decrease in the concentrations of these components to those for standard reaction mixture (condition 1, see ‘Materials and Methods’ section) led to formation of a remarkable amount of dimeric and tetrameric forms of the enzyme. In addition, keeping of the enzyme (∼1.5–2.5 mg/ml) in condition 1 during several hours led to a formation of an insoluble precipitate.
We have compared the level of the enzyme activation after its preincubation under the previously described (20,21) condition 1 (see Methods) and the new condition 2 described earlier. We have revealed a decrease in the previously observed activation effect of ss 21-GT (100%) and ss 19-CA (225%) (20,21) to ∼35–40 and ∼80–90%, respectively. Under conditions 2 according to the SAXS data the formation of the enzyme oligomers occurred only after IN preincubation with specific ODNs (see later) leading to an increase in the enzyme activity.
It is known, that IN-dependent reactions proceed in the presence of Mn2+ or Mg2+ ions (1). In addition, preincubation of IN in the presence of Mn2+ ions leads to an activation of the enzyme in the 3′-processing reaction (1,20,21). It was reported that, in the presence of Mn2+ ions, tetramer formation increases even in the presence of DNA (13). However, a fluorescence anisotropy assay carried out in the presence and in the absence of Mg2+ have shown that DNA-binding activity of IN is not strictly dependent on Mg2+ (22), consistent with other studies (33–35). According to (16) IN aggregation is favoured by the absence of Me2+ ions. At the same time, IN catalyses the 3′-processing and integration reactions only in the presence of Me2+ ions (1).
The enzyme activation after preincubation with MnCl2 was remarkably lower that that for specific ODNs (20,21). Preincubation of IN under conditions 1 simultaneously with ODNs and Mn2+ ions led to a small increase in the ODN effect on the enzyme activity (21). Similar results were obtained under conditions 2, the activation effect increasing to 45–50 and 95–105% for 21-GT and 19-CA, respectively. These data speak in favour that in the presence of specific ODNs Me2+ ions do not play a crucial role in the enzyme activation and formation of its catalytically active oligomeric forms. At the same time, during storage the addition of Mn2+ or Mg2+ ions to solutions corresponding to conditions 2 led to a decrease in the stability of the enzyme solutions and to a precipitate formation. Therefore, Me2+ ions were excluded from the reaction mixture. After the enzyme preincubation under conditions 2 in the absence of specific ODNs, the relative catalytic activity of IN in the 3′-processing reaction (after dilution ∼103-fold) was decreased as compared with the non-preincubated enzyme only ∼2.5–3-fold. This indicates that the standard non-preincubated solution of IN contains preformed catalytically active oligomeric forms but the concentration of these forms is relatively low. At the same time, the efficient catalysis of the 3′-processing reaction by the enzyme preincubated under conditions 2 shows that after the IN dilution and a significant decrease in the concentration of non-ionic detergents and salts, IN monomers are capable to form catalytically active oligomeric forms even at significantly lower concentrations of the enzyme. Thus, we have found specific condition under which there was no ODN-independent formation of typical IN dimeric, tetrameric and aggregative forms, and all interactions between the enzyme monomers were ODN-dependent. These specific model conditions 2 were used to study the regularities of the dynamic equilibrium between IN monomeric and oligomeric forms by the SAXS technique.
Interaction of HIV-1 IN with DNA analysed by SAXS
Macromolecular interactions of IN with ODNs were investigated using the SAXS technique. In this approach, the intensity of X-ray scattering values are correlated with the average sizes of scattering particles by the Guinier equation [36]:
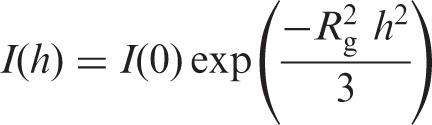 |
1 |
where I(h) is the scattered intensity, I(0) is the extrapolated scattered intensity at the zero scattering angle of diffraction (h = 0), and Rg is the radius of gyration of the scattering particles. At the zero angle of diffraction the dependence of I(0) on the concentration of the scattering particles is described by the equation:
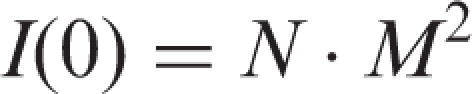 |
2 |
where M is the molecular mass and N is the number (or concentration) of the scattering particles. Using these equations it is possible to estimate the fractional composition of particles of any size, the concentrations of different components and their radii. The Rg values for particles homogeneous in density directly characterized their size. In the case of the particles non-homogeneous in density (maldistribution of the density within particle volume) the Rg values characterize mainly the size of a more compact part of the particles. But in any case, a change in the Rg values of analysed particles, for example proteins, after their interaction with different ligands usually characterizes a change in the size and/or density redistribution within the complex in comparison with that in the initial protein globule(s).
The interaction of IN with DNA was analysed by SAXS using different specific and non-specific ss and ds ODNs (see ‘Materials and Methods’ section).
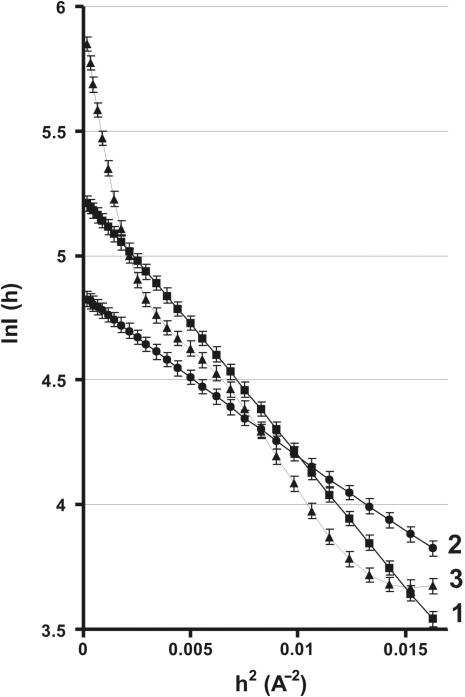
A SAXS roentgenogram in the Guinier coordinates is shown in Figure 1. In order to reach an equilibrium before measuring the X-ray scattering, either IN alone, ODN alone, or a mixture of IN plus ODN were preincubated for 30 min. We first obtained the roentgenogram of IN alone (Figure 1, curve 1) and used standard approaches to SAXS roentgenogram analysis (31). The typical smooth curvature of this line indicated that the solution contained protein particles of uniform size. Standard computer fitting of SAXS data to equations (1) and (2) together (31,36,37) employing numerical methods of optimization (32) showed that the IN solution contained only one form corresponding to a monomer with the gyration radius 16.8 ± 0.5 Å (see further for more details).
Figure 1.
