Abstract
Mung bean yellow mosaic India virus (MYMIV) is a member of genus begomoviridae and its genome comprises of bipartite (two components, namely DNA-A and DNA-B), single-stranded, circular DNA of about 2.7 kb. During rolling circle replication (RCR) of the DNA, the stability of the genome and maintenance of the stem–loop structure of the replication origin is crucial. Hence the role of host single-stranded DNA-binding protein, Replication protein A (RPA), in the RCR of MYMIV was examined. Two RPA subunits, namely the RPA70 kDa and RPA32 kDa, were isolated from pea and their roles were validated in a yeast system in which MYMIV DNA replication has been modelled. Here, we present evidences that only the RPA32 kDa subunit directly interacted with the carboxy terminus of MYMIV-Rep both in vitro as well as in yeast two-hybrid system. RPA32 modulated the functions of Rep by enhancing its ATPase and down regulating its nicking and closing activities. The possible role of these modulations in the context of viral DNA replication has been discussed. Finally, we showed the positive involvement of RPA32 in transient replication of the plasmid DNA bearing MYMIV replication origin using an in planta based assay.
INTRODUCTION
Geminiviruses constitute an important group of plant pathogens infecting a broad variety of crops and pose serious threat to these crop plants throughout the world. It is a diverse group having circular, single-stranded DNA (ss DNA) genome of varying length (∼2.5–3.0 kb) and comprises of either monopartite or bipartite genome structure. Mung bean yellow mosaic India virus (MYMIV), previously known as IMYMV (1), has a bipartite genome and belongs to the genus begomoviridae of the family geminiviridae. These viruses are prevalent in the northern part of India causing severe economic losses to leguminous crops (2). Geminiviruses replicate via rolling circle mode of DNA replication [rolling circle replication (RCR)] inside the nucleus of the host cell and encode only few viral factors for its replication (3). Geminiviruses predominantly infect the terminally differentiated cells of the host where the replicative proteins are present in negligible amount (4). However, they reprogram the cell cycle machinery towards their own growth as a result of complex mechanism of interaction between the viral factors and various cell cycle regulatory proteins of the host e.g. retinoblastoma related protein (pRBR) (5–9). During early G1 phase, pRBR remains bound to one of the important transcription factors, E2F, which controls the transcription of many genes involved in G1/S phase transition and S phase progression. Once pRBR gets phosphorylated at the late G1 phase, it releases the E2F factor, which perhaps might turn on the expression of many genes e.g. PCNA (10–12). The binding of geminiviral Rep protein to pRBR also releases the E2F factor and might force the cell to enter into S phase like scenario, a condition prevalent also in many animal cells infected with tumour viruses.
Replication initiator protein (Rep) of MYMIV is the major viral protein responsible for viral DNA replication. It is a multifunctional, oligomeric protein having site-specific DNA-binding (13), nicking and ligation, ATP dependent topoisomerase I (14) and ATPase activities (15). Similar to the Rep proteins of other begomoviruses, the MYMIV-Rep also cleaves the viral genome at a specific and conserved nonamer site i.e. TAATATT↓AC and initiates the RCR replication (16). Although, the geminivirus-Rep is a crucial protein for viral replication, it requires the help of various other host factors for efficient viral DNA replication. It interacts with a number of host factors, such as pRBR as mentioned earlier, RF-C, which might help in loading of PCNA (17), PCNA for putative initiation and control of DNA replication (15), Histone H3 for putative removal of nucleosomal block and efficient transcription and replication, GRIMP (kinesin) and GRIK (a Ser/Thr kinase) which might prevent the cell from undergoing mitosis (18). Although the role of ssDNA-binding protein [Replication protein A (RPA)] has been widely documented in chromosomal, plasmid as well as viral DNA replication (19) of animal systems, its role in geminivirus DNA replication has not been reported yet.
RPA is a eukaryotic, multifunctional, ssDNA-binding, heterotrimeric protein that plays critical role in 3R′s of DNA metabolism, namely, replication, repair and recombination (19). The protein is reasonably well conserved across species among all the eukaryotes. It is composed of three subunits, viz., 70, 32 and 14 kDa. RPA binds to and stabilizes the ssDNA during DNA replication (20). Genetic studies in yeast reveal that all the three subunits are essential for cell viability and alterations in any of the genes arrest the cell growth (21). RPA70 kDa is the major subunit of the complex having two strong ssDNA-binding domains in the middle of the subunit, while 32 and 14 kDa subunits share one weak DNA-binding domain each (22,23). RPA32 kDa subunit shows regulatory role and once bound to DNA, it gets phosphorylated in a cell cycle dependent manner by cell cycle dependent kinases and DNA dependent protein kinases (24–26). Recent evidences suggest that, in Xenopus, hypo-phosphorylated form of RPA32 is a specific component of the pre-initiation complex for replication (27). It has been shown in case of human that once it gets hyper-phosphorylated, it is unable to bind to the replicative centre in cellular milieu (28). RPA interacts with many key proteins involved in DNA replication like DNA polymerase α, RF-C, PCNA and DNA helicase (29,30). During DNA replication, RPA binds to the replicative centres (forks), stabilizes the ssDNA and facilitate the nascent strand synthesis by interacting and stimulating the replicative DNA polymerase α and δ (29,31). It also interacts with and recruits various key viral proteins of different systems, such as SV 40 large T antigen (29), EBNA1 proteins of Epstein–Barr virus (32), E1 proteins of papilloma virus (33,34) and NS1 proteins of parvovirus (35) at the initiation site of DNA replication and helps in the formation of replication forks in these viruses. Although the roles of RPA in the initiation phase, i.e. during pre-RC to RC formation and in the elongation phase of chromosomal DNA replication are understood in the animal system, its role in the rolling circle mode of DNA replication and in plant viral DNA replication has not been investigated.
The hypothetical issue of the stability of the ssDNA genome of the geminivirus inside the plant nucleus and the structural integrity of the stem–loop conformation of the replication origin led us to examine the contribution of RPA in geminiviral DNA replication. We wanted to investigate whether the plant RPA plays any role during the initiation and elongation phases of MYMIV DNA replication. Since the independent studies revealed that the MYMIV replicon could transiently replicate in pea, French bean, mungbean, tobacco and Arabidopsis [ICGEB activity report (2004), PhD thesis of M. N. Islam, unpublished data], the genes of RPA subunits were isolated from pea as representatives and their biochemical behaviour were studied. The yeast model based experiments revealed that both the subunits were important for MYMIV DNA replication. Here, we present evidences that the RPA32 kDa subunit directly interacted with MYMIV-Rep both in vitro as well as in vivo through a novel interacting site present at the C-terminus of the Rep. RPA32 also modulated the functions of Rep by enhancing its intrinsic ATPase activity and down regulating its intrinsic nicking and ligation activity. Finally, we show that pea RPA32 upregulated the transient replication of the MYMIV-amplicon in the plant-based assay. This is the first report of requirement of RPA in geminivirus DNA replication.
MATERIALS AND METHODS
Plant growth
Pea (Pisum sativum) plant was grown in germination paper or vermiculite in green house under controlled condition of 25°C and 16 h daylight.
Isolation of full-length genes of pea RPA32 and 70 genes by 5′ and 3′ random amplification of cDNA ends (RACE) technique
RNA was isolated from pea leaves by standard guanidium isothiocyanate method and mRNA was separated from total RNA with the help of biotinylated oligo dT primers and streptavidin linked paramagnetic beads (Roche, USA). mRNA was used to synthesize first strand cDNA using the first strand cDNA synthesis kit (Invitrogen, USA). The reaction mixture containing 5 μg of mRNA, 500 ng of oligo dT primer having adapter at the 5′ end and 1 μl of 10 mM dNTP mix in a total volume of 14 μl was heated at 65°C for 5 min. To this mixture 4 μl of 5× first strand buffer [250 mM Tris–HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2 and 1 μl 0.1 M DTT] and 1 μl of Superscript III Reverse Transcriptase (200 U/μl) was added to a final volume of 20 μl. The mixture was incubated at 50°C for 60 min and the reaction was terminated by heat inactivation at 70°C for 15 min. The RNA complementary to the cDNA was removed by adding 1 μl of RNase A (10 mg/ml stock) to the mixture. The single-stranded cDNA was purified through quick Qiagen DNA purification column. This cDNA was then used for RACE–PCR.
The closely related plant homologues of RPA70 and 32 genes were multiple aligned using Mac vector V. 6.0 program (Oxford). Degenerate primers were designed for both the genes from conserved regions. The PCR was carried out in 50 μl reaction volume containing 5 μl 10× buffer [100 mM Tris–HCl (pH 8.8), 500 mM KCl, 15 mM MgCl2, 0.1% gelatin, 0.05% Tween-20 and 0.05% NP-40], 8 μl of 1.25 mM dNTP mix, 100 ng each of degenerate primers and 2.5 U of Taq DNA polymerase, at very low stringency conditions (annealing at 40°C). The isolated PCR fragments were gel purified and cloned in pGEMT Easy vector (Promega, USA) followed by manual or automated sequencing.
To isolate the full-length PsRPA70 and 32 genes, both 5′ and 3′ RACE were performed. The 5′ end of the genes were amplified by PCR using adapter specific 5′ RACE forward primer and a gene specific reverse primer. The 3′ RACE was carried out with gene specific forward primers and reverse primer complementary to the oligo dT adapter primer. The sequences of the primers used for isolation of RPA70 and RPA32 kDa genes are given in Table 1.
Table 1.
