Abstract
Holliday junctions are a central intermediate in diverse pathways of DNA repair and recombination. The isomerization of a junction determines the directionality of the recombination event. Previous studies have shown that the identity of the central sequence of the junction may favor one of the two isomers, in turn controlling the direction of the pathway. Here we demonstrate that, in the absence of DNA sequence-mediated isomer preference, polycations are the major contributor to biasing strand cleavage during junction resolution. In the case of wild-type phage λ excision junctions, spermidine plays the dominant role in controlling the isomerization state of the junction and increases the rate of junction resolution. Spermidine also counteracts the sequence-imposed bias on resolution. The spermidine-induced bias is seen equally on supercoiled and linear excisive recombination junction intermediates, and thus is not just an artefact of in vitro recombination conditions. The contribution of spermidine requires the presence of accessory factors, and results in the repositioning of Int's core-binding domains on junctions, perhaps due to DNA-spermidine–protein interactions, or by influencing DNA conformation in the core region. Our results lead us to propose that spermidine together with accessory factors promotes the formation of the second junction isomer. We propose that this rearrangement triggers the activation of the second pair of Int active sites necessary to resolve Holliday junctions during phage λ Int-mediated recombination.
INTRODUCTION
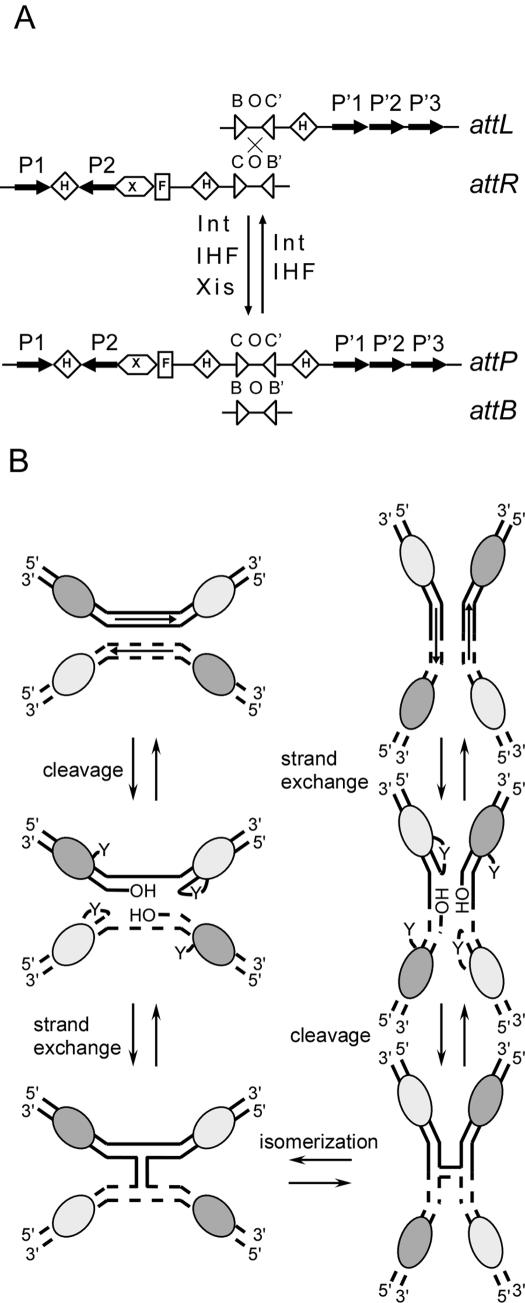
Site-specific recombination mediated by phage λ integrase (Int) was the first DNA recombination reaction reconstituted in vitro (1) and has been the subject of intense study ever since [reviewed by (2)]. Nevertheless, some basic aspects of the recombination reaction remain poorly understood. Int is a multi-functional enzyme, which binds to its recombination substrates, the att sites (Figure 1A), with different domains—the arm-binding domains interact with the so-called arm sites (P1, P2, P′1–P′3), while the core-binding and catalytic domains interact with the core sites (B, B′, C and C′). Int juxtaposes the att sites in synaptic complexes in preparation for two rounds of single-strand DNA cleavage, exchange and ligation (Figure 1B). The first strand exchange event generates a Holliday junction intermediate which is resolved by the second strand exchange event (Figure 1B). Recombination between the phage attP site and bacterial attB site results in integration of the phage into the bacterial chromosome and requires the host-encoded protein IHF. Recombination between attL and attR, which flank the integrated prophage, excises the phage in preparation for re-entry into the lytic cycle and requires the phage encoded Xis protein and the host-encoded Fis protein in addition to IHF. Int performs catalysis using a type IB topoisomerase mechanism, and the substrates and products of the reaction are isoenergetic. While recombination requires no high-energy cofactors, it is stimulated 5–10× by polycations (1,3).