Guinier plot representation of the SAXS data. The logarithmic dependencies of the intensity of X-ray scattering, ln I(h) on the diffraction angle, h2, are presented. Analysis of data was performed using the equation I(h) = I(0) exp(−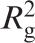 h2/3). Curve 1: 67.4 µM IN in the absence of ODNs; curve 2: 30.8 µM 21-COM alone; curve 3: 66.9 µM IN after preincubation with 30.8 µM 21-COM.
h2/3). Curve 1: 67.4 µM IN in the absence of ODNs; curve 2: 30.8 µM 21-COM alone; curve 3: 66.9 µM IN after preincubation with 30.8 µM 21-COM.
We next analysed the SAXS data obtained with a free ODN. Figure 1 (curve 2) shows the results with the specific ss 21-COM ODN. In this particular case, a specific line with smooth curvature corresponding to the presence of only one type of scattering particles was obtained. Similar results were obtained for all ODNs analysed. The different slopes of the curves 1 and 2 correspond to differences in the sizes of the X-rays scattering particles.
The effect of ligand-induced changes on IN was then investigated by preincubating the enzyme with ODNs. The SAXS roentgenogram gave a very different profile in this case, with a sharp increase in the slope of the initial part of the curve (Figure 1, curve 3). In addition to this sharp increase, various changes in the slopes were observed. All these slope changes indicated the formation of several new particles of different sizes after complexation of the enzyme with the ODN. Similar results were obtained for all other ODNs showing that different nucleoprotein complexes were formed.
More complicated SAXS roentgenograms corresponding to IN after its preincubation with different specific ODNs were analysed in the same way taking into account the SAXS data for IN and ODN incubated separately. The first step of fitting of the SAXS data was analysis of the monomer distribution between the particles with different or comparable average Rg values. For example, using SAXS roentgenogram for IN (34.7 µM; 100%) and specific GCAGT (133.3 µM) at their fixed concentrations it was found that the solution contained the following particles of different sizes which can in principle be presented as various IN forms bound to one or several molecules of the ligand (S): 82.78% monomeric form (E + ES), 2.5% dimeric form (E2S + E2S2) and 8.37% tetrameric form (E4S + E4S2 + E4S3 + E4S4).
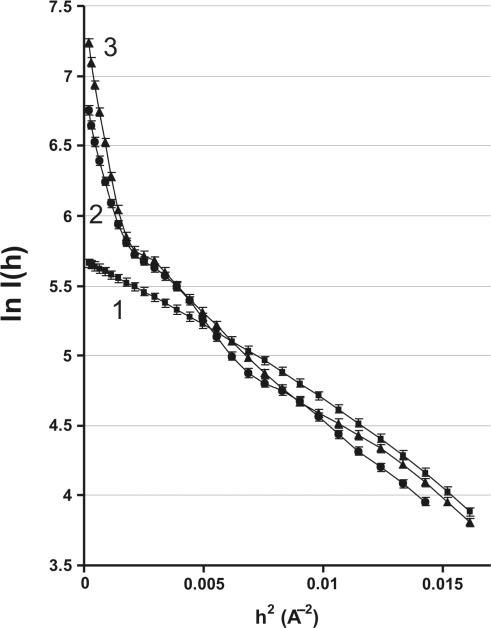
The average residual error of all calculated parameters with the experimental SAXS data corresponding to all mixtures analysed was within ∼3–4%. According to this analysis of the SAXS data the solution did not contain detectable amount of IN trimeric E3 forms (E3S, E3S2 or E3S3). Some of these IN E2 and E4 particles bound with ODNs, for example some of E4 species (E4S, E4S2, E4S3 or E4S4), in principle can either be formed or not depending on a specific pathway of their formation. Elucidating how many molecules of substrate are bound to E4 or another IN oligomeric form demands carrying out additional experiments on the dependencies of SAXS roentgenograms upon the concentration of ODNs and analysing of their agreement with possible kinetic schemes of formation of different types of IN particles and their ODN-bound forms. Therefore, at the next step we have analysed the dependencies of SAXS data on the concentration of different ss and ds specific and non-specific ODNs. Figure 2 demonstrates SAXS data for GCAGT (see futher for other details).
Figure 2.
The SAXS roentgenograms in Guinier coordinates. Curve 1: 41 µM IN in the absence of ODNs; curve 2: 40 µM IN preincubated with 30.8 µM GCAGT; curve 3: 29.8 µM IN preincubated with 228.6 µM GCAGT.
Kinetic scheme of IN oligomerization
To describe the formation of the IN oligomeric forms we have suggested several alternative and formally possible kinetic schemes taking into account different possibilities including even an effective interaction between two molecules of free IN with a formation of IN2, formation of different trimeric E3S, E3S2 and E3S3 forms, as well as different ways of a formation of various dimeric and tetrameric forms of IN bound to different numbers of ligand molecules.
The maximum stoichiometry of different forms of IN (E0–m) with different numbers of ODN molecules (S0–n) was estimated using the SAXS experimental data from the same mixtures obtained by a stepwise increase of ligand concentration and computer fitting to the highly cooperative equilibrium scheme described earlier (28,29):
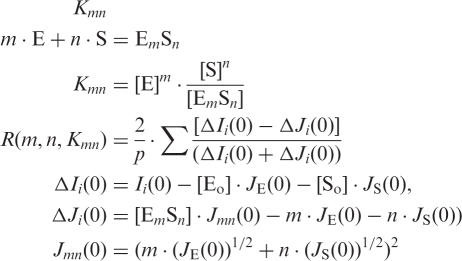 |
3 |
where Kmn is the dissociation constant for the EmSn complex; [Eo], [So], [E], [S], [EmSn] and JE(0), JS(0), Jmn(0) are the initial and equilibrium concentrations, and intensities (at h = 0) of enzyme, ODN and complex, respectively; Ii(0) is the SAXS intensity from mixture (i) of titration; ΔIi(0) and ΔJi(0) are experimental and model differential SAXS intensity (i) to the zero angle (h = 0); p is the amount of the titration mixtures; R is the optimization criterion.
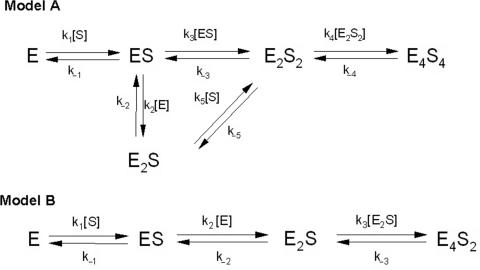
The function minimization using standard computer program (28–32) was carried out and the obtained Kd values corresponding to different kinetic schemes were compared with the experimental Kd values obtained earlier (20,21). It was shown that many kinetic models including ones taking into account a possibility of a formation of IN2 or different trimeric forms of IN (E3S, E3S2, E3S3) lead to the Kd values which are very far from the experimental Kd values. We have found that the analysed processes can be better described by only two of all kinetic models analysed, Models A and B (Figure 3). The Kd values corresponding to Model A and Model B are given in the Table 1. The Kd values corresponding only to non-specific ss T5, ds T21/A21 and short specific GCAGT calculated according to Model A were comparable with the experimental Kd values (Table 1). The Kd values calculated using the Model B for all specific ss and ds ODNs were 103–105 times higher than the experimental ones (Table 1). However, for all specific and non-specific ss and ds ODNs the Kd values calculated according to Model A were practically the same as experimental values (Table 1). Thus, it was obvious that the interaction of IN with specific ODNs and formation of oligomeric forms can be better described by the kinetic Model A, while in the case of non-specific ones one cannot exclude both ways of the complex formation (see later).