Primers used for isolation of RPA32 and RPA70 genes
| Orientationa | Sequence of primer (5′→3′)b | Usage |
|---|---|---|
| S | TTY RYK VTT GAY GAY GGH ACM GG | Partial RPA32 |
| A | CTC RTC SAW WGT YGA GTA TAT KHR MCC | |
| S | CCG GAG CTC GAG CGC GCT AGC | 5′ RPA32 |
| A | CACAG GCC TGA CAG ACA AGG C | |
| S | GGA ATA CAT GTT GAA GAA CTA GC | 3′ RPA32 |
| A | GTC GTC GAC ATA TGA TCA AGC TT | |
| S | ATG GAT CCA TGT TCT CCA GCT CCC AAT TTG AC | RPA32 full-length |
| A | TTG AGT CGA CTC AAG CTT GTT TGT AGT GGG AGT C | |
| S | TCA ARR GGA ARY TTG ARR CCT GC | Partial RPA70 |
| A | CCR WTC ATS WWA GGR CAA GC | |
| S | CTT ACG AGC CTG GTT TGA TCA AG | 5′ RPA70 |
| A | GTC ACA GGC CTG ACA GAC AAG GC | |
| S | GGA ATA CAT GTT GAA GAA CTA GC | 3′ RPA70 |
| A | GAA GGA TTC ACA GAT GTC ACA ACC | |
| S | CAT GGA TCC ATG TCG GTG AAT CTC ACG GCG AG | RPA70 full-length |
| A | TTG AGT CGA CTT ACT TCC TAC CGA ACT TGG AAA TC |
aS, sense; A, antisense.
bThe primers underlined contained BamHI restriction site in the forward primer and SalI in the reverse primer preceded by a stop codon.
Yeast complementation assay
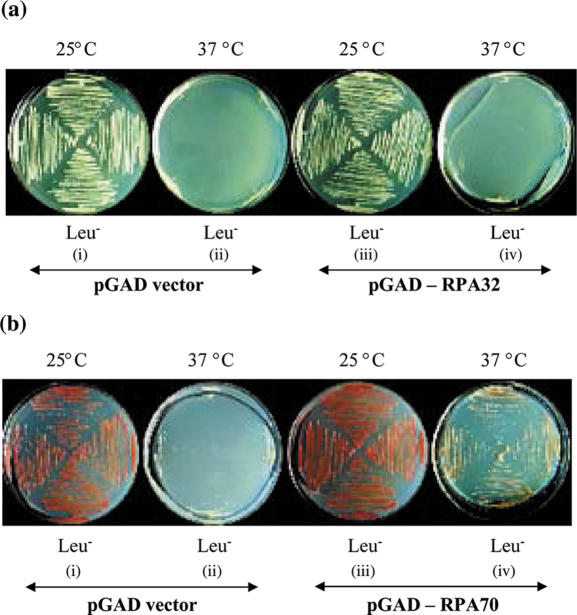
PsRPA32 and 70 genes were cloned at BamHI and SalI restriction sites in yeast shuttle vector pGAD. The cloning was confirmed by restriction digestion. The pGAD vector and pGAD vector harbouring either PsRPA32 or PsRPA70 genes were transformed in the temperature sensitive (ts) mutant strains for respective genes. The transformants were selected on synthetic SD plates lacking leucine at 25°C. The individual colonies were then streaked on SD Leu− plates and allowed to grow for 3 days either at 25°C (permissive) or 37°C (restrictive) conditions. The complementation was checked by the growth of transformed ts mutants at restrictive temperature (37°C).
Replication assay in yeast
The wild-type yeast strain W303a and ts mutant strains of yeast for RPA32 and RPA70 were transformed with either YCp50 vector or YCpO−-2A construct (1). The transformants were selected on SD media lacking uracil at 25°C. The individual colonies were then streaked on SD Ura− plates and allowed to grow either at 33°C (permissive) or 35°C (semi-restrictive) temperatures for RPA32 ts mutant and similarly at 30 and 33°C for RPA70 ts mutant. The plates were analysed after 4 days of growth.
Plasmid retention assay
The procedure was adopted essentially from Marahrens et al. (36). Briefly, the ts mutants of yeast for RPA32 and RPA70 genes were transformed either with control YCp50 or YCpO−-2A plasmids and the transformants were selected on Ura− plates. Single colonies were inoculated in 5 ml of Ura− broth and grown overnight at 25°C. About 1% of the culture was inoculated into 25 ml Ura− broth and grown till the OD600 nm reached 0.6. Again, 1% of the grown culture were inoculated into 25 ml of YPD medium and incubated for 8–10 generations with constant shaking at different sub-lethal temperatures (22,28,30 and 33°C). The cultures were serially diluted and plated on Ura− as well as YPD plates with different dilutions and incubated at the corresponding sub-lethal temperatures for 3 days for colony formation. The colonies were counted and the percent stability of the plasmid were calculated by the ratio of the number of colonies on Ura− plates to the number of colonies on YPD plates and multiplied by 100.
DNA constructs and purification of the recombinant proteins
The full-length gene of PsRPA32 was PCR amplified from pea cDNA using gene specific primers containing suitable restriction sites and cloned in pGEMT Easy vector (Table 1). The gene cloning was confirmed by restriction digestion and sequencing. The fragment was then digested with BamHI and SalI followed by recloning in pGEX-4T-2 vector (Amersham Biosciences, USA) to obtain glutathione S-transferase (GST) tagged fusion protein. The pGEX-4T-2 vector harbouring PsRPA32 gene was used to transform Escherichia coli BL-21 (DE3) cells. The PsRPA32 was overexpressed at 30°C by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The protein was purified to near homogeneity following manufacturer's protocols (Amersham Biosciences, USA). The tagged protein was further purified using the heparin sepharose CL-6B column (Amersham Biosciences, USA).
The recombinant clone pET28a-Rep (14) was used as a template to PCR-amplify various deletion mutants of Rep, with primers containing suitable restriction sites at the N- and C-termini, respectively. The amplified fragments were cloned in pGEMT Easy vector followed by recloning in pET28a (Novagen, Germany) using same restriction sites. Point mutants of Rep were constructed by using Quick-change site-directed mutagenesis kit (Stratagene, USA) following manufacturer's protocol. PCR amplification was carried out with complementary mutagenesis primers and the full-length wild-type pET28a-Rep as template. The mutations were confirmed by sequencing. The full-length and various mutants of Rep were overexpressed in E.coli BL-21 (DE3) strain and the proteins were purified to near homogeneity under nondenaturing conditions as described previously (37). The biochemical activities of recombinant wild-type as well as mutant Rep proteins were checked by in vitro site-specific cleavage and/or ATPase activity (15).
GST pull down assay
Purified GST–PsRPA32 fusion protein was incubated with indicated amounts of wild-type or mutant proteins of His- or MBP-Rep in binding buffer [25 mM Tris–HCl (pH 8.0), 75 mM NaCl, 2.5 mM EDTA, 5 mM MgCl2, 2.5 mM DTT and 1% NP-40] at 37°C for 30 min. To this complex, pre-washed and binding buffer equilibrated glutathione S-sepharose resin was added. The mixture was slowly mixed for another half an hour at room temperature. The unbound protein fraction was separated from the resins by centrifugation at 3000g for 3 min. The resin containing the bound protein was washed with increasing concentrations of NaCl (100–400 mM) in binding buffer. Equal amount of 2X sample buffer was then added to the resin, boiled for 5 min, centrifuged briefly and the supernatant was analysed by SDS–PAGE. The protein bands were visualized by either Coomassie blue or silver staining.
Yeast two-hybrid analysis
The MYMIV-Rep was excised from the BamHI and XhoI sites of pET28a-Rep and recloned into BamHI and SalI sites of pGAD-C1 and pGBD-C1 vectors (38). Similarly, the full-length PsRPA32 and 70 kDa genes was excised from the BamHI and SalI sites of pGEMT Easy vector and recloned into the BamHI and SalI sites of pGAD-C1 and pGBD-C1 vectors. This cloning resulted in an N-terminal in frame fusion of the GAL4 activation domain and the DNA-binding domain with Rep, PsRPA32 and PsRPA70 genes. All the constructs were verified by restriction digestion and sequencing.
The yeast two-hybrid assay was performed using reporter yeast strain AH109, which was transformed with the appropriate plasmids and grown on SD plates in the absence of Trp and Leu to select for co-transformants. Protein interaction analysis was performed on SD plates lacking Leu, Trp and His (SD Leu−, Trp−, His−). After 3 days of growth at 30°C, individual colonies were streaked out and tested for β-galactosidase activity in a filter lift assay (1). With the help of a forcep, a 3 mm Whatman filter (in 85 mm plate) was placed over the surface of the streaked colonies. The filter was carefully lifted and completely submerged in liquid nitrogen for 10 s and allowed to thaw at room temperature. This freeze/thaw step was repeated 3–5 times, after which the filter was carefully placed on filter paper presoaked in 5 ml buffer Z [0.1 M Na-phosphate buffer (pH 7.0), 0.01 M KCl and 0.001 M MgSO4], 20 μl 20% X-gal and 8 μl β-mercaptoethanol. The filter was then wrapped in aluminium foil and kept at 30°C. The development of blue colour was checked after 6–8 h.
ATPase assay
The protocol was essentially adapted from Fukuda et al. (39). The ATPase reaction were carried out in a reaction buffer containing 20 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 100 mM KCl, 8 mM DTT and 80 μg of BSA/ml. Briefly, 1 μl (10 μCi) of [γ-32P]ATP (6000 Ci/mmol) was diluted 50-fold with 5 mM ATP. Diluted radiolabelled ATP (1 μl) was mixed with desired amounts of proteins (Rep and/or RPA32) and incubated at 37°C for 30 min. After the reaction, 1 μl of the reaction mix was spotted on PEI-TLC plate (Sigma–Aldrich, USA), air-dried and chromatographed using 0.5 M LiCl and 1M HCOOH as the running solvent. Following completion of chromatography, TLC paper was dried and autoradiographed (15). For time course kinetics, the samples were withdrawn after different time intervals as indicated. The relative intensities of the released Pi were estimated by densitometric scanning using Typhoon 9210 scanner and analysed by ImageQuant TL software (Amersham Biosciences, USA).