Figure 1.
Phage λ Int-mediated site-specific recombination. (A) Schematic representation of att sites indicating protein binding sites and cofactor requirements for excisive (attL × attR) and integrative (attP × attB) recombination. B, B′, C and C′ are binding sites for Int's core and catalytic domains. O denotes the overlap region between the loci of strand cleavage, 7 bp of identity between the two partner substrates. P1, P2 and P′1–P′3 denote the binding sites for Int's arm-binding domain. H denotes IHF binding sites. X denotes at least two Xis binding sites. F denotes a Fis binding site. (B) Catalytic events mediated by Int during site-specific recombination [adapted from Ref. (17)]. The Int protein together with appropriate accessory factors juxtaposes the two recombination substrates in a synaptic complex, indicated upper left. The active site tyrosine of two of the four Int monomers (light grey ovals) attacks the phosphodiester backbone and forms a transient 3′ phosphotyrosyl bond between the enzyme and the TS of each DNA substrate. Ligation occurs when the resulting free 5′-OH group from a partner substrate (or from the original substrate) acts as a nucleophile at this phosphotyrosyl linkage. Since the two strands of each DNA substrate are cleaved independently, a Holliday junction is generated during recombination. The junction complex undergoes isomerization, which positions the previously inactive Int monomers into the appropriate conformation for subsequent catalytic steps. The Holliday junction is resolved by a repetition of the previous DNA cleavage, strand exchange and ligation steps occurring on the BS of each DNA substrate, resulting in two recombinant DNA molecules.
A major question remaining in the analysis of tyrosine recombinases is what drives the reaction forward. The tyrosine recombinases appear equally likely to bind substrates or products and assemble nucleoprotein complexes given the presence of the appropriate accessory proteins and recombination partner. In the case of λ Int, a certain asymmetry has been found with respect to binding preference—in both integrative and excisive recombination, one att site (attP and attL, respectively) serves as the strong binding partner and donor of Int molecules to the weak binding partner (attB and attR, respectively) (4,5). These interactions serve to align the substrates in their appropriate synaptic complexes [reviewed in (2,6)]. The crystal structure of the synaptic complex in Cre-lox recombination showed that the two activated catalytic sites are positioned differently with respect to the scissile phosphates and each other than the not-yet-active catalytic sites, and that the trajectories of the cleaved strands differ (7). This asymmetry is achieved by preferred bending at one side of the overlap region, the region between the sites of top (TS) and bottom (BS) strand exchange (Figure 1A); Flp recombinase also bends its substrate (8–13). In the case of Int, the order of strand exchanges is determined by the accessory factors (14,15). While accessory factors also influence the second strand cleavage event (16), it is not clear how the decision between different cleavage loci is made in the context of the Holliday junction.
Crystallographic studies of both the Cre-lox and the Flp-frt junction complexes have shown that the Holliday junction complexes are 2-fold symmetric—two of the junction arms are closer to each other than the remaining pair [reviewed in (6,13)]. Based on these crystal structures, junction isomerization has been proposed to activate the second pair of Int monomers to perform cleavage, strand exchange and ligation on the second set of DNA strands (17). What promotes isomerization to the second isomer of the Holliday junction, thereby activating cleavage, is not known. Several new crystal structures of a tetramer of full-length Int have been solved, in which the core-binding domains interact with either double-stranded DNA or HJ composed of core sites (one ligated, one not) while the arm-binding domains interact with two identical arm-site-containing molecules (18). The new crystal forms show an unexpected arrangement of Int's arm domains with respect to the other two, in which one arm domain communicates with the core-binding domain of a neighboring Int and is thus poised to receive and transmit information allosterically (18–20). While these crystals are very informative and suggestive, they do not reveal the dynamic events occurring during junction isomerization, especially those involving accessory proteins, since the crystals do not include the accessory proteins or their binding sites.