Figure 3.
Kinetic schemes of IN interaction with ODNs. Model A and B represent two possible kinetic pathways leading to formation of different nucleoprotein complexes. Oligomeric states of integrase are represented as E: monomer; E2: dimer; E4: tetramer. S corresponds to ODN substrate.
Table 1.
Kd values determined from inhibition studies, or calculated from SAXS data using models A and B
| Oligonucleotide | Kda (µM) Experimental | Kdb (µM) Model A | Kdb (µM) Model B |
|---|---|---|---|
| T5 | 43.0 ± 15 | 58.2 ± 20.0 | 70 ± 20 |
| GCAGT | 13.0 ± 4.0 | 7.1 ± 0.7 | 3.1 ± 1.6 |
| A21 · T21 | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.09 ± 0.03 |
| 19-CA | 0.03 ± 0.01 | 0.031 ± 0.015 | 273 ± 19 |
| 21-GT | 0.01 ± 0.003 | 0.065 ± 0.029 | 153 ± 17 |
| (21-GT) · (21-COM) | 0.0013 ± 0.001 | 0.0023 ± 0.001 | 315 ± 44 |
The data speak in favour of that dimeric molecules of E2S2 interact mainly to each other forming a tetramer E4S4 as dimer of dimers while a possible formation of trimeric forms, like E3S3, does not play a significant role in the analysed pathways. Interestingly, ds 21-GT and ds 19-CA are poor substrates of the integration reaction, catalysed by IN (38), and therefore they can, to some extent, serve as analogues of specific HIV DNA and non-specific host human DNA. Therefore, a formation of E4S4 complex with these ds ODNs can, to some extent, modulate the formation of IN integration complex with viral and host DNAs. The ss 19-CA and ss 21-GT are not substrates of IN-dependent reactions. At the same time, one cannot exclude that E4S4 form complexed with ss 19-CA and ss 21-GT also can modulate to some extent the integration complex formation.
Analysis of the different steps in IN oligomerization
We have analysed the dependencies of SAXS data upon the concentration of different ss and ds specific ODNs. The function minimization in the case of an established kinetic scheme makes it possible to calculate not only the Kd value for the first step of enzyme interaction with its substrate (E + S = ES, characterized by Kd in Table 1, or Kd(1) in Model A), but also to make a complete quantitative description of all steps of enzyme interaction with ligands and the formation of various oligomeric forms of the protein. Using computer fitting in agreement with the kinetic equations and the optimization approaches described previously (31,32) we have calculated the Kd, Rg and equilibrium concentrations of all forms of the enzyme characterizing the IN interaction with various ODNs. All calculated parameters for ss 21-GT are summarized in Table 2. Similar fitting was performed for all ODNs used (Tables 3–5) and in all cases the average residual error of all calculated parameters with the experimental SAXS data corresponding to all mixtures analysed was ∼3–6%. The Kd values characterizing all steps of IN interaction with various ODNs are reported in Table 3.
Step E + S = ES, Kd(1) = k1/k−1. At this step, monomeric IN demonstrated very high affinity for long specific ODNs and especially for ds 21-GT (Kd(1) of 2.2 nM). The affinity of the E form for specific long ODNs was significantly higher than for short ODNs. In addition, the calculated Kd values (Table 3) were the same as experimental Kd values (20,21) within the error in determination for all specific and non-specific ODNs.
Step ES + E = E2S, Kd(2). The complexation of ES monomer with E (free of ODNs) is characterized by the comparable Kd values for all long specific ss and ds ODNs (Table 2). However, the affinities of non-specific A5, T21 · A21, or short specific GCAGT are approximately one order of magnitude lower than those for long specific ODNs. This can indicate different changes in the IN monomer structure after its complexation with long specific and non-specific (or short specific) ODNs. In addition, the data of Table 3 suggest that although of GCAGT is a specific activator of IN, this ODN effects the IN oligomerization rather as a non-specific than specific DNA ligand. These data correlate with a remarkably lower activation effect of GCAGT as compared with long specific ODNs (20,21).
Step ES + ES = E2S2, Kd(3). As mentioned earlier, under the model conditions IN monomers free of specific ODNs do not interact forming IN2 dimers. Interestingly, even after the complexation of these monomers with long specific ss and ds or non-specific ODNs, the affinity of ES forms to each other are low and comparable for specific and non-specific ODNs.
Step E2S2 + E2S2 = E4S4, Kd(4). For specific ODNs, this step was characterized by extremely high Kd values, showing that the formation of E4S4 from E2S2 dimers is a very inefficient process. In contrast, a completely different result was obtained for non-specific ODNs. In this case, the Kd(4) values were significantly lower than those observed for specific ODNs, suggesting that the formation of E4S4 from E2S2 is more efficient for non-specific than for specific ODNs.
Step E2S + S = E2S2, Kd(5). In this case, Kd(5) values were extremely different for specific and non-specific ODNs. Specific ODNs showed high affinity for the E2S form compared with non-specific ODNs. This can reflect high sensitivity of the IN monomer (free of ODN) within E2S to ODNs with different structures. The affinities of specific ss 21-GT and specific ds ODNs to E form (E + S = ES) are comparable with or lower than those in the case of E2S form (SE − E + S = SE − ES; Table 3). This finding can speak in favour of a possibility of a change in the conformation of the IN monomer after its complexation with ES (ES + E = ES · E = ES · E*; where E is not equal to E*). This result was obtained under model conditions preventing the formation of E2. At the same time, under standard conditions, the formation of E2 form is a favoured process. Therefore, one cannot exclude that the formation of E2S complex (E2 + S = E2S) can also lead to a significant change in the properties of the second subunit after E2 dimer complexation with one molecule of ODN.
Table 2.