Nicking and ligation activity
The oligonucleotide T1 (5′-CGACTCAGCTATAATATTACCTGAGT-3′), used for the nicking assay, was 5′ end-labelled with T4 polynucleotide kinase (Promega, USA). The oligonucleotide T2 (5′-CTATAATATTACCTGAGTGCCCCGCG-3′), employed for the ligation assay, was unlabelled and used in 50-fold higher molar excess over the T1 oligonucleotide. Approximately, 1 ng T1 (specific activity ∼1.0 × 109 c.p.m./μg) was incubated with 500 ng purified Rep, in absence or presence of increasing concentrations of PsRPA32 in 50 μl reaction volume containing 25 mM Tris–HCl (pH 8.0), 75 mM NaCl, 2.5 mM EDTA, 5.0 mM MgCl2, 2.5 mM DTT at 37°C for 30 min. The nicking reaction was terminated by adding 6 μl loading buffer (1% SDS, 25 mM EDTA, 10% glycerol) and heating at 90°C for 2 min. For ligation reaction, about 50 ng of T2 oligonucleotide was added to the nicking reaction. The products were resolved on a 15% polyacrylamide–urea gel and visualized by autoradiography (37). The relative intensities of the bands were estimated by densitometric scanning using Typhoon 9210 scanner and analysed by ImageQuant TL software (Amersham Biosciences, USA).
DNA-binding assay
In the presence of PsRPA32, the ori-DNA-binding activity of Rep1–362 was carried out using nitrocellulose filters (DAWP 02500, 0.65μ, Millipore, USA). The reaction mixture (50 μl) consisted of one ng of 5′ 32P-labelled MYMIV-A ori DNA (216 bp), Rep1–362 and/or PsRPA32 in 1× binding buffer containing 20 mM Tris–HCl (pH 8.0), 25 mM KCl, 1 mM DTT, 100 μg/ml BSA, 0.02% poly(dI–dC) and 10% dimethyl sulfoxide (DMSO). After 30 min of incubation at 37°C, the reaction mixture was filtered through the pre-washed nitrocellulose filters in the manifold system (Millipore, USA), the filter was washed with 10 vol of 1× binding buffer and dried under a lamp. The extent of ori-DNA bound by Rep protein in presence or absence of PsRPA32 protein was calculated from the Cerenkov counts of filters measured in Beckman multi-purpose scintillation counter (LS 6500).
Transient replication assay in plant system
The plamid (MYMIV based amplicon) [ICGEB activity report (2004), PhD thesis of M. N. Islam (2005)] as well as the amplicon along with plasmid harbouring pea RPA32 gene were introduced into agrobacterium strain LBA 4404 and incubated at 30°C. Single colonies were then inoculated into YEM media having appropriate selection markers and grown at 30°C for 36 h. The cultures were then infiltrated into Nicotiana xanthi leaves. After 7 days post-infiltration the leaves were collected and genomic DNA were isolated by standard (CTAB) method. The genomic DNA was digested with ScaI enzyme overnight and subjected to Southern analysis. The bands were detected by radiolabelled green fluorescent protein (GFP) probe followed by autoradiography. The PCR methodology was also used for the detection of episomal (VA) circular DNA. The sequences of the pair of primers were as follows:
5′-GCTCTAGACCATGGCAAGTAAAGGAGAAGAACTT- 3′ (at the GFP site)
5′-AGA AGCTTCTATGCGTCGTTGGCAGATTG- 3′ (at the N-terminus of Rep).
RESULTS
Isolation of RPA70 and RPA32 genes from P.sativum (pea)
We used rapid amplification of cDNA ends (RACE) technique to isolate two full-length genes of the subunits of RPA heterotrimeric complex, i.e. PsRPA70 and PsRPA32 from pea. The open reading frames of PsRPA70 and PsRPA32 encode predicted products of 638 amino acid residues with a molecular mass of 71.5 kDa and of 274 amino acid residues with a molecular mass of 30 kDa, respectively (NCBI GeneBank with accession nos. AY289132 and AY299688 for PsRPA70 and PsRPA32, respectively). The deduced amino acid sequences of PsRPA70 and PsRPA32 genes were compared with other known plant RPA70 and 32 homologues and similarities were observed (Supplementary Figures S1a and S2a). The multiple alignment of PsRPA32 gene showed 48 and 40% identities at amino acid level and 60 and 52% identities at the nucleotide level with the Arabidopsis and rice homologues, respectively. On the other hand, PsRPA70 gene revealed 68 and 57% identities at amino acid level and 69 and 61% identities at nucleotide level with Arabidopsis and rice homologues respectively (Supplementary Figures S1b and S2b). To determine the phylogenetic relationships of the PsRPA70 and PsRPA32 genes with other eukaryotic homologues, an unrooted tree was drawn based on the alignment using ClustalW program of Mac vector V. 6.0 (Supplementary Figures S1c and S2c). The dendrogram revealed that the plants RPAs are more closely related to each other in comparison with RPAs from other eukaryotic organisms.
Complementation of yeast ts mutant strains by pea RPA32 and RPA70 kDa genes
It has been reported that the interaction between RPA and cellular factors is species-specific and yeast RPA is unable to replace the function of human RPA in complete SV40 DNA replication system (40). In spite of such limitations, studies in genetic complementation provide the fast and easy approach to test the functionality of the isolated genes. The PsRPA32 and 70 genes were individually cloned in frame with GAL4 activation domain of the yeast shuttle vector, pGAD. The pGAD vector (control) as well as the pGAD vector harbouring PsRPA32 gene were used to transform the ts mutant of yeast RPA32 (HMY345) and selected on synthetic media lacking leucine at 25°C. The colonies thus obtained were then streaked in replica on Leu− plates. One plate was incubated at 25°C and the other at 37°C. The yeast cells harbouring both the vector as well as the vector with pea RPA32 gene were able to form colonies at 25°C (permissive) but not at 37°C (restrictive) temperature [Figure 1a (i–iv)], suggesting that PsRPA32 could not complement for the yeast RPA32 function. Such inability is in accordance with the earlier report that human RPA32 failed to complement the yeast rpa2 mutant (41). It has been reported that the activities of the 32 and 14 kDa subunits of RPA are species-specific and their ssDNA-binding property is one of the determinants of the species-specificity (42). This could be a reason for the failure of complementation by pea RPA32. Similarly, we carried out the complementation assay for PsRPA70 gene in the ts mutant strain of yeast RPA70 (W303-1A/rfa1-t6). In this case, the vector DNA harbouring PsRPA70 gene was able to replicate at 37°C (restrictive) temperature [Figure 1b (i–iv)], suggesting that PsRPA70 gene can complement for yeast RPA70 gene function. It was reported earlier that the replacement of yeast SBD-A and SBD-B present in RPA70 kDa subunit with human or rice SBD-A and B could rescue a yeast rpa1 mutant (43,44). Our results also suggest that the function of RPA70 subunit is highly conserved across species.
Figure 1.
Complementation of pea RPA32 and RPA70 genes in the ts mutant strains of yeast. Pea RPA32 and RPA70 genes were cloned in frame with yeast pGAD vector and transformed in the ts mutant of yeast for RPA32 (HMY345) and RPA70 (W303-1a/rfa1-t6) genes respectively. The transformants were selected on synthetic yeast media lacking leucine at 25°C. Panel (a) shows independent colonies of the yeast ts mutant strain of RPA32 transformed either with pGAD vector [(i) and (ii)] or with pGAD-PsRPA32 construct [(iii) and (iv)] and grown at 25°C (permissive) and 37°C (restrictive). Similarly, panel (b) shows the yeast ts mutant strain of RPA70 transformed either with pGAD vector [(i) and (ii)] or with pGAD-PsRPA70 construct [(iii) and (iv)], grown at 25° and 37°C.
Interaction between PsRPA32 and PsRPA70 ex vivo
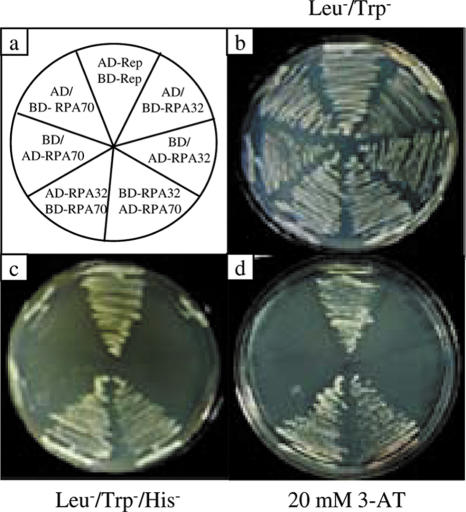
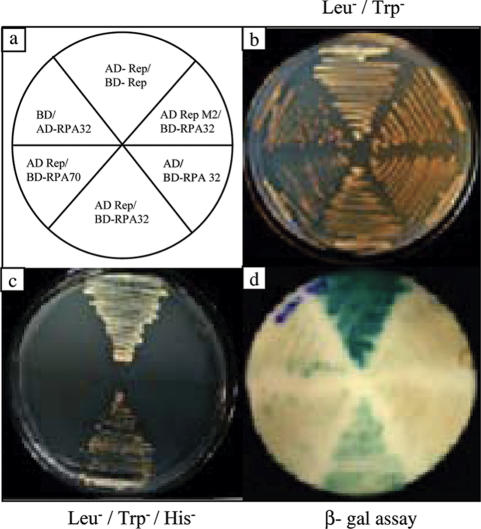
In order to establish that the isolated RPA32 from pea is a true functional homologue, we examined the interaction between pea RPA32 and pea RPA70 genes using the yeast two-hybrid system. Both the PsRPA32 and PsRPA70 genes were cloned in frame with yeast GAL4 activation domain (pGAD) and GAL4 DNA-binding domain (pGBD) vector. The constructs were co-transformed in different combinations in a yeast reporter strain AH109 and the co-transformants were selected on synthetic media lacking leucine and tryptophan (Figure 2a and b). The results showed that PsRPA32 protein interacted with PsRPA70 protein in yeast cells as evidenced from the growth of auxotrophic AH109 strain on triple dropout plate (SD Leu−, Trp−, His−) (Figure 2c). The interaction between the two proteins was very strong as the colonies shown in Figure 2c could tolerate high-concentrations (20 mM) of 3-AT (Figure 2d). The MYMIV-Rep- Rep interaction was taken as the positive control for this assay (37). The interaction between the two proteins was also confirmed by β-galactosidase assay (data not shown).
Figure 2.