Currently it is not known if the probability of the junction occupying each of the two isomeric states is equal or if the two isomeric states are energetically different. If the former is true, for example in the case of the simpler Cre-lox recombination reaction, the reaction would have an equal likelihood of proceeding forward towards the ultimate reaction products or backward towards the starting recombination substrates. In the case of the Int-att reaction, one might predict that the directionality of the reaction at the Holliday junction stage would be determined by the accessory factors, just as earlier in the reaction. Previous studies from the Landy lab have looked at the isomerization of att site junctions and found little if any preference for one isomer over the other (21). However, for practical purposes these studies were done with minimal junctions that included only the core-binding sites for the Int protein and lacked the accessory protein binding sites (Figure 1). We have now reexamined this issue from two perspectives—first, we have used Holliday junctions containing all of the sequences normally required for a complete recombination reaction and compared junction resolution using wild-type Int protein alone or together with accessory factors, to test the effect of the latter on the directionality of resolution. Second, we have examined the influence on isomerization of polycations, including spermidine. As mentioned above, spermidine has long been known to stimulate Int-mediated recombination reactions ∼5- to 10-fold (3). However, its role in the reaction is still mysterious.
Polyamines are a subset of polycationic compounds that are present in all prokaryotes and eukaryotes (22–24). Most steps in polyamine biosynthesis are shared among all organisms. The most common polyamines are putrescine, spermidine and spermine, which are divalent, trivalent and tetravalent, respectively. In Escherichia coli, the total intracellular concentration of putrescine is 20 mM, spermidine is 6 mM (similar to the magnesium concentration), and spermine is lacking [(25,26); reviewed in (23)]. Polyamines are involved in many central metabolic processes in vivo, including RNA and protein biosynthesis, and the elongation stage of DNA replication. Not only are polyamines crucial for bacteria (27) but for bacteriophages as well. While the T series phages use polyamines to various extents, bacteriophage λ does not grow at all in polyamine deficient hosts, either after induction of a lysogen or in a lytic infection (28).
In vitro studies showed that spermidine is required for the integration of λ DNA into chromosomal DNA (1) and that it stimulates the excision of λ prophage ∼5- to 10-fold (29). Spermidine can be replaced to some extent by some divalent and trivalent cations, including MgCl2 and hexammine cobalt (3). Subsequent work showed that spermidine is required for a step after synapsis but before strand exchange (30–32). The dispensability of spermidine for synapsis was surprising, since spermidine's role as a counterion would have been a logical stimulator of substrate juxtaposition.
We have investigated the role of spermidine during resolution of Holliday junctions and found that spermidine affects the directionality of recombination. In the absence of spermidine, Holliday junctions are resolved roughly equally in either direction. This is also the case when Int is performing the resolution of junctions by itself, with or without spermidine. In contrast, the presence of spermidine together with accessory proteins induces directional resolution towards reaction products. This directional resolution is most evident when the overlap region consists of mixed purines and pyrimidines, as occurs in the wild-type overlaps of the phage and prophage att sites.
MATERIALS AND METHODS
DNA substrates and proteins
DNA substrates were generated by PCR using plasmid templates containing cloned attL, attR, attB or lox sites. Substrates were radiolabeled at the 5′ end with [γ-32P]ATP (Perkin-Elmer Life Sciences) using T4 polynucleotide kinase (New England Biolabs). Supercoiled plasmids encoding attR (pHN868) or attP (pHN894) were purified using the Promega Wizard Plus SV Midiprep kit. Oligo substrates were obtained from Integrated DNA Technologies, Inc., and gel-purified. Int, IHF and Xis proteins were purified as described previously (33). The following generous gifts are acknowledged: IntF [IntY342F; (34)] from Dr Howard Nash (NIH); Cre protein from Dr Gregory Van Duyne (University of Pennsylvania); and a loxP-containing plasmid from Dr Alex Burgin (deCode Biostructures Inc.).
Recombination reactions and Holliday junction accumulation
Excision reactions contained 1 nM radiolabeled attL site (179 bp), 4 nM unlabeled attR site (136 bp), 250 ng salmon sperm DNA as a non-specific competitor, 44.5 mM Tris–borate (pH 8.3), 7.5 mM Tris–HCl (pH 8.0), 1.15 mM EDTA, 5 mM spermidine, 60 mM KCl, 0.275 mg/ml BSA and 15% (v/v) glycerol. Int, IHF and Xis were present at 50, 35 and 50 nM final concentrations, respectively. The reactions were incubated 60 min, or as indicated, at 37°C and then stopped by the addition of 0.4% SDS containing xylene cyanol. Excision reactions for the accumulation of supercoiled junctions were performed as above except using 2 nM double end-labeled 900 bp attL and 400 ng supercoiled attR-containing pHN868 in 500 μl. Int, IHF and Xis were present at 100, 70 and 100 nM final concentration, respectively. Integration reactions were performed as above, except that they contained 8 nM labeled attB (634 bp), unlabeled 50 μg/ml (0.5 μg/reaction) supercoiled attP plasmid (pHN894), 50 nM Int and 35 nM IHF.