Equilibrium concentrations of all components of the reaction mixture and another parameters characterizing the IN interaction with single-ss 21-GT ODN
| Initial concentration (µM) | Equilibrium concentration (µM)a | ||||||
|---|---|---|---|---|---|---|---|
| E0 | S0 | E | S | ES | E2S | E2S2 | E4S4 |
| 67.4 | 31.3 | 33.1 | 0.007 | 23.1 | 3.0 | 2.57 | 0.0148 |
| 63.8 | 73.0 | 0.05 | 9.26 | 44.1 | 0.009 | 9.41 | 0.198 |
| 58.4 | 134.0 | 0.006 | 75.6 | 41.3 | 0.001 | 8.24 | 0.152 |
| 50.1 | 229.5 | 0.002 | 179.4 | 36.7 | 0.0003 | 6.51 | 0.095 |
| Gyration radii (Å) | |||||||
| E | S | ES | E2S | E2S2 | E4S4 | ||
| 16.8 ± 0.5 | 14.0 ± 0.3 | 18.5 ± 0.3 | 19.3 ± 0.2 | 33.8 ± 0.9 | 61.0 ± 0.4 | ||
| Kd values (µM) | |||||||
| Kd(1) | Kd(2) | Kd(3) | Kd(4) | Kd(5) | |||
| 0.0106 ± 0.01 | 253.6 ± 40 | 206.0 ± 4.0 | 447.0 ± 65.0 | 0.0087 ± 0.0004 | |||
aThe average error in compliance of all calculated parameters with experimental SAXS data corresponding to all mixtures analysed was ∼3.3%.
Table 3.
Kd values characterizing the interaction between the various oligomeric forms of IN during complex formation with different ODNs
| Oligonucleotide | Kd(1)a (µM) | Kd(2)a (µM) | Kd(3)a (µM) | Kd(4)a (µM) | Kd(5)a (µM) |
|---|---|---|---|---|---|
| ss A5 | 2.6 ± 0.4 | 12.0 ± 2.0 | 180 ± 86 | 10.0 ± 2.0 | 120 ± 4 |
| ss GCAGT | 7.1 ± 0.7 | 13.3 ± 1.1 | 340 ± 76 | 24.9 ± 5.6 | 183 ± 16 |
| ds A21 · T21 | 0.040 ± 0.010 | 18.0 ± 4.0 | 250 ± 80 | 17.0 ± 3.0 | 100 ± 40 |
| Ss 19-CA | 0.030 ± 0.015 | 131 ± 34 | 277 ± 41 | 2300 ± 850 | 0.66 ± 0.13 |
| Ss 21-GT | 0.0106 ± 0.01 | 253.6 ± 40 | 206.0 ± 4.0 | 447.0 ± 65.0 | 0.0087 ± 0.0004 |
| ds 19-CA or (19-CA) · (21-COM) | 0.017 ± 0.007 | 133 ± 16 | 127 ± 8 | 15090 ± 469 | 0.010 ± 0.003 |
| ds 21-GT or (21-GT) · (21-COM) | 0.0022 ± 0.001 | 209 ± 83 | 138 ± 7 | 4475 ± 154 | 0.015 ± 0.005 |
aEach Kd value corresponds to a different stage of IN oligomerization according to Model A. The average error in compliance of all calculated parameters with experimental SAXS data corresponding to all mixtures analysed was ∼3–6%.
Table 4.
Radii of gyration of different IN forms in the presence or in the absence of ODNs
| Radius of gyrationa (Å) | ||||||
|---|---|---|---|---|---|---|
| ODN | E | S | ES | E2S | E2S2 | E4S4 |
| ODNs | 16.8 ± 0.5 | – | – | – | – | – |
| Ss A5 | 16.8 ± 0.5 | 4.2 ± 0.1 | 18.6 ± 1.5 | 26.3 ± 0.5 | 46.1 ± 1.8 | 66.3 ± 4.5 |
| Ss GCAGT | 16.8 ± 0.5 | 4.5 ± 0.1 | 18.4 ± 1.6 | 25.2 ± 0.5 | 45.7 ± 1.8 | 65.2 ± 8.5 |
| Ds A21 · T21 | 16.8 ± 0.5 | 15.3 ± 0.1 | 18.5 ± 1.5 | 26.7 ± 1.5 | 48.2 ± 2.0 | 68.8 ± 4.5 |
| Ss 19-CA | 16.8 ± 0.5 | 15.9 ± 0.4 | 18.7 ± 0.4 | 20.4 ± 0.3 | 26.5 ± 0.7 | 51.2 ± 0.4 |
| Ss 21-GT | 16.8 ± 0.5 | 14.0 ± 0.3 | 17.0 ± 0.4 | 19.4 ± 0.3 | 30.6 ± 1.3 | 61.6 ± 0.5 |
| Ss 21-GT | 16.8 ± 0.5 | 14.0 ± 0.3 | 18.5 ± 0.3 | 19.3 ± 0.2 | 33.8 ± 0.9 | 61.0 ± 0.4 |
| Ss 21-COM | 16.8 ± 0.5 | 13.8 ± 0.3 | 16.9 ± 0.2 | 18.0 ± 0.2 | 32.2 ± 1.7 | 56.8 ± 1.1 |
| Ds 21-GT | 16.8 ± 0.5 | 15.2 ± 0.3 | 16.9 ± 0.2 | 17.1 ± 0.2 | 36.5 ± 0.8 | 56.8 ± 1.8 |
| Ds 19-CA | 16.8 ± 0.5 | 15.2 ± 0.3 | 16.9 ± 0.2 | 17.1 ± 0.2 | 36.5 ± 0.8 | 56.8 ± 1.8 |
aThe radius of gyration was calculated from the SAXS data. The adequacy of computer fitting of all Rg values for various ODNs was according to model A. The average error in compliance of all calculated parameters with experimental SAXS data corresponding to all mixtures analysed was within ∼3–6%.
Table 5.