Yeast two-hybrid interactions between pea RPA32 and RPA70 genes. Panel (a) represents the schematics of combinations of fusion constructs used for yeast two-hybrid interaction study. The growth of the reporter AH109 strain harbouring different fusion constructs as represented in (a) on synthetic yeast media lacking leucine/ tryptophan and leucine/ tryptophan/ histidine are shown in (b) and (c) respectively. The growth of the replica colonies on triple dropout media containing 20 mM 3-AT is shown in (d).
Expression of PsRPA32 gene in developmentally differing leaf tissues of pea
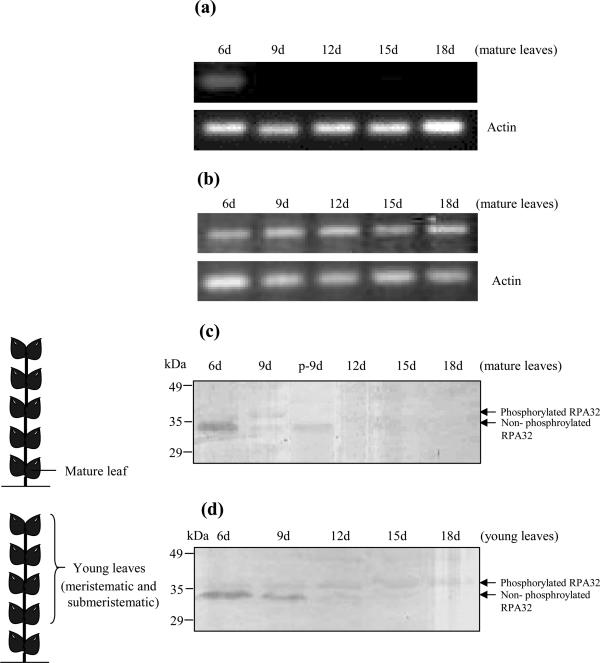
In many hosts, geminiviruses infect terminally differentiated cells that lack the DNA replication factors (4). With this fact in mind, we wanted to investigate the presence of RPA subunits in various uninfected tissues of pea. To examine the developmental regulation of RPA32, we isolated RNA from the lower most (mature) leaf of the pea at different time intervals. The transcript analysis was carried out by RT–PCR. The results revealed that the RPA32 transcripts were abundantly expressed in 6 days old leaves and reduced sharply from 9 day onward to almost undetectable levels (Figure 3a). However, the RPA70 transcript level remained constant over the period of observation (Figure 3b). The findings of RPA32 transcript analysis were also corroborated with western analysis of the total pea protein using the recombinant pea RPA32 specific polyclonal antibodies (raised in our lab). The PsRPA32 kDa protein was detected in the mature leaf of 6 days old seedlings and reduced thereafter, as the leaves grew older (Figure 3c). We were able to detect two bands in 9 days old leaves, which represented the phosphorylated (upper band) and non-phosphorylated (lower band) forms of RPA32. To confirm the phosphorylated nature of upper band, the proteins of 9d sample were treated with λ-phosphatase and the presence of the upper band was sharply reduced (Figure 3c, lane p-9d) suggesting the presence of phosphorylated form of the RPA32 in the 9d old sample. In contrast, the western analysis of the young (meristematic and sub-meristematic) leaves showed the gradual decrease in the level of non-phosphorylated form and increase in the level of phosphorylated form of RPA32 over the same period of observation (Figure 3d). It has been reported that RPA32 gets phosphorylated in a cell cycle dependent manner and that the phosphorylated form is unable to bind to the replicative centres in vivo and are no longer involved in DNA replication (28). Our data also suggested that, as the leaf tissue progressed towards developmental maturity, RPA32 was phosphorylated and thus conceivably became limiting for DNA replication. These results clearly indicated that RPA32 expressed only in the young proliferating tissues.
Figure 3.
Developmental pattern of expression of RPA32 kDa gene in pea leaf. (a) RT–PCR analysis of the RPA32 (panel a) and RPA70 (panel b) transcripts from mature (lower most) pea leaves isolated at indicated time intervals. The RNA was isolated from lower most leaves after 6, 9, 12, 15 and 18 days interval and about 5 μg of RNA was used to prepare cDNA with reverse transcriptase. The PCR were carried out with these cDNA samples for 28 cycles and 10 μl of the PCR product was resolved on 1.4% agarose gel and analysed. Lower panels show the amplification of actin as a loading control. Panels (c) and (d) show the western blot analyses of the total protein isolated from pea leaves reflecting the developmental pattern of RPA32 protein. About 30 μg of the total protein extracted from 6, 9, 12, 15 and 18 days old mature (lower most) leaves (panel c) and young (meristematic and sub-meristematic) leaves (panel d) from pea were resolved on 10% SDS–PAGE gel and subjected to western blot analysis using the rabbit polyclonal antibodies against recombinant pea RPA32 kDa protein and the blots were visualized by alkaline phosphatase linked secondary antibodies (BCIP/NBT). The schematic illustrations of the leaves, used for isolation and extraction, have been made on the left side of data panels of Figure (c) and (d). Two bands in the lane 9d (panel c) and lanes 9d–15d (panel d) corresponds to phosphorylated (upper) and non-phosphorylated (lower) forms of RPA32. The third column (p-9d) of panel c shows the 9d proteins that were treated with 10 U of λ-phosphatase. Standard molecular weight markers are indicated.
Bioassay of viral DNA replication in yeast ts mutant strains of RPA32 and RPA70
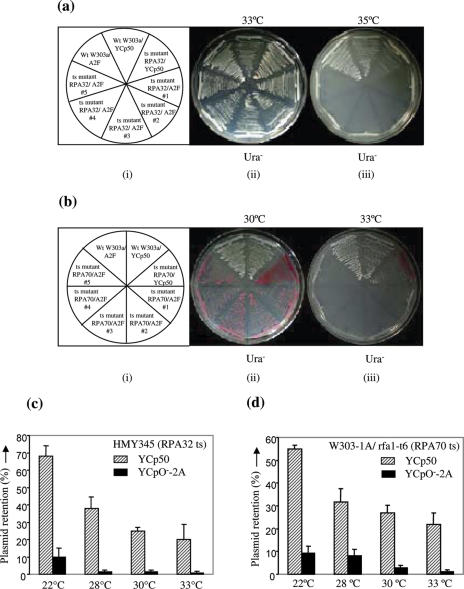
We used budding yeast as a model system to elucidate the role of RPA32 and 70 kDa subunits in geminiviral DNA replication. Earlier studies had shown that budding yeast supported geminiviral DNA replication and that the plasmid YCpO−-2A, harbouring two tandem copies of DNA-A component of MYMIV bipartite genome, replicated in yeast by using specifically the viral replication origin and virus encoded factors (1). The plasmid YCpO−-2A was used to transform the yeast ts mutant strains of RPA32 and RPA70 and growth of the transformed yeast strains were monitored at permissive as well as sub-lethal (or, semi-permissive) temperatures. The wild-type W303a yeast strain as well as ts mutants of RPA32 and RPA70 were also transformed with the control YCp50 plasmid vector bearing the yeast ARS sequences. All the transformants were selected on media lacking uracil. The result showed that the wild-type W303a strain harbouring either YCp50 or YCpO−-2A construct was able to grow at 35°C (sub-lethal) temperature. The ts mutant of RPA32 harbouring the YCp50 vector was able to survive at 35°C, albeit with reduced efficiency. However, the ts mutant of RPA32 that harboured the YCpO−-2A construct failed to survive at this sub-lethal temperature (Figure 4a). This suggested that RPA32 was the limiting factor for growth of plasmid having specifically the geminiviral origin. Similar results were also obtained for RPA70 and in this case the sub-lethal temperature was found to be 33°C (Figure 4b). These results were further validated by the plasmid retention assay in which the relative replication efficiencies in terms of stability of YCp50 (control) as well as YCpO−-2A plasmid were calculated at different sub-lethal temperatures in ts mutants of yeast for RPA32 and RPA70 (Figures 4c and d). The result showed that the percent stability of YCpO−-2A plasmid in the ts mutant of yeast RPA32 decreased several folds in comparison to the control YCp50 plasmid. For example, a shift to semi-permissive (28°C) temperature from permissive (22°C) temperature caused the decrease of stability of YCp50 and YCpO−-2A plasmid by <2-fold and 5- to 7-fold, respectively (Figure 4c). Similar preferential loss of stability of plasmid with geminivirus origin of replication was also observed in the RPA70 ts mutant of yeast (Figure 4d). However, the losses of YCpO−-2A plasmids were more severe with the ts defects in yeast RPA32 kDa protein than with the RPA70 kDa protein.
Figure 4.
Colony forming ability of yeast plasmids with or without geminiviral replicon at temperatures semi-restrictive for expression of RPA. [(a) (i)] and [(b) (i)] represent the sector diagram of the combinations of the yeast strains (W303a or ts mutants of RPA32 and 70) and transforming yeast plasmids (YCp50 or YCpO−-2A). The wild-type W303a yeast strain as well as ts mutants of RPA32 and RPA70 were transformed with either YCp50 vector (control) or with YCpO−-2A construct. The transformants were streaked on Ura− plates and allowed to grow at 33°C (permissive) (a, ii) and 35°C (semi-restrictive) (a, iii) temperatures in the case of RPA32 ts mutant. Similarly, the transformants were grown in Ura− plates at 30°C (b, ii) and 33°C (b, iii) temperatures for RPA70 ts mutant strains. Different colonies of ts mutant RPA32/A2F construct have been marked by # n. (c) and (d) Shows the percent stability of the plasmid YCp50 (control) and YCpO−-2A in the ts mutants of yeast for RPA32 and RPA70 respectively at different semi-restrictive temperatures.  – represent YCp50 and
– represent YCp50 and  represent YCpO−-2A.
represent YCpO−-2A.
Together, these results suggested that RPA32 and RPA70 kDa subunits are more critical for replication of plasmid DNA having the geminiviral replication origin compared to the plasmids with yeast origin of replication (ARS).