To accumulate Holliday junctions, peptide WRWYCR was included at a final concentration of 0.5–1 μM (33). Typically, ∼8 of these reactions were needed to provide sufficient HJ substrate for several experiments. Reactions were stopped with the addition of 0.5% SDS and 0.5 mg/ml proteinase K, and incubated an additional 30 min at 37°C. Short HJ between linear substrates were isolated by running the reactions on a Tris–SDS 5% polyacrylamide gel (29:1), exposing to film and excising the appropriate band. The DNA was isolated from the gel using crush/soak passive elution into TE/0.1% SDS in the absence of any cations. The DNA was precipitated using standard methods, resuspended in TE, and quantified by spectrophotometry at OD260. Supercoiled HJ for either excisive or integrative recombination were isolated on a 0.8% agarose gel electrophoresed in 1× TBE for 5 h at 80 V. The gels were stained with EtBr and the appropriate peptide-dependent bands were excised and DNA purified from the gel using the MoBio UltraClean 15 kit and quantitated by OD260. Resolution assays of supercoiled junctions were assembled as the linear junctions (except that the reaction mix contained no glycerol) and were incubated for 1 h at 37°C, then stopped with 0.5% SDS and 0.5 mg/ml proteinase K for 30 min at 37°C. Ficoll based loading dye was added and reactions were electrophoresed on 0.8% agarose gels in 1× TBE, for 3 h at 80 V. Gels were dried and exposed to a phosphorimager cassette and quantitated using Molecular Dynamics ImageQuant v. 5.2.
Assembling ‘variant core’ Holliday junctions
Six sets of linear substrates consisting of a double-stranded attL site along with the matching double-stranded attR site were assembled using overlap extension [see Table 1 for oligo sequences]. Equimolar amounts (∼400 ng) of upper and lower strand oligos (attL and attR were assembled separately) were combined in React2 buffer (Invitrogen) for downstream ‘fill-in’), boiled for 5 min and slowly cooled to room temperature over 3 to 4 h. Then dNTP's (0.2 mM—Roche) and Klenow polymerase (4 U—Invitrogen) were added. Reactions were incubated for 60 min at room temperature, then phenol extracted and gel-purified on 8% PAA in 0.5× TBE. Gels were stained with ethidium bromide, the appropriate bands were excised and crush/soaked from the gel slice and precipitated as above. These six attL and attR partner sets were recombined in the presence of peptide WRWYCR in the conditions described above, and HJs were isolated and purified as above. These HJs comprise full-length att sites, complete with accessory protein binding sites and both core and arm Int binding sites.
Table 1.
Sequence of oligomers used to construct att sites with alternate overlap sequences
| Name | att site | Strand | Sequence (5′–3′) |
|---|---|---|---|
| WT1 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTTATACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATTa |
| bsb | AATCAAATAATGATTTTATTTTGACTGATAGTGACCTGTTCGTTGCAACAAATTGATAAGCAATGCT | ||
| attR | tsc | TTCATAGTGACTGCATATGTTGTGTTTTACAGTATTATGTAGTCTGTTTTTTATGCAAAATCTAATTTAATATATT | |
| bs | CCCGTTTCGCTCAAGTTAGTATAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATT | ||
| WT2 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTTGTACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATT |
| attR | bs | CCCGTTTCGCTCAAGTTAGTACAAAAAAGCTGAACGAGAAACGTAAAATG ATATAAATATCAATATATTAAATTAGATT | |
| TS1 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTAAGACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATT |
| attR | bs | CCCGTTTCGCTCAAGTTAGTCTTAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATT | |
| TS2 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTAAAACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATT |
| attR | bs | CCCGTTTCGCTCAAGTTAGTTTTAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATT | |
| BS1 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTTTTACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATT |
| attR | bs | CCCGTTTCGCTCAAGTTAGTAAAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATT | |
| BS2 | attL | ts | AATGAATCCGTTGAAGCCTGCTTTTTTCTACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATT |
| attR | bs | CCCGTTTCGCTCAAGTTAGTAGAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATT |
aBases indicated in bold letters represent the overlap region while bases in italics show the complementary region between top and BS sites.
bThe sequence shown in WT1 for attL bs is used for all attL BS sites.
cThe sequence shown in WT1 for attR ts is used for all attR TS sites.