Relative amounts of different IN monomeric and oligomeric forms in the presence of specific and non-specific ODNs
| Initial concentrations (µM) | Relative amount of different IN forms in equilibriuma (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Ligand | E0 | S0 | E | ES | E2S | E2S2 | E4S4 | Activationb (%) |
| ss A5 | 40.0 | 10.4 | 18.7 | 70.1 | 0.6 | 0.7 | 9.9 | (Inhibitor) |
| 39.0 | 24.6 | 8.9 | 76.5 | 0.7 | 0.9 | 13.0 | ||
| 34.0 | 48.9 | 4.5 | 79.0 | 1.2 | 1.3 | 14.0 | ||
| 29.0 | 83.2 | 3.8 | 78.4 | 1.3 | 1.5 | 15.0 | ||
| ds A21 · T21 | 40.0 | 0.17 | 19.0 | 69.4 | 0.7 | 0.9 | 10.0 | (Inhibitor) |
| 38.0 | 0.41 | 8.9 | 75.1 | 0.9 | 1.1 | 14.0 | ||
| 34.0 | 0.76 | 5.0 | 75.9 | 1.4 | 1.7 | 16.0 | ||
| 29.0 | 1.30 | 3.0 | 75.1 | 1.6 | 2.0 | 18.3 | ||
| ss GCAGT | 40.1 | 30.8 | 18.7 | 66.8 | 0.5 | 0.6 | 9.8 | 40 |
| 37.9 | 72.7 | 9.0 | 76.5 | 0.7 | 0.9 | 12.9 | ||
| 34.7 | 133.3 | 5.1 | 77.7 | 1.2 | 1.3 | 14.7 | ||
| 29.8 | 228.6 | 3.0 | 80.7 | 1.0 | 1.1 | 14.3 | ||
| ss 19-CA | 59.62 | 39.5 | 39.00 | 47.46 | 8.42 | 4.85 | 0.27 | 225 |
| 55.78 | 92.1 | 0.90 | 84.15 | 0.32 | 14.30 | 0.39 | ||
| 51.71 | 169.1 | 0.24 | 85.25 | 0.079 | 13.56 | 0.89 | ||
| 45.05 | 289.6 | 0.11 | 86.60 | 0.033 | 12.19 | 1.09 | ||
| ss 21-GT | 61.78 | 31.3 | 53.60 | 37.4 | 4.87 | 4.17 | 0.024 | 100 |
| 53.77 | 73.0 | 0.093 | 82.0 | 0.017 | 17.50 | 0.370 | ||
| 49.70 | 143.0 | 0.012 | 83.1 | 0.002 | 16.58 | 0.300 | ||
| 43.30 | 229.0 | 0.005 | 84.76 | 0.007 | 15.00 | 0.220 | ||
| ds 19-CA | 64.20 | 14.0 | 79.38 | 15.69 | 3.82 | 3.82 | ∼0.0 | 250c |
| 57.15 | 32.6 | 49.35 | 49.35 | 5.30 | 5.30 | 0.005 | ||
| 48.10 | 59.9 | 0.89 | 77.83 | 0.16 | 21.10 | 0.050 | ||
| 41.85 | 102.6 | 0.036 | 80.34 | 0.0055 | 19.60 | 0.024 | ||
| ds 21-GT | 62.92 | 15.1 | 77.23 | 15.76 | 5.78 | 1.23 | ∼0.0 | 210c |
| 55.47 | 35.3 | 43.80 | 41.20 | 7.53 | 7.43 | ∼0.0 | ||
| 47.67 | 64.7 | 0.13 | 77.37 | 0.036 | 22.47 | 0.0016 | ||
| 41.54 | 110.8 | 0.014 | 79.38 | 0.0034 | 20.60 | 0.0012 | ||
All these results indicate that after complexation with IN monomers, both specific and non-specific ODNs stimulate the formation of oligomeric forms, although these forms are different in their affinities for ODNs and for each other.
Estimation of the gyration radius of IN · ODN oligomeric forms
SAXS data depend on the size of X-ray scattering protein globules and thus each mixture containing proteins of various sizes and in different amounts gives distinct curves. The interaction of enzymes with specific ligands can lead to more or less compact structures of protein globules, as well as to changes of their forms (sphere, ellipsoid, etc.) (28–31,36,37).
We tried to find out whether different behaviour of various IN forms (Tables 2 and 3) correlated with changes in the structural properties and the radius of gyration were calculated as described earlier, reported in Table 4. In the absence and in the presence of ODNs, the IN monomer (E, free of ODN) gave an Rg of 16.8 Å. Complexation of IN monomers with ODNs led to the formation of ES species with Rg values within a similar range (16.8–18.7 Å), suggesting that all ODNs (specific or non-specific) interacted with the monomer inside its globule. When going from ES to higher nucleoprotein complexes, E2S, E2S2 and E4S4, the Rg values gradually increased, but in all cases higher values were obtained with non-specific ODNs. These results indicate that nucleoprotein complexes are more compact when IN is in the complex with specific ODNs.
Variations in Rg values can reflect a change in the size and/or a redistribution of the relative density within the complexes in comparison with those of the initial protein globule. It can be hypothesized that during interaction of both E + S = ES and E2S + S = E2S2, the Rg values of the newly formed complexes could change mainly due to the addition of one molecule of the S ligand. But the different results obtained with both groups of ODNs showed that: (i) for specific ODNs, the change when going from E to ES, gave an increase in Rg values of only 0.6–1.2%, while (ii) an interaction of E2S with S resulted in an increase of 57–113%. This suggests that specific ODNs can probably interact with IN monomers inside the enzyme globule, while a transition of the complex from E2S to E2S2 might be associated with significant structural changes of the protein structure within the dimer.
Different alternative hypotheses can be proposed to explain the increase in Rg with the successive formation of E2S, E2S2 and E4S4: (i) formation of longer ellipsoid-like structures for both subunits of the dimer; (ii) a significant movement of structural parts of the oligomers leading to a redistribution of protein density within a protein globule(s) to form less compact structures; (iii) transformation of the DNA binding site of IN in such a way that it becomes more shallow and thus ODNs can be positioned partially outside of the enzyme surface; (iv) in principle, one cannot exclude that there may be a combination of relatively small changes of IN oligomers structure specific for each complex according to all ways mentioned earlier.
For non-specific ODNs (as well as for the short specific GCAGT) their interaction with the monomeric form, E, was associated with a higher increase in Rg (ΔRg = 8.7–9.7%) as compared with specific ODNs (ΔRg = 0.6–1.2%). The complexation of E2S with the second ODN molecule (E2S + S = E2S2) resulted in an increase of 75–81%. The E2S2 dimeric forms in the case of non-specific ODNs were characterized by Rg values up to ∼82% larger than those for long specific ODNs. These data suggest that interaction of IN with non-specific ODNs led to the formation of dimeric forms of IN that are less compact, probably longer ellipsoid-like structures.
These differences in the Rg values of the ODN-induced complexes are associated with the differences in the affinity of ODNs and different IN-forms for each other (Table 3). The formation of E2S from ES in the case of non-specific ODNs occurs easier than in the case of specific ODNs. In addition, the second protein monomer within the E2S IN dimer demonstrates lower affinity for GCAGT and A5 than the first one, while the affinity of E and E2S to specific ODNs was nearly the same (Table 3). Thus, the differences in the behaviour of the E2S dimeric form correlate well with different changes in Rg values (Table 4) obtained for E2S with non-specific (25–26 Å) or specific ds ODNs (17 Å). These results provide additional evidence that the interaction of IN with specific ODNs proceeds differently than with non-specific ODNs throughout all steps of the enzyme oligomerization process.
Estimation of the relative amount of IN oligomeric forms
Table 2 gives the results obtained when IN was titrated with ss 21-GT. We observed that ES represented the main form following interaction of IN with 21-GT, and that under these special conditions the formation of E2S2 most probably arose from two ES molecules, better than through the E2S intermediate. These data also show that E4S4 was present at very low concentrations. The same experiment was performed for all other ODNs, thus allowing calculation of the relative amount (%) of all forms of IN after reaching equilibrium (Table 5). Even though in the presence of all ODNs the main form was ES, one very important difference was found between E2S2 and E4S4 depending on the ODN sequences.