Interaction between MYMIV-Rep and PsRPA32 employing the yeast two-hybrid system
To understand how pea RPA helped in MYMIV DNA replication, the interactions between MYMIV-Rep and the subunits of RPA were examined since Rep is the major player in MYMIV DNA replication. The full size Rep i.e. Rep1–362, RPA32 and RPA70 genes were cloned in frame either with yeast GAL4 activation domain (pGAD) or the GAL4 DNA-binding domain (pGBD) vectors. The constructs were co-transformed in a yeast reporter strain AH109 in different combinations (Figure 5a) and the co-transformants were selected on synthetic media lacking leucine and tryptophan (Figure 5b). Yeast cells harbouring Rep- and RPA32-fusion proteins were able to grow on Leu−, Trp− and His− plates (Figure 5c) suggesting that the two proteins interacted with each other ex vivo. These cells also exhibited β-galactosidase activities (Figure 5d). None of the plasmids encoding the fusion proteins in combination with empty plasmid vector was able to induce His3 or LacZ expression in yeast cells. Various deletion as well as point mutants of the full-length MYMIV-Rep were engineered to characterize the domain important for its interaction with RPA32 (Figure 6e). The mutant Rep1–362K(227)E (or RepM2), which failed to interact with RPA32 in the in vitro pull down assay (Figure 6d, lane 8), was cloned in frame with the yeast GAL4 activation domain (pGAD) vector and its interaction was also checked as mentioned above. The mutant failed to interact with pea RPA32 in yeast two-hybrid assay (Figure 5c). Similarly, the interaction between PsRPA70 and MYMIV-Rep was also checked but the two proteins did not interact (Figure 5c). Together, these results showed that only the PsRPA32 kDa subunit interacted with MYMIV-Rep within the yeast cells.
Figure 5.
Yeast two-hybrid interaction of pea RPA32 and RPA70 with MYMIV-Rep1–362. PsRPA32/RPA70 and wild-type Rep1–362 as well as its mutant Rep1–362 M2 (K227E substitution) were cloned in frame with activation domain (AD) and DNA-binding domain (BD) vectors and the combination of plasmids shown in panel (a) were co-transformed in yeast reporter strain AH109. The transformants were selected on synthetic media lacking leucine and tryptophan. (b) and (c) Show the growth of co-transformants on synthetic media lacking leucine/tryptophan and leucine/tryptophan/histidine respectively. β-Galactosidase assay of replica colonies of (c) on theYPD plate are shown in (d).
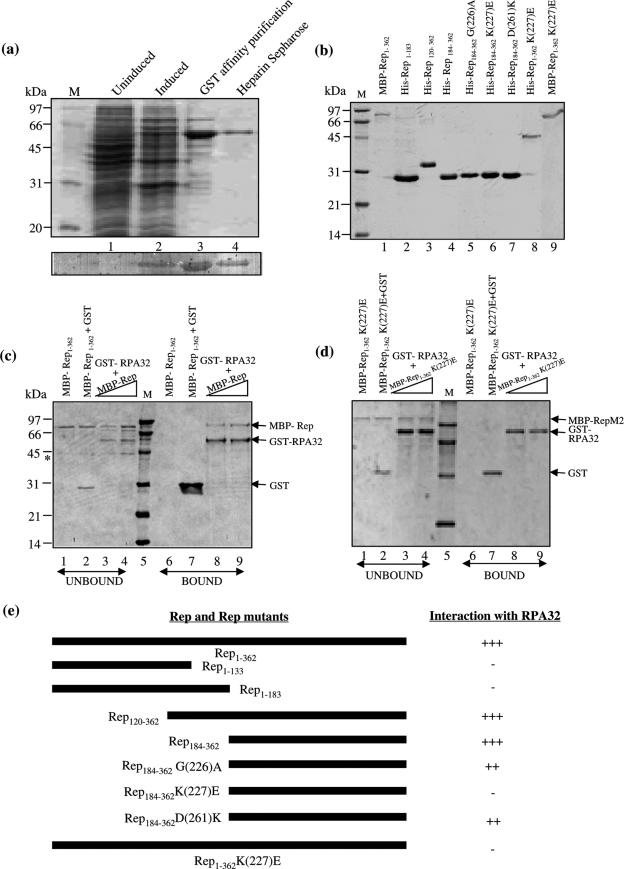
Figure 6.
In vitro interactions of pea RPA32 kDa protein with MYMIV-Rep protein. (a) Overexpression and purification of GST tagged RPA32 kDa protein. The GST tagged RPA32 was overexpressed in E.coli BL-21(DE3) cells with 0.5 mM IPTG. The protein samples were resolved on 12% SDS–polyacrylamide gel and visualized by Coomassie staining. Lane 1 shows uninduced and lane 2 shows induced total protein. The soluble fractions of the induced proteins were affinity purified through Glutathione S-transferase (GST) beads (lane 3), which were further subjected to purification through heparin sepharose CL-6B column chromatography (lane 4) for the removal of contaminating proteins. Lower panel shows the western blot analysis of the same gel with pea RPA32 polyclonal antibody. M—protein markers. (b) Wild-type Rep1–362 as well as various mutated or truncated versions of Rep protein purified through different affinity columns. (c) In vitro interactions of GST-RPA32 with MBP tagged MYMIV Rep1–362. Interaction study between GST-RPA32 and MBP-Rep1–362 was carried out by GST pull down assay. The unbound (input) fractions are shown in lanes 1–4 and corresponding bound fractions are shown in lanes 6–9. As controls, MBP-Rep1–362 alone and MBP-Rep1–362 alongwith purified GST protein were incubated with GST beads as shown in lanes 1, 2 and the bead eluates are shown in lanes 6, 7, respectively. Lanes 3, 4 and 8, 9 show the presence of GST-RPA32 with increasing concentrations of MBP-Rep1–362. (*) represents the MBP protein that gets co-eluted during purification. M represents protein markers. (d) In vitro interactions of GST-RPA32 with MBP tagged MYMIV Rep1–362K(227)E mutant (M2). The experimental protocol was similar to that mentioned in (c) except that the MBP-Rep1–362K(227)E mutant protein was used instead of MBP-Rep1–362 wild-type protein. (e) Schematic representation of interactions between GST-RPA32 and Rep1–362 wild-type as well as its mutants. Numeric letters designate the co-ordinates of the amino acids, +++ represents strong, ++ represents mild positive interactions and − represents negative interactions.
Interaction between recombinant MYMIV-Rep and PsRPA32 kDa in vitro
In order to confirm the above data, we examined the interaction between pea RPA32 and MYMIV-Rep in vitro and for this purpose the recombinant proteins of various mutants were made. The GST tagged PsRPA32 protein was overexpressed in E.coli BL-21 (DE3) cells (Figure 6a, lanes 1 and 2) and the overexpressed protein was chromatographed through the glutathione S-sepharose beads (lane 3). The tagged protein was further purified using the heparin sepharose CL-6B column (lane 4). The recombinant MYMIV-Rep1–362 and its various truncations were produced in bacteria and purified using suitable affinity chromatographic techniques (Figure 6b). The biochemical activities of each of the Rep mutants were also verified.
The in vitro interactions between MYMIV MBP-Rep1–362 and GST-PsRPA32 were carried out by GST pull down technique. The two proteins were mixed along with the GST beads and incubated in a binding buffer followed by gradual washing with increasing salt concentration (100–400 mM NaCl). The bound proteins were analysed by 12% SDS–PAGE. The results showed that PsRPA32 interacted with MBP-Rep1–362 and the interaction is stable even in presence of >400 mM of NaCl (Figure 6c). As a negative control, the MBP-Rep1–362 was incubated with purified recombinant GST protein (Figure 6c, lanes 2 and 7). Using the 6X His tagged Rep-truncations in this study, the C-terminal Rep (i.e. Rep184–362) but not the N-terminal Rep (i.e. Rep1–183) was found to interact with PsRPA32 kDa protein (Supplementary Figure S3a–e). To further map the site of interaction on the C-terminal Rep, we changed various amino acids in the C-terminal fragment of Rep as well as in the full-length Rep by site-directed mutagenesis. The targeted sites were Walker-A (including K227 and G226), Walker B motifs (including D261 and D264) and the extreme carboxy terminal regions. The pull down experiments with various mutant proteins showed that the amino acid ‘K’ at 227th position of Rep was critical for its interaction with RPA32 kDa protein (Supplementary Figure S3f–h and 6d). The loss of interaction with Rep (K227E) was also confirmed in the yeast two-hybrid system (Figure 5c). The summary of the results of interaction between PsRPA32 and wild-type or truncated version of Rep is presented in Figure 6e. The figure revealed that although many deletion and substitution mutants of Rep were included in the studies of interaction with pea RPA32 kDa protein, the null-interaction resulted with only one kind of Rep-mutant.
Taken together, these results showed that recombinant PsRPA32 interacted with recombinant MYMIV-Rep protein both in vitro and ex vivo. PsRPA32 interacted with Rep through a novel interacting site present in the C-terminus of the Rep protein. However, PsRPA70 was not able to interact with Rep in yeast two-hybrid assay (Figure 5c and d). Since this result differs from the ones with animal viruses, the experiments were carried out several times both in presence and absence of 3-AT, a histidine biosynthesis inhibitor. The reproducible negative results indicated the novel mode of interaction of RPA protein with MYMIV replication initiator.