HJ resolution assays
Resolution reactions were assembled like the recombination reactions except HJ were used as substrate DNA (1 nM or as specified). The specified divalent, trivalent or tetravalent cation was added at the concentration indicated. The specified proteins were added at concentrations listed above or as indicated. For determining the rates of resolution, the reactions that required longer time points (15 to 60 min) were assembled on ice and were initiated by transfer to permissive temperature (either room temperature or 37°C). Those reactions that required short time points (5 s–5 min) were assembled in thin wall PCR tubes without proteins, placed in a thermocycler at the specified temperature, and initiated by the addition of proteins. All reactions were stopped with the addition of 1% SDS. Products were separated on a Tris–SDS 5% polyacrylamide gel and exposed to a phosphorimager screen and analyzed using Molecular Dynamics ImageQuant v. 5.2 software.
Band shift assays
Resolution reactions (above) were assembled on ice, incubated at 30°C for 30 min, and loaded directly onto a native 5% polyacrylamide gel. In order to prevent cleavage of the junctions, we replaced Intwt with the IntF protein, which lacks the active site nucleophile. IntF was present at 100 nM.
KMnO4 footprinting
Reactions (40 μl) contained 7.5 mM Tris–HCl (pH 8.0), 0.05 μg sonicated salmon sperm DNA, 60 mM KCl, 44.5 mM Tris–borate (pH 8.9), 1 mM EDTA, and 2 nM single end-labeled excision Holliday junction. IntF (100 nM), IHF (35 nM), Xis (100 nM) and spermidine (5 mM) were present in the reactions as indicated. Reactions were incubated for 25 min at 30°C to assemble complexes. Thymine modification was performed by adding KMnO4 to a final concentration of 10 mM and incubating at 30°C for 2.5 min. Reactions were quenched with 3 μl of 14.3 M β-mercaptoethanol. Salmon sperm DNA (4 μg) was added as a carrier and the DNA was precipitated with ethanol. To cleave the modified bases, 150 μl of 1 M piperidine was added to each sample and incubated at 90°C for 30 min. DNA was precipitated with 1-butanol and residual piperidine was removed under vacuum. A + G ladder was prepared with the appropriate labeled excision junction using the standard Maxam and Gilbert sequencing reaction (35). Pellets were resuspended in formamide loading buffer and run on a 4.5 or 8% sequencing gel (7 M urea, 19:1 acrylamide: bis-acrylamide), depending on which strand was labeled.
RESULTS
Spermidine is required for directional Holliday junction resolution
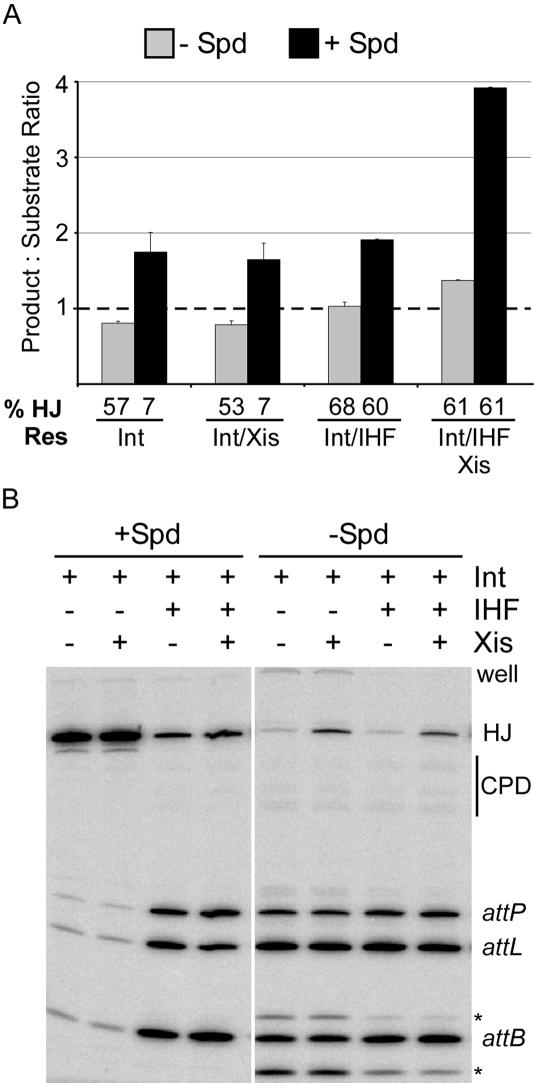
Strand exchange in the presence of spermidine is stimulated weakly by Xis and more strongly by IHF, and the two proteins have additive behaviors [Figure 2A; (21)]. The Landy lab had shown that spermidine inhibited junction resolution mediated by Int in the presence of Xis and IHF, which reaches nearly 100% in the absence of spermidine [(21); also see Figure 5B]. We found two surprises—first, HJ resolution in the absence of spermidine does not require any accessory proteins; Int by itself or with any combination of accessory factors is very efficient (Figure 2). Second, in the absence of spermidine, resolution occurs with equal likelihood towards recombination substrates as towards products. In consequence, recombination would be expected to be no more than half as efficient in the absence versus the presence of spermidine. In contrast, junction resolution in the presence of spermidine as well as both host factors occurs in favor of attP and attB 4× more than in favor of attL and attR (Figure 2). Since the junctions were isolated as the intermediates in attL × attR recombination, this represents a resolution bias towards the products of the recombination reaction. Based on this and other data, we conclude that spermidine affects strand exchange in the context of HJ.