In the presence of the specific ODNs, ES and E2S2 were the major species (Table 5). Their relative amount increased with concentration, length and structure of each ODN. However, an unexpected result was observed with E4S4, since extremely low concentrations of the tetrameric form were detected, suggesting that E2S2 accumulated and no fast transition to E4S4 took place.
A completely different pattern was obtained with non-specific ODNs. In this case, the relative amounts of E2S2 were very low, while notable concentrations of E4S4 were detected. Most probably E2S2 was faster transformed into E4S4 in accordance with the higher affinity of E2S2 dimers to each other [see Kd(4) in Table 3]; this may be a result of different structural properties of the E2S2 form (Rg values) for non-specific and specific ODNs (Table 4).
These data correlate with our previous results concerning IN activation by specific ODNs (20,21). In that work we found that the activity of IN was stimulated after preincubating the enzyme with low concentrations of specific ODNs and that the level of activation increased with the length of the ODNs. Moreover, specific ODNs containing CA at the 3′-end were better activators than GT-containing ODNs and the stimulation was even higher in the presence of specific ds ODNs. At the same time, preincubation of IN with non-specific ODNs led to the enzyme inhibition. It is important to note that in spite of some experimental differences (IN concentrations and detergents, see earlier) there was a close correlation between the relative amount of dimeric forms, E2S2, and the specific level of enzyme activation after preincubation with specific ODNs (Table 5).
From these results it can be concluded that preincubation of IN with specific ODNs under special conditions preventing ODN-independent formation of oligomers leads mainly to the formation of a dimeric form, E2S2. This enzymic form is most probably the one that catalyses the 3′-processing reaction and that is responsible for the enzyme activation previously described by us (20,21).
DISCUSSION
HIV-1 IN is a highly conformationally dynamic enzyme. The activity of recombinant IN in solution is related to an equilibrium between different oligomeric forms and depends on various factors such as detergents, cations and in particular the presence of DNA substrates (16,17,20–24). In this work, we used the SAXS approach to investigate the effect of specific or non-specific ODNs on IN oligomerization. Under standard conditions of IN-dependent reactions, IN monomers can form not only dimeric, but also tetrameric complexes and other different non-specific associates (16,22). It should be emphasized that under the special model conditions (see earlier) oligomerization of IN monomers in the absence of ODNs is prevented and the enzyme exists only as monomers. At the same time, a comparison of enzyme behaviour under different conditions can help to elucidate a possible contribution of specific sequence-dependent processes of IN oligomerization to a formation of catalytically active forms.
Our data revealed that under the special conditions the formation of different IN oligomeric forms was a strictly DNA-dependent process leading, after preincubation of the enzyme with specific ODNs, to its activation, while non-specific ODNs yielded catalytically inactive oligomeric species (20,21). At the same time, addition of any non-specific ss or ds d(pN)n (n ≥ 2) to the 3′-processing reaction mixture with ds 21-GT leads to a competition of specific and non-specific ligands for the same DNA-binding site and the affinity of non-specific ODNs is only ∼10–20-fold lower than that of specific ss and ds ligands of the same length (20,21). The main contribution to the total affinity of the enzyme for all specific and non-specific ODNs was observed from their interaction with the first three to four 3′-terminal nucleotide units of GT or CA-containing ss and ds d(pN)n. In particular, differences in these tri–tetranucleotide sequences mainly provide the difference in the affinity for specific and non-specific ODNs (20,21). At the same time, the level of the enzyme activation depends not only on the sequence of these terminal nucleotides, but also increases with the ODN lengths to 20–21 nucleotide units. Our studies of IN interaction with different ss and ds ODNs show that IN first binds DNA in a non-specific manner and that subsequent reorganization of the enzyme including its oligomerization can lead to a specific and catalytically competent complex before catalysis and display strong selectivity for catalysis proceeding only with specific DNA. This conclusion is in agreement with the literature data, where non-specific manner of IN interaction with different DNAs was also shown (18). In addition, we have shown that IN preincubation with specific and non-specific DNA determines various types of IN reorganization leading to IN oligomers with different Rg values (Table 4), which are capable or not of catalysis of the 3′-processing reaction.
Interaction of IN with specific ODNs led mainly to the formation of E2S2 while E4S4 were scarce. All ODNs containing CA and GT at their 3′-end induced formation of catalytically active E2S2 dimers (Table 5). However, the level of enzyme activation was variable. It increased with ODN length, the nature of the dinucleotide (CA or GT) at the 3′-end, and with the transition from ss to ds ODNs, suggesting conformation changes of IN monomers within the E2S2 dimeric form.
Non-specific ODN inhibitors, in contrast, gave a completely different result. While preincubation of IN with these ODNs led to extremely low equilibrium concentrations of dimeric forms, tetramers were present at a higher concentration and a displacement of the equilibrium to these forms occurred with an increase in ODN concentrations (Table 5).
According to the SAXS data the solution did not contain detectable amounts of IN trimeric forms either in the presence of specific or non-specific ODNs, in an agreement with the absence of trimeric forms from the data on IN chemical crosslinking (17). The formation of a trimeric form does not seem to be a thermodynamically favourable process.
Concerning the extremely low concentration of tetrameric forms observed in the presence of specific ODNs, it is important to note that the formation of E4S4 from two E2S2 dimers is not a favourable process in the case of specific long ODNs (Tables 2 and 5). The affinity of E2S2 dimers for each other is lower in the case of ds long ODNs (Tables 2 and 5). As a consequence, the relative concentrations of E4S4, when specific ds ODNs were present in the preincubation, did not exceed 0.05% (Table 5). For non-specific ODNs, in contrast, the formation of E4S4 tetramers from E2S2 dimers occurs 102–104-fold more easily, and the fraction of E4S4 forms for A5 and A21 · T21 reached ∼15–18% (Table 5).
All these data show that different types of IN oligomeric complexes are formed depending on the specific sequence of ODNs preincubated with the enzyme. In addition, the features of formation and functioning of these oligomeric forms followed different pathways leading to either active or inactive oligomers with different Rg values (Table 4).