Modulation of the biochemical activities of Rep by PsRPA32
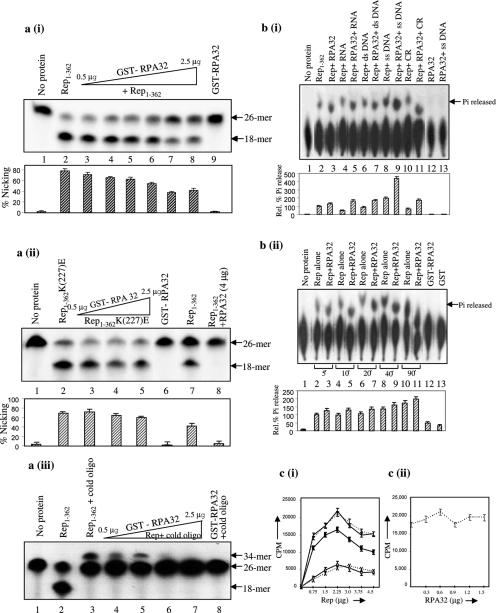
In order to elucidate the functional significance of the interaction between PsRPA32 and Rep, we checked whether PsRPA32 could modulate the intrinsic biochemical activities of Rep. First, we investigated the effect of RPA32 on nicking and ligation activity of MYMIV-Rep. A 5′ 32P-labelled 26mer nucleotide substrate was incubated with 500 ng of Rep and the 18mer cleaved product was analysed on 15% Urea–PAGE. The nicking activity of Rep was inhibited in presence of increasing concentrations of PsRPA32 [Figure 7a (i)]. However, the nicking activity of Rep1–362K(227)E mutant (∼0.5 μg) remained unchanged in presence of PsRPA32 [Figure 7a (ii)]. It is worth noting that the Rep (K227E) mutation does not cause any global disruption of the structure of the Rep protein as confirmed by ‘Rasmol’ structure analysis programme. We also checked the ligation activity of Rep in presence of PsRPA32. The ligation activity was also similarly reduced with increasing concentrations of PsRPA32 [Figure 7a (iii)]. This could be due to reduced availability of ligation substrates as a result of reduced nicking in presence of the RPA32 kDa protein. These results suggest that RPA32 is likely to have role in limiting the initiation of RCR. Such activity might help either in restricting the viral DNA copy number or shifting the replication reaction from the initiation mode to the elongation one.
Figure 7.
Modulation of intrinsic biochemical activities of Rep by PsRPA32. (a) The autoradiogram of the 15% polyacrylamide-urea gel showing nicking and ligation activities. (i) The 5′ 32P-labelled 26mer oligonucleotide substrate (lane 1), cleaved product i.e. 18mer generated by ∼500 ng of Rep1–362 alone (lane 2) and in presence of increasing concentrations (0.5–2.5 μg) of PsRPA32 (lanes 3–7) respectively are shown. Labelled substrate incubated with purified RPA32 (lane 8) is shown as control. The bar graph shown below represents the percent nicking for each lane. Experiments were performed in triplicate and the error bars indicate SEMs. The data were obtained by densitometric scanning of each lane of Figure (a) (i). (ii) Experiments performed are similar to (i) except that the Rep1–362(K227E) mutant protein was used instead of Rep1–362 wild-type protein (lanes 2–5). The cleaved products of Rep1–362 wild-type and in presence of RPA32 are shown in lanes 7 and 8, respectively. The amount of the Rep1–362(K227E) protein used for nicking reaction was ∼500 ng; and 0.5–2.5 μg of RPA32 was used. Lane 6 represents the nicking activity of RPA32 alone. The bar graph shown below represents the percent nicking for each lane with error bars indicating SEMs of the three independent experiments. (iii) Ligation activity of Rep in presence of PsRPA32. The 5′ 32P-labelled substrate and cleaved product by Rep1–362 alone are shown in lanes 1 and 2, respectively. The Rep-mediated ligated product of 5′-32P-labelled oligonucleotide and the unlabelled 26mer oligonucleotide (lane 3) and in presence of increasing concentrations (0.5–2.5 μg) of PsRPA32 (lanes 4–7) are shown. (b) Autoradiogram showing the ATPase activity of Rep with or without RPA32 protein. (i) Autoradiogram shows the ATP hydrolysing activity of Rep1–362 to release free phosphate (Pi) from ATP (lane 2), in presence of different modulators like RNA (lane 4), ds DNA (lane 6), ssDNA (lane 8) and CR (lane 10). The intrinsic ATPase activity of Rep in presence of RPA32 kDa protein and different modulators are shown in lanes 3, 5, 7, 9 and 11. The ATPase activity of PsRPA32 protein, without or with ssDNA is shown in lanes 12 and 13 as control. Densitometric scanning quantitated the amount of Pi released. Bar graph below represents the percent Pi released for each lane with error bars indicating SEMs of three independent experiments. Percent Pi released by Rep was arbitrarily assigned as 100% and other values were relative to this scale. Lane 1 represents control without any protein. (ii) Time course kinetics of intrinsic ATPase activity of Rep1–362 in presence or absence of RPA32 after 5, 10, 20, 40 and 90 min time intervals of incubation (lanes 2–11). The percent Pi release for each lane is shown in bar diagram below. The error bars for each lane indicate SEMs. Lanes 1, 12 and 13 represent control without any protein, with RPA32 and with GST respectively. (c) Panel (i) represents the DNA-binding ability of increasing concentrations of Rep1–362 (0.75–4.5 μg) to common region (CR) without or with ∼1 μg of RPA32 [— for Rep, ----- for Rep + ∼ 1 μg RPA32], in presence of 10 ng unlabelled CR (10× specific competitor) [ Rep, ----X--- - Rep +1 μg RPA32] and in presence of non-specific DNA molecule [50× non-specific competitor,
Rep, ----X--- - Rep +1 μg RPA32] and in presence of non-specific DNA molecule [50× non-specific competitor,  ]. The DNA-binding of 1.5 μg of Rep in presence of increasing concentrations of RPA32 (0.3–1.5 μg) is shown in panel (ii). Error bars indicate SEMs.
]. The DNA-binding of 1.5 μg of Rep in presence of increasing concentrations of RPA32 (0.3–1.5 μg) is shown in panel (ii). Error bars indicate SEMs.
Besides nicking and closing activity, the ATPase activity of Rep is also essential for viral DNA replication. Hence, we investigated the role of PsRPA32 on Rep-mediated ATPase activity in presence or absence of different modulators [Figure 7b (i)]. The ATPase activity of Rep increased to ∼2-fold in presence of either PsRPA32 or ssDNA individually [Figure 7b (i), lanes 2, 3 and 8]. The ATPase activity of Rep in presence of both ssDNA and RPA32 kDa protein was about 4–5 times the normal activity of Rep [compare lane 2 with lane 9 of Figure 7b (i)], suggesting that RPA32 kDa might stabilize the Rep–ssDNA interaction. Neither RNA nor dsDNA could influence the Rep-mediated ATPase activity [Figure 7b (i), lanes 4 and 6]. As a control, the ATPase activity of Rep was also checked in presence of purified GST protein and GST alone did not affect the activity at all (data not shown). Time course kinetics of the ATPase activity of Rep with or without RPA32 showed that the activity increased up to about 20 min and reached plateau thereafter [Figure 7b (ii)]. These results also suggest that RPA32 might help Rep protein in its post-initiation activities of viral DNA replication where the ATPase activities play crucial role(s).
We further checked the DNA-binding activity of Rep to the common region (MYMIV-CR, the origin of RCR) in presence of PsRPA32 (Figure 7c). RPA32 did not have any effect on the binding of Rep to ‘CR’ DNA. In the presence of 10-fold excess of specific competitor DNA, the CR-binding to Rep (1.5 μg) dropped down by about 4.5-fold whereas the similar binding reduced only 1.3-fold in presence of 50-fold excess of non-specific competitor, suggesting specific binding nature of Rep to ‘CR’ DNA. The specific reduction of Rep-binding was not influenced at all in presence of RPA32 kDa protein [Figure 7c (ii)].
Combining the yeast-data with the results obtained in vitro on the RPA32-induced up regulation of the Rep-mediated ATPase activity, it appears that RPA32 protein could be a positive regulator of post-initiation activity in viral DNA replication. To examine the effect of PsRPA32 kDa protein on viral replication in planta, we resorted to transient replication and expression assays in the leaves of N.xanthi plant.
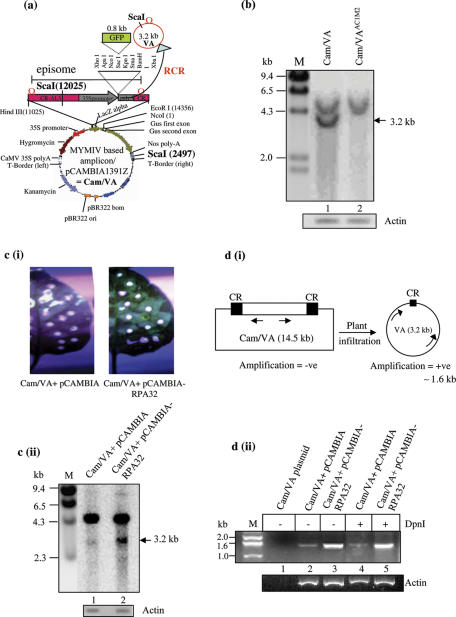
RPA32kDa induced enhanced replication of MYMIV based amplicon in Nicotiana
Recent studies in our laboratory have shown that a MYMIV based amplicon is competent to replicate in the host as well as non-host plants [ICGEB activity report (2003); PhD thesis of M. N. Islam, (2005)]. A segment of MYMIV DNA ‘A’, viz., CR-AL3 can act as a functional replicon in the yeast model. The CR-AL3 segment includes replication origin (CR) and the DNA encoding the replication proteins, namely Rep and the replication enhancer (AL3). Hence, the binary construct (Figure 8a) bearing two copies of CR is capable of releasing an episomal DNA of 3.2 kb within the injected plant during RCR (45). The release of the portion of DNA (3.2 kb) flanked by two CRs results because of the RCR which ensues following the site-specific nicking of Rep at the CR. The replicated DNA could be visualized as a band of 3.2 kb in Southern blot following digestion of the isolated DNA with ScaI restriction enzyme and under similar circumstances, the unreplicated and unreleased DNA would be detected as a band of about 4.8 kb. The amplicon was agro-infiltrated into the N.xanthi leaves and the replicated DNA of 3.2 kb was visualized by Southern analysis at 14 days post inoculation (Figure 8b, lane 1). The replicated band of 3.2 kb was found to be Rep dependent, since the mutant Rep (K227E, a mutation in the Walker-A motif of the helicase domain) failed to release the episomal DNA of 3.2 kb (Figure 8b, lane 2).
Figure 8.