Figure 2.
Effect of spermidine and protein combinations on HJ resolution directionality and efficiency in excisive recombination. (A) Directionality is defined in context to the pathway from which the HJ substrates were obtained. For excision, attL and attR are (original) substrates while attP and attB are products. A ratio of 1 (dotted line) represents an equal amount of ‘substrates’ (attL/attR) and products (attP/attB) and indicates no bias; a ratio of >1 represents more products than substrates, indicating ‘forward’ resolution bias, while a ratio of <1 represents fewer products than substrates, indicating ‘reverse’ resolution bias. % Res refers to % resolution, and indicates the extent of HJ converted to substrates and products in the presence of the specified proteins. (B) Example of the actual data used in (A). The attL substrate used to make the Holliday junctions was double end-labeled, while the attR substrate was unlabeled; thus, only 3 of the 4 resolution products are visible. The ratio of products to substrates is determined as the (% attP+attB counts/% attL counts). The * represent uncharacterized side products.
One trivial possibility was that, in the absence of spermidine, Int is present at higher effective concentration because spermidine competes with Int binding. The low bias of resolution may have been a consequence of this. However, lowering the concentration of Int in the absence of spermidine does not increase the bias of resolution above 1 in favor of products (7, 8, 10, 17 and 25 nM Int concentrations were tested), although resolution drops to 2–3% at 10 nM Int and below (data not shown). In contrast, in the presence of spermidine, the bias of resolution remains above 4× at the same Int concentrations (data not shown).
Spermidine alters Holliday junction conformation
We tested whether the presence of spermidine affected the conformation of free Holliday junctions and of nucleoprotein complexes assembled on junction substrates. Spermidine is known to bind and compact DNA, and has long been used as a DNA precipitation agent. To eliminate or at least reduce the non-specific condensation effects of spermidine, our reactions included 100 ng of non-specific double-stranded DNA in addition to 8 ng of Holliday junction substrates. In order to prevent cleavage of the junctions, we used the catalytically-inactive IntF protein [IntY342F; (34)].
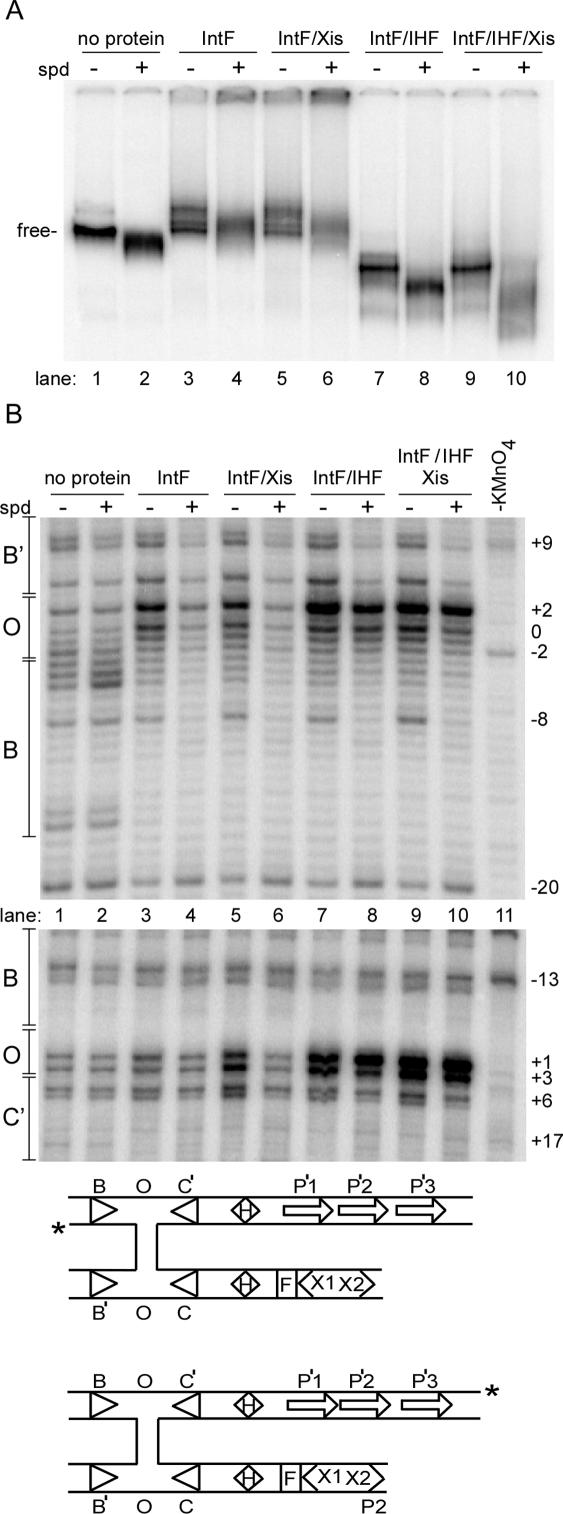
Spermidine increases the mobility of free junctions in gels, albeit subtly (Figure 3A, lane 1 versus lane 2), in agreement with previous findings that cations which neutralize charge repulsion cause junctions to assume a stacked-X conformation that is more compact than the square-planar conformation (36). Although spermidine is in great molar excess, all our reactions also have 100 ng of non-specific salmon sperm DNA. Int alone or together with Xis retards the mobility of the junctions (Figure 3A). In contrast, the junctions containing Int and IHF migrate significantly faster than those that do not contain IHF, presumably due to a more compact structure resulting from the bending of the junction arms. Spermidine further compacts the junctions (Figure 3A lane 8 versus lane 7). Because IntF has altered DNA binding properties in at least some circumstances (32), we also tested the effect of spermidine on complexes assembled with the Intwt. Although fewer junction complexes are seen since many of the complexes resolve to products, the Intwt junction complexes are essentially identical in mobility to the IntF complexes, and are affected by spermidine in the same way (data not shown). In summary, our data shows that spermidine changes the conformation of the junction complexes, despite the large size of the complex containing the junction substrate (∼205 kDa), four Int molecules (∼160 kDa total), two IHF heterodimers (∼44 kDa total) and at least two Xis monomers (14 kDa total), totaling 423 kDa.