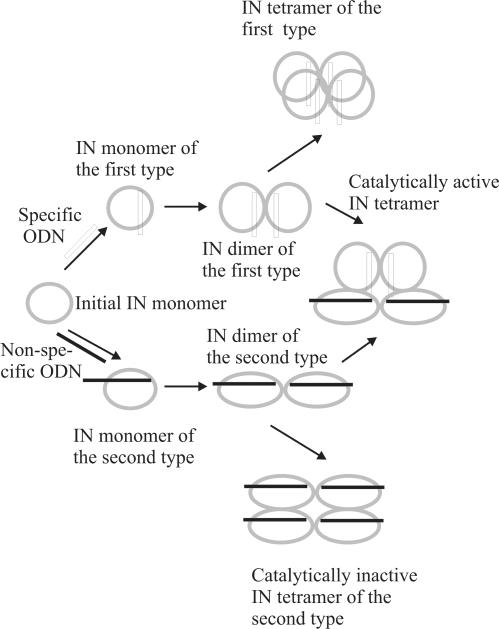
When interpreting our data, it should be taken into account that in cells and in solutions containing no specific components preventing DNA-independent oligomerization, IN can exist as E, E2 and other forms. In addition, under the conditions used in SAXS experiments, long specific ODNs interact with two molecules of IN monomers independently. Since viral DNA contains two LTR terminal sequences within one molecule, the interaction of LTR–DNA–LTR molecule with the enzyme E and E2 forms should be different as compared with two separated specific ODNs mimicking the LTR sequences. Thus, the affinity of LTR–DNA–LTR for the separated enzyme monomers can be higher since including two terminal ends to one molecule changes a kinetic pathway of the complexation process. In addition, the interaction of LTR–DNA–LTR with a preformed E2 dimer should be more efficient since the formation of contacts between the second LTR and the second monomer of E2 form will proceed as an intermolecular process. Thus, the formation of the E · LTR–DNA–LTR · E dimeric form (the interaction between two E monomers of this dimer are not shown) in normal conditions can proceed easier than that for two 3′-terminal long specific ds ODNs: LTR · E · E · LTR. However, we suppose that a preformed LTR · E · E · LTR complex can in some extent modulate a natural E · LTR-DNA-LTR · E complex and formation of tetrameric form of both of them may be an inefficient process because of a specific structure of these dimeric forms (Table 5). This complex of E2 with two molecules of specific ODNs mimicking viral DNA (LTR · E · E · LTR) and demonstrating specific kinetic and structural characteristics (Tables 3–5) will be termed below as a complex of the first type. Formation of a complex of the first type is schematically shown in Figure 4.
Figure 4.
Schematic presentation of IN monomers interaction with specific and non-specific ODNs. The scheme shows the interaction of initial IN monomers with ODNs leading to a formation of the enzyme dimers of two types and a formation of catalytically active tetramers of two different dimers.
The interaction of IN with non-specific ODNs leads to the formation of another type of ODN · E · E · ODN dimer with higher Rg values (Table 4) and different characteristics of the affinity of enzyme monomer and dimer for each other during the formation of oligomeric forms (Table 3). These dimers of the second type form the enzyme E4S4 forms more efficient, but the tetramers are catalytically inactive (Figure 4).
According to our data the dimeric form of IN is active in the 3′-processing reaction. This conclusion is an agreement with the ability of the Pt2+-crosslinked E2 form to catalyse the 3′-processing and one LTR end integration reaction (17). Time-resolved fluorescence anisotropy studies also showed that the most competent form for catalysis corresponds to E2 dimer-bound viral DNA, whereas higher-order complexes predominate at higher concentrations of IN, accounting for a decrease in the 3′-processing activity after complexation with DNA containing two LTRs (18). There were several reports that the integration reaction is catalysed by the IN tetrameric form (16–18). Interestingly, the tetrameric form of IN catalyse full-site concerted integration but is unable to catalyse the 3′-processing reaction (17). Thus, it is obvious, that the IN tetrameric form is not a simple sum of two dimers retaining the properties of free dimeric forms.
The dimers of the first type in the complex with specific long ds ODNs can catalyse the 3′-processing reaction. As mentioned earlier, specific ds ODNs are very poor substrates in the integration reaction (38), and, according to (18), the efficiency of integration of these ds ODNs do not exceed 5% as compared with that of the processing reaction (100%). Therefore, one cannot exclude that in in vitro experiments the integration reaction may be a consequence of an incorrect side way of association of two specific ODN molecules, when the second specific ds ODN can interact with one monomer of dimeric E2S form not as a specific, but as a non-specific ODN mimicking the human host non-specific DNA. At the same time, the formation of the complex E · LTR–DNA–LTR · E of the first type can proceed in cells by a single way due to the intermolecular process of natural viral LTR–DNA–LTR interaction with E2 dimer, and the significant increase in the DNA affinity may be a result of an additive contribution of two termini of two LTRs to the total affinity of the E2 dimer to the natural viral DNA. From our point of view, the ODN · E · E · ODN complexes formed at high concentration of specific ds ODN can imitate the natural E · LTR–DNA–LTR · E complex, and these complexes demonstrate low affinity for each other. It is possible to explain some literature data and our findings about different structures of dimers in the complex with specific and non-specific ODNs proposing that only tetramers consisting of dimes of the first and the second type may be a catalytically active as tetramers catalysing the full-site concerted integration (Figure 4). Taking into account that under natural conditions not preventing the E2 dimer formation, one can propose several alternative pathways of different dimers formation. First, the E · LTR–DNA–LTR · E dimer of the first type can interact with a preformed E2 complex, which can have a structure of the dimer of the second type (E · E · LTR–DNA–LTR · E · E). In addition, in the absence of free 3′-terminal ends of viral LTR–DNA–LTR, which are bound to the active sites of dimers of the first type (E · LTR–DNA–LTR · E), the non-specific central part of LTR–DNA–LTR sequence can interact with the E2 dimer (or even in series with two E monomers) promoting formation of the dimer of the second type before E2 association with the dimer of the first type. Non-specific DNAs posses lower affinity for the enzyme as compared with the specific ones. But at increased concentrations of DNA non-specific fragments of some molecules of viral DNA free from the enzyme can interact with E2 dimers in a non-specific way and also promote the formation of dimers of the second type and later their association with dimers of the first type. Interestingly, preincubation with ss and ds 19-CA and 21-GT stimulates the formation of E2S2 with comparable efficiencies (Table 5). At the same time, the formation of the E4S4 tetramer in the presence of ss and ds 19-CA is ∼5–20-fold more effective than that for ss and ds 21-GT (Table 5). These data suggest that the removal of 3′-terminal GT nucleotide may have an important role during E4S4 tetramer formation from E2S2 dimers. One cannot exclude that a specific change in the IN dimer structure after removal of 3′-terminal GT results in the IN forms which are unable to catalyse the 3′-processing reaction, and therefore the Pt2+-crosslinked IN tetramer catalyses full-site concerted integration but unable to catalyse the 3′-processing reaction (17).
The SAXS data suggest that the conformation of the oligomeric form of IN is highly sensitive to DNA sequences, and also that, depending on the ODN used, there are individual specific types of conformation changes of IN monomers within the oligomeric form (Table 4). These changes may allow the enzyme to catalyse the consecutive reactions involved in the integration: (a) complexation with specific or non-specific DNA, (b) formation of dimeric forms, (c) catalysis of the 3′-processing reaction, (d) formation of tetramers due to the interaction of dimers of the first and the second types including the interaction of IN oligomers bound to viral DNA with non-specific host DNA, (e) integration of the viral DNA. Such stepwise conformational checking of the DNA structure by IN can therefore be a way for the enzyme to distinguish between the specific viral and non-specific host DNA. Thus, several steps of the reaction may provide the enzyme specificity. In contrast to non-specific DNA, specific DNAs can direct the assembly of catalytically active oligomeric forms of IN dimers and then activate the enzyme following structural changes providing a formation tetramer from the dimers of the first and the second type. In summary, our results establish a series of steps of IN and DNA assembly that finally can provide integration proficient complex.