Enhanced replication of MYMIV based amplicon in presence of RPA32. (a) The panel shows the schematic view of the MYMIV based amplicon (Cam-VA). A horizontal line bordered by two vertical lines indicates the portion of DNA, flanked by two CRs. The release of sub-viral episomal DNA [VA circle, also in 8(d)] of 3.2 kb following RCR is indicated by curved arrows and circles. The ScaI site, used for Southern analysis, is shown. (b) Southern blot analysis of the genomic DNA. The DNA was isolated from the leaves of N.xanthi infiltrated either with MYMIV based amplicon (lane 1) or with amplicon mutant (lane 2) at 14 days post-infiltration. About 50 μg of the genomic DNA was digested with ScaI and resolved on 0.8% agarose gel followed by Southern blot analysis with the radiolabelled GFP probe. The ScaI digestion of the amplicon results in a non-replicated band (Cam/VA) of 4.8 kb and the replicated band (VA) of 3.2 kb respectively. (c) (i) shows the GFP expression in the leaves of Nicotiana infiltrated either with the amplicon along with empty pCAMBIA vector or amplicon plus pCAMBIA-RPA32 at 7 days post-infiltration. The Southern blot analyses of MYMIV-amplicon alone and amplicon plus RPA32 are shown in (c) (ii). The horizontal arrow marks the replicated DNA (3.2 kb). Radiolabelled GFP was used as the probe. The M lane shows the molecular weight standards in kb. The Southern blot was reprobed with actin for loading control. (d) The PCR based analysis to distinguish between the replicated (VA) and unreplicated (Cam-VA) DNA. Panel (i) shows the schematics of the experiment. Arrows represent the primers designed for the PCR, panel ii, the plasmid DNA of the amplicon (Cam-VA), isolated from E.coli, was taken as a negative control (lane 1). The genomic DNA isolated from amplicon (lane 2) or amplicon plus RPA32 (lane 3) samples after 7 days post-infiltration was taken as template for the PCR using primers as mentioned. The PCR was carried out for 23 cycles only. Lanes 4 and 5 show the PCR analysis of the same DNA after DpnI digestion. Lower panel shows the amplification of actin as a loading control.
We argued that if RPA32 kDa protein was important for MYMIV DNA replication, the ectopic over expression of RPA32 kDa protein in the transient expression system would enhance the release of episomal DNA in the infiltrated leaves. The amplicon and the amplicon plus RPA32 gene provided in trans were agro-infiltrated into the N.xanthi leaves and the protein was allowed to be ectopically overexpressed for 7 days. The replication efficiency was subsequently assayed by Southern blot analysis of total DNA isolated from the infiltrated zone using the GFP radiolabelled probe at 7 days post-infiltration. We observed that the replication of the episomal DNA was enhanced in presence of PsRPA32 as evidenced by the increased accumulation of 3.2 kb DNA in Southern analysis [Figure 8c (ii), lanes 1 and 2]. The enhanced replication was also reflected in the increased accumulation of the GFP protein in the RPA32 overexpressing leaves when observed under UV [Figure 8c (i)]. The DNA extraction time from the infiltrated leaves was set at the 7th days post-infiltration as the relative RPA32-induced effect on episomal replication was found maximal at this point of time. The Southern blot was reprobed with the actin DNA to reveal the uniformity in the amount of DNA loading. Similar results were also derived following infiltration in the pea plants (data not shown).
In order to ascertain that 3.2 kb band was a true replicated product, we also devised a PCR based method for high-resolution detection. Two divergent primers [shown in Figure 8d (i)] were designed in such a manner that a PCR product of 1.6 kb would be obtained only when the circular, episomal DNA is released from the vector during RCR. However, there would not be any PCR amplification with the unreplicated plasmid (Cam/VA) DNA template under such conditions because of the low processivity of the DNA Polymerase (otherwise the amplification would have resulted in a 12.9 kb DNA band). The PCR primers (sequences shown in the Materials and Methods section) were located in the GFP and the N-terminus of Rep, respectively and were about 1.6 kb apart. The total DNA isolated from the amplicon and the amplicon plus RPA32 infiltrated leaves were used as the template for the PCR [Figure 8d (ii)]. The PCR(s) were carried out for 23 cycles only at which the DNA bands of the amplicon alone sample were just visible by ethidium staining. The results showed that a PCR product of 1.6 kb was obtained in amplicon as well as amplicon plus RPA32 infiltrated samples [Figure 8d (ii), lanes 2 and 3]. The band intensity in the amplicon plus RPA32 sample was higher in comparison to amplicon alone sample suggesting enhanced replication in presence of RPA32 protein. This PCR-result corroborated well with the data derived from the Southern analyses [Figure 8c (ii)]. The supplementary data presented in Figure S4 show that the enhancement of replication was specific for the RPA32 protein, as the ectopic expression of GUS protein (as a non-specific control) from the pBI121 plasmid did not enhance the replication characteristics of amplicon plasmid. The plasmid DNA of the amplicon isolated from E.coli was taken as a negative control [Figure 8d (ii), lane 1]. As the nascent DNA is resistant to DpnI digestion, the DNA from the amplicon and amplicon plus RPA32 infiltrated samples were treated with DpnI enzyme before PCR to distinguish the replicated episomal DNA from the parental DNA. The presence of 1.6 kb band in these samples further confirmed that the episomal DNAs are the true products of replication [Figure 8d (ii), lanes 4 and 5].
DISCUSSION
We have investigated the role of pea RPA, especially of the subunit RPA32, in the RCR of MYMIV DNA ‘A’ component. Two major subunits of RPA, viz., RPA70 and RPA32 genes were isolated from pea and the biochemical interactions of the respective proteins were examined with MYMIV-Rep. The RPA of pea plant was chosen in this study, since it has been shown independently that MYMIV DNA can replicate transiently in pea following agroinoculation (M. N. Islam et al., unpublished data). Earlier investigations from our laboratory revealed that a plasmid vector harbouring geminiviral origin of replication could replicate in budding yeast (1). Hence, we used the yeast model system to elucidate the role of RPA32 and 70 kDa subunits in geminiviral DNA replication. The replication assay at the sub-lethal temperature using the ts mutants of yeast RPA70 and 32 revealed that both the subunits are essential for the replication of the plasmid vector bearing geminiviral origin (Figure 4). Moreover, it is clear from the in planta based transient replication assay that the RPA32 kDa protein up regulates MYMIV DNA replication (Figure 8). These results strongly suggested that both RPA32 and 70 might be involved in geminiviral DNA replication. This is the first report of the involvement of RPA32 and 70 kDa genes in the geminiviral DNA replication.
Multiple alignments of pea RPA32 and 70 kDa genes with their respective homologues of eukaryotic organisms suggest that pea RPA32 and 70 kDa genes are much closer to their plant homologues (Supplementary Figures S1 and S2). The complementation assay with the ts strains of yeast RPA32 and RPA70 by pea RPA32 and 70 kDa genes suggested that only RPA70 is able to complement for the function of yeast RPA70. The failure of pea RPA32 to complement for yeast RPA32 function (Figure 1) is not very surprising. This result is in accordance with the earlier reports that human RPA32 failed to complement the yeast rpa2 mutant (41) and the RPA32 and 14 kDa subunits are presumably responsible for species-specific interaction with ssDNA (42). However, there are at least two lines of evidence to suggest that the isolated PsRPA32 is a true functional homologue of the RPA32s reported from other systems. First, pea RPA32 interacted strongly with pea RPA70 in the yeast two-hybrid assay (Figure 2). Second, the polyclonal antibodies raised against the recombinant pea RPA32 proteins recognised the proteins of correct molecular size from the total protein extract of pea in the western analysis (Figure 3c).
We found that RPA32 was able to interact with MYMIV-Rep both in the yeast two-hybrid and in the in vitro pull down assays (Figures 5 and 6). Surprisingly, RPA70, the major subunit of RPA heterotrimeric complex did not show any interaction with MYMIV-Rep in yeast two-hybrid assay. This result is in contrast with the earlier reports that the initiator proteins of different animal viruses, e.g. large T antigens of SV40, E1 of papillomavirus, EBNA1 of Epstein–Barr virus etc., make direct contacts with the RPA70 kDa subunit of RPA complex (32,34,46). This could be due to either the different strategies that the plant DNA viruses adopt to interact with host proteins or the RCR mode of DNA replication of geminiviruses that differs from the ‘Cairns’ (θ) mode of replication for most of animal DNA viruses. Interestingly, RPA32 interacted with a novel site present in the C-terminal of the Rep protein. This is significant since most of the host proteins e.g. PCNA (15), coat protein (37), pRBR (6), GRIK and GRIMP (18) etc. interact with Rep through the oligomerization domain of the Rep and only a few proteins [namely, GRAB1 and GRAB2, (47)] are known to interact with C-terminal domain of WDV RepA. Although RPA70 was not in direct contact with Rep as evident from the yeast two-hybrid data, it could still be associated with Rep as an integral component of the RPA complex. Many key proteins of DNA replication like DNA pol α, PCNA etc. make direct contact with RPA70 kDa subunit of RPA complex and get recruited at the site of their respective functions. However, the possibility of RPA70 making contact with viral Rep protein and modulating its functions could not be totally ruled out in vivo, since the binding mode of the RPA complex could be different from that of RPA70 alone. Eventually, it would be more significant to understand how the activity of Rep is altered in presence of RPA heterotrimeric complex rather than with the individual subunits.
As a result of Rep–RPA32 interaction, the nicking and closing activity of Rep was down regulated (Figure 7a). The fact that RPA32 interacted with C-terminal of the Rep and affected the nicking-closing activity that is located at the N-terminus is significant and interesting. It is likely that the binding of RPA32 to the C-terminal segment of Rep caused allosteric changes in the Rep protein in such a manner that Rep was rendered less accessible to the substrate DNA molecule. This interpretation corroborates with our earlier observation of ‘cross-talk’ between N- and C-terminal of the Rep protein (14). However, the exact relationship between the inhibition of nicking and enhanced replication of the viral amplicon remains very puzzling at the present moment. The DNA synthesis might enhance if Rep disengages from nicking-closing to fork extending activities. Moreover, the observation of inhibition of nicking-closing was an in vitro datum while the replication enhancement was an in vivo effect. At the replication origin many proteins besides RPA might assemble in vivo, thus making the in vivo nicking status very difficult to predict.