Figure 3.
Conformational changes induced by spermidine in Holliday junctions assembled with various combinations of proteins. (A) An EMSA using wild-type excision HJ substrates in the presence or absence of spermidine and the indicated proteins. IntF is the catalytically defective mutant Int Y342F protein. Multiple bands within a lane most likely indicate either alternate conformation of equivalent complexes and/or differing stoichiometries of Int. (B) KMnO4 footprinting of the same excision HJ complexes shown in (A). In the upper gel are the labeled B′ OB strands of the junctions, while in the lower gel are shown the labeled BOC' strands of the junctions (see the schematic below the gels). The numbering of the bases is relative to the point of cleavage, where TS cleavage occurs between the bases at −3 and −2, while BS cleavage occurs between bases at +4 and +5.
The effect of spermidine can be seen not only on the gross structure of the excisive junction complex, but also on its fine structure (Figure 3B). In the absence of any proteins, spermidine increases the accessibility to potassium permanganate of the thymine nucleotide at position −5 about 2× (Int cleaves the DNA between T −2 and T −3; Figure 3B upper panel, lane 2 versus lane 1). Binding by IntF alone or IntF/Xis in the absence of spermidine sensitizes the T-rich overlap (O) region to permanganate modification (Figure 3B lanes 3 and 5), concordant with the open conformation of the junctions seen in the Cre and Flp crystal structures (17,37). The thymine bases most sensitive to modification are at the 0 and +2 positions of the attL continuous strand (Fig. 3B, top) and at the +1 and +2 positions of the attR crossover strand (Fig. 3B, bottom). However, the junction center (the overlap region) is protected when spermidine is added to junctions bound by IntF or IntF and Xis (Figure 3B, lanes 4 and 6 versus lanes 3 and 5). The Int protein remains bound in these complexes, as indicated by protection of the B and B′ core sites (Figure 3B). This spermidine-dependent stacking and ‘occlusion’ of the junction center correlates with decreased recombination efficiency in these same conditions (Figure 2). Thus, the open conformation of the junction (shown by sensitivity to potassium permanganate) is required for efficient recombination. The addition of IHF to junctions bound by IntF or IntF and Xis in the presence of spermidine counteracts the effect of spermidine—the overlap region again becomes sensitive to potassium permanganate (i.e. accessible to solvent; Figure 3B, lanes 8 and 10 versus lanes 4 and 6). The best explanation for this conformational change is that IHF re-positions the catalytic domains of Int in a way that results in the opening of the junction center. This conformational change in the overlap region is perfectly correlated with efficient recombination. The change in cleavage pattern in the presence and absence of spermidine indicates that spermidine affects the isomerization state of the Holliday junction, although Int in the presence of accessory factors can overcome the stacking of the junction to a significant extent.