ACKNOWLEDGEMENTS
This work was supported in part by the Molecular and Cell Biology Program of the Presidium of the Russian Academy of Sciences (project no. 10.5), the Russian Foundation for Basic Research (project no. 03-04-49781), the Siberian Division of the Russian Academy of Sciences, the French Agency for Research against AIDS (ANRS), the Centre National de la Recherche Scientifique (CNRS) and the University of Bordeaux II.
REFERENCES
- 1.Asante-Appiah E, Skalka AM. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 2.Katzman M, Sudol M. Mapping viral DNA specificity to the central region of integrase by using functional HIV-1/Visna virus chimeric proteins. J. Virol. 1998;72:1744–1753. doi: 10.1128/jvi.72.3.1744-1753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth MJ, Schwartzberg PL, Goff SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 4.Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization and enhances catalytic activity. Proc. Natl. Acad. Sci. USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heuer TS, Brown PO. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-crosslinking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 6.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conerved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992;12:2321–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 8.Lutzke RA, Plasterk RH. Structure-based mutational analysis of the C-terminal DNA- binding of HIV-1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyltransferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 10.Chen JC, Kucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral binding. Proc. Natl. Acad. Sci., USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and rertrovira integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 13.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of Avian Sarcoma Virus integrase. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 14.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Brochon JC. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- 17.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet J-F, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J. Biol. Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 19.Katzman M, Katz RA. Substrate recognition by retroviral integrases. Adv. Virus Res. 1999;52:371–395. doi: 10.1016/s0065-3527(08)60307-3. [DOI] [PubMed] [Google Scholar]
- 20.Caumont A, Jamieson G, de Soultrait VR, Parissi V, Fournier M, Zakharova OD, Bayandin R, Litvak S, Tarrago-Litvak L, Nevinsky GA. High affinity interaction of HIV-1 integrase with specific and nonspecific single-stranded short oligonucleotides. FEBS Lett. 1999;455:154–158. doi: 10.1016/s0014-5793(99)00859-5. [DOI] [PubMed] [Google Scholar]
- 21.Bugreev DV, Baranova S, Zakharova OD, Parissi V, Desjobert C, Sottofattori E, Balbi A, Litvak S, Tarrago-Litvak L, Nevinsky GA. Dynamic, thermodynamic, and kinetic basis for recognition and transformation of DNA by HIV-1 integrase. Biochemistry. 2003;42:9235–9247. doi: 10.1021/bi0300480. [DOI] [PubMed] [Google Scholar]
- 22.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Hawkins ME, Brochon JC. DNA binding induces dissociation of the multimeric form of HIV-1 integrase: a time-resolved fluorescence anisotropy study. Proc. Natl. Acad. Sci., USA. 2001;98:10090–10095. doi: 10.1073/pnas.181024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bera S, Vora AC, Chiu R, Heyduk T, Grandgenett DP. Synaptic complex formation of two retrovirus DNA attachment sites by integrase: a fluorescnce energy transfer study. Biochemistry. 2005;44:15106–15114. doi: 10.1021/bi0508340. [DOI] [PubMed] [Google Scholar]
- 24.Vercammen J, Maertens G, Gerard M, De Clercq E, Debyser Z, Engelborghs Y. DNA-induced polymerization of HIV-1 integrase analyzed with fluorescence fluctuation spectroscopy. J. Biol. Chem. 2002;277:38045–38052. doi: 10.1074/jbc.M205842200. [DOI] [PubMed] [Google Scholar]
- 25.Ishchenko AA, Bulychev NV, Zharkov DO, Maksakova GA, Johnson F, Nevinsky GA. Isolation of 8-oxoguanine-DNA glycosylase from Escherichia coli and studies on its substrate specificity. Molecular Biology (Mosc) 1997;31:278–283. [Google Scholar]
- 26.Fasman GD. Handbook of Biochemistry and Molecular Biology: Nucleic Acids. Vol. 1. Florida: CRC, Boca Raton; 1975. p. 589. [Google Scholar]
- 27.Calmels C, de Soultrait VR, Caumont A, Desjobert C, Faure A, Fournier M, Tarrago-Litvak L, Parissi V. Biochemical and random mutagenesis analysis of the region carrying the catalytic E152 amino acid of HIV-1 integrase. Nucleic Acids Res. 2004;32:1527–1538. doi: 10.1093/nar/gkh298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuzikov FV, Zinoviev VV, Vavilin VI, Malygin EG. Small-angle X-ray scattering study of enzyme-substrate interaction in solution. Studia Biophisica. 1988;125:169–172. [Google Scholar]
- 29.Tuzikov FV, Zinoviev VV, Vavilin VI, Malygin EG. Application of the small-angle X-ray scattering technique for the study of the two-step equilibrium enzyme-substrate interactions. Biopolymers. 1996;38:131–139. doi: 10.1002/(sici)1097-0282(199602)38:2<131::aid-bip1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 30.Tuzikov FV, Tuzikova NA, Galimov RV, Panin LE, Nevinsky GA. General model to describe the structure and dynamic balance between different human serum lipoproteins and its practical application. Med. Sci. Monit. 2002;8:MT79–88. [PubMed] [Google Scholar]
- 31.Feigin LA, Svergun DI. Structure Analysis by Small-Angle X-Ray and Neutron Scattering. New York and London: Plenum Press; 1987. [Google Scholar]
- 32.Gill PE, Murray W, Wrighe MH. Practical Optimization. London, New York, Toronto, Sydney, San Francisco: Academic Press; 1981. [Google Scholar]
- 33.Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazuda DJ, Wolfe AL, Hastings JC, Robbins HL, Graham PL, LaFemina RL, Emini EA. Viral long terminal repeat substrate binding characteristics of the human immunodeficiency virus type 1 integrase. J. Biol. Chem. 1994;269:3999–4004. [PubMed] [Google Scholar]
- 35.Vink C, Lutzke RA, Plasterk RH. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA. Nucleic Acids Res. 1994;22:4103–4110. doi: 10.1093/nar/22.20.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guinier A, Fournet G. Small Angle Scattering of X-ray, New York: Wiley; 1955. [Google Scholar]
- 37.Feigin LA, Svergun DI. Structure Analysis by Small-angle X-ray and Neutron Scattering. New York: Plenum Press; 1987. [Google Scholar]