Since RPA32 interacted with the C-terminal domain of the Rep, we investigated the impact of this interaction by studying the biochemical alteration of the characteristics resident in the C-terminal domain of the Rep. Thus we found that the interaction led to the enhancement of intrinsic ATPase activity of Rep, which in turn was further boosted by single-stranded DNA (Figure 7b). Our preliminary data indicate that the helicase activity of Rep could also be up regulated by the presence of RPA32 (D. K. Singh et al., unpublished data). Such up regulation of ATPase and the helicase activity could also enhance the replication of viral amplicon.
It is not difficult to envisage that the RPA32 mediated enhancement of ATPase activity of Rep would be essential for the post-initiation step of viral DNA replication. In case of parvoviruses (35), the interaction of RPA with initiator protein NS1 leads to the energy-requiring extensive unwinding of the nicked origin. A similar scenario could also be envisaged for MYMIV DNA replication where the Rep–RPA complex would lead to extensive unwinding of the origin, thus facilitating the formation and progression of the replication fork.
The in vivo role of RPA proteins in geminivirus DNA replication has been highlighted from studies in the yeast model system at the semi-permissive temperatures and transient replication in plants. The limiting amount of RPA32 kDa protein adversely affected MYMIV- based plasmid DNA replication and caused severe plasmid loss resulting in the disability of colony formation in yeast. It would be important to map the domain of RPA32 kDa protein that interacts with geminivirus-Rep and the domain(s) that control viral DNA replication. Such studies would indicate how the physical interaction leads to the control of viral DNA replication. At present, in planta based studies are difficult to carry out because of lack of availability of various allelic mutants of RPA32. However, the use of VIGS vectors might be helpful in answering some of the relevant questions by knocking out the proteins in a transient manner. Similar studies are also necessary for the RPA70 and 14 kDa subunits. However, it is clear from Figure 8 that in planta over expression of RPA32 leads to enhanced replication of the viral amplicon. Unfortunately, the mechanism of RPA32-induced stimulation of replication is not clear yet as several mechanisms of stimulation could be hypothesized. The ectopic over expression of RPA32 might increase the total pool of trimeric RPA as our data (Figure 3) suggest that resident RPA32 was limiting whereas RPA70 was not (at least at the transcript level). Moreover, it is not clear how the abundance of RPA32 could control the level of other subunits of RPA. Additionally, some individual subunits like RPA32 could control viral replication independently. Further experiments need to be carried out to unravel the exact mechanism of RPA32-induced stimulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors would like to thank Dr S. J. Brill (Rutgers University, Piscataway, NJ) for providing yeast RPA32 mutant strain, Dr Richard D. Kolodner (Ludwig Institute for Cancer research, La Jolla, CA) and Keiko Umezu (Nara Institute of Science and Technology, Nara, Japan) for yeast RPA70 mutant strains. The authors also thank Dr Marc S. Wold (University of Iowa, Iowa city, IA) for his suggestions. The assistance of Dr M. K. Reddy for RACE technique is greatly appreciated. The financial help to D.K.S. and S.K. from CSIR, New Delhi, India is duly acknowledged. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Raghavan V., Malik P.S., Choudhury N.R., Mukherjee S.K. The DNA-A component of a plant geminivirus (Indian mung bean yellow mosaic virus) replicates in budding cells. J. Virol. 2004;78:2405–2413. doi: 10.1128/JVI.78.5.2405-2413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varma A., Malathi V.G. Emerging threats of geminiviruses: a serious threat to crop production. Ann. Appl. Biol. 2003;142:145–164. [Google Scholar]
- 3.Saunders K., Lucy A., Stanley J. DNA forms of the geminivirus Africa cassava mosaic virus consistent with the rolling circle mechanism of replication. Nucleic Acids Res. 1991;19:2325–2330. doi: 10.1093/nar/19.9.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagar S., Pedersen T.J., Carrick K.M., Hanley-Bowdoin L., Robertson D. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cell. Plant Cell. 1995;7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ach R.A., Durfee T., Miller A.B., Taranto P., Hanley-Bowdoin L., Zamryski P.C., Gruissem W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arguello-Astorga G., Lopez-Ochoa L., Kong L.J., Orozco B.M., Settlage S.B., Hanley-Bowdoin L. A novel motif in geminiviral replication proteins interacts with the plant retinoblastoma related protein. J. Virol. 2004;78:4817–4826. doi: 10.1128/JVI.78.9.4817-4826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong L.J., Orozco B.M., Roe J.L., Nagar S., Ou S., Feiler H.S., Durfee T., Miller A.B., Gruissem W., Robertson D., et al. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 2000;19:3485–3495. doi: 10.1093/emboj/19.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Q., Sanz-Burgos A.P., Hannon G.J., Gutierrez C. Plant cell contains a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Q., Saurez-Lopez P., Gutierrez C. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J. 1995;14:4073–4082. doi: 10.1002/j.1460-2075.1995.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egelkrout E.M., Robertson D., Hanley-Bowdoin L. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell. 2001;13:1437–1452. doi: 10.1105/tpc.13.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez C. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 2000;19:792–799. doi: 10.1093/emboj/19.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez C. Geminiviruses and the plant cell cycle. Plant Mol. Biol. 2000;43:763–772. doi: 10.1023/a:1006462028363. [DOI] [PubMed] [Google Scholar]
- 13.Fontes E.P.B., Luckow V.A., Hanley-Bowdoin L. A geminiviral replication protein is a sequence-specific DNA-binding protein. Plant Cell. 1992;4:597–608. doi: 10.1105/tpc.4.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pant V., Gupta D., Choudhury N.R., Malathi V.G., Varma A., Mukherjee S.K. Molecular characterization of the Rep protein of the blackgram isolate of Indian mung bean yellow mosaic virus. J. Gen. Virol. 2001;82:2559–2567. doi: 10.1099/0022-1317-82-10-2559. [DOI] [PubMed] [Google Scholar]
- 15.Bagewadi B., Chen S., Lal S.K., Choudhury N.R., Mukherjee S.K. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J. Virol. 2004;78:11890–11903. doi: 10.1128/JVI.78.21.11890-11903.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley J. Analysis of Africa cassava mosiac virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology. 1995;206:707–712. doi: 10.1016/s0042-6822(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 17.Luque A., Sanz-Burgos A.P., Ramirez-Parra E., Castellano M.M., Gutierrez C. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology. 2002;302:83–94. doi: 10.1006/viro.2002.1599. [DOI] [PubMed] [Google Scholar]
- 18.Kong L.J., Hanley-Bowdoin L. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression pattern during plant development and infection. Plant Cell. 2002;14:1817–1832. doi: 10.1105/tpc.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA replication. Ann. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 20.Fairman M.P., Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brill S.J., Stillman B. Replication protein-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 22.Bochkarev A., Bochkareva E., Frappier L., Edwards A.M. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA-binding. EMBO J. 1999;16:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkarev A., Pfuetzner R.A., Edwards A.M., Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 24.Dutta A., Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fotedar R., Roberts J.M. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J. 1992;11:2177–2187. doi: 10.1002/j.1460-2075.1992.tb05277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henricksen L.A., Carter T., Dutta A., Wold M.S. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 1996;24:3107–3112. doi: 10.1093/nar/24.15.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francon P., Lemaitre J., Dreyer C., Maiorano D., Cuvier O. A hypophosphorylated form of RPA34 is a specific component of pre-replication centres. J. Cell Sci. 2004;117:4909–4920. doi: 10.1242/jcs.01361. [DOI] [PubMed] [Google Scholar]
- 28.Vassin M.V., Wold M.S., Borowiec J.A. Replication protein A (RPA) phosphorylation prevents RPA association with replication centres. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dornreiter I., Erdile L.F., Gilbert I.U., Winkler D., Kelly T.J., Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loor G., Zhang S.J., Zhang P., Toomey N.L., Lee M.Y.W.T. Identification of DNA replication and cell cycle proteins that interacts with PCNA. Nucleic Acids Res. 1997;25:5041–5046. doi: 10.1093/nar/25.24.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny M.K., Lee S.H., Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: Single-strand stabilization and stimulation of DNA polymerase α and δ. Proc. Natl Acad. Sci. USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D., Frappier L., Gibbs E., Hurwitz J., O'Donnell M. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein–Barr virus. Nucleic Acids Res. 1998;26:631–637. doi: 10.1093/nar/26.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Y., Loo Y.M., Militello K.T., Melendy T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 1999;73:4899–4907. doi: 10.1128/jvi.73.6.4899-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo Y.M., Melendy T. Recruitment of Replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 2004;78:1605–1615. doi: 10.1128/JVI.78.4.1605-1615.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen J., Tattersall P. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 2002;76:6518–6531. doi: 10.1128/JVI.76.13.6518-6531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 37.Malik P.S., Kumar V., Bagewadi B., Mukherjee S.K. Interaction between coat protein and replication initiation protein of Mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology. 2005;337:273–283. doi: 10.1016/j.virol.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 38.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda K., Morioka H., Imajou S., Ikeda S., Ohtsuka E., Tsurimoto T. Structure function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J. Biol. Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 40.Brill S.J., Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 41.Brill S.J., Stillman B. Replication protein-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 42.Sibenaller Z.A., Sorensen B.R., Wold M.S. The 32- and 14-kilodalton subunit of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 43.Philipova D., Mullen J.R., Maniar H.S., Lu J., Gu C., Brill S.J. A hierarchy of SSB protomers in replication protein A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 44.Knaap E., Jagoueix S., Kende H. Expression of an ortholog of replication protein A1 (RPA1) is induced by gibberellin in deepwater rice. Proc. Natl Acad. Sci. USA. 1997;94:9979–9983. doi: 10.1073/pnas.94.18.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenger D.C., Revington G.N., Stevenson M.C., Bisaro D.M. Replication release of geminivirus genomes from tandemly repeated copies: Evidence for rolling circle replication of plant viral DNA. Proc. Natl Acad. Sci. USA. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M., Park J.S., Ishiai M., Hurwitz J., Lee S.H. Species specificity of human RPA in simian virus 40 DNA replication lies in T-antigen-dependent RNA primer synthesis. Nucleic Acids Res. 2000;28:4742–4749. doi: 10.1093/nar/28.23.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Q., Sanz-Burgos A.P., Guo H., Garcia J.A., Gutierrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 1999;39:647–656. doi: 10.1023/a:1006138221874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.