Spermidine effects on supercoiled excisive recombination junctions
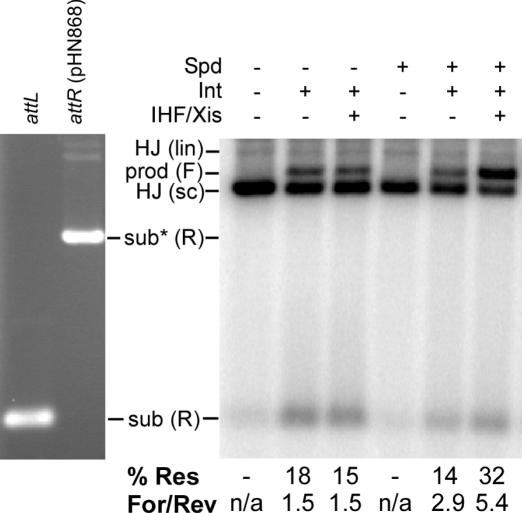
Because in vivo DNA is negatively supercoiled, we tested whether the spermidine effect holds on supercoiled HJ. However, we were unable to distinguish supercoiled ‘figure 8’ junctions from a supercoiled substrate plasmid containing both attL and attR substrates. Instead, we started with a 900 bp labeled attL site and a supercoiled attP site, and trapped the supercoiled ‘alpha’ structure HJ using peptide WRWYCR as described in Materials and Methods (33). The purified supercoiled HJ were then used as the substrate for junction resolution. The results (Figure 4) show that spermidine has the same effect on supercoiled alpha-structure HJ as it did on ‘linear’ junctions—it inhibits recombination by Int alone, but stimulates recombination by Int in the presence of IHF and Xis. Moreover, the bias of resolution without spermidine is the same in the case of Int alone or Int in the presence of accessory factors (∼1.5), but addition of spermidine biases resolution towards products about 5× in the presence of IHF and Xis. Spermidine also biases the resolution of supercoiled junctions a bit more toward products in the absence of accessory factors (3×), as if supercoiling somewhat replaces bending IHF and Xis with respect to resolution bias, although not extent of recombination.
Figure 4.
The effect of spermidine on the resolution of supercoiled alpha-structure excisive junctions. The substrates used to generate the junctions are shown at left (EtBr-stained panel), and are a supercoiled plasmid with attR and a double end-labeled 900 bp fragment with an attL site. The extent of resolution is indicated below the gel (%Res) along with the ratio of forward to reverse resolution products (For/Rev). Forward resolution results in a long linear molecule, labeled ‘prod (F)’. Reverse resolution results in the same molecules that were used to make the junction, only one of which is labeled [the 900 bp fragment labeled ‘sub (R)’]. The second substrate, marked with a *, is unlabeled, and thus invisible on the phosphorimage. This gel is representative of three independent experiments.
Spermidine effects on integrative recombination junctions
We also tested the effect of spermidine on junction resolution during integrative recombination. To isolate the substrates, we used a labeled attB fragment and a supercoiled attP substrate, and trapped the HJ with peptide WRWYCR as above. Spermidine inhibited resolution of junctions when Int alone performs integrative recombination (55% resolution without spermidine and 19% resolution with spermidine), but this inhibition was lessened by IHF (35% resolution with spermidine). Spermidine increased the directionality bias, from about 1.2 to nearly 3 in favor of products, of integrative junctions when they were resolved by Int alone; this was similar to resolution on supercoiled excisive HJ. The directionality of resolution was fairly similar in the presence of IHF whether spermidine is present or absent (a bias of 1.9 in the absence versus 2.4 in the presence of spermidine). In the case of the directionality of resolution of integrative pathway junctions, supercoiling together with IHF may lessen the importance of spermidine. Conversely, spermidine and Int alone resolve supercoiled junctions with a forward bias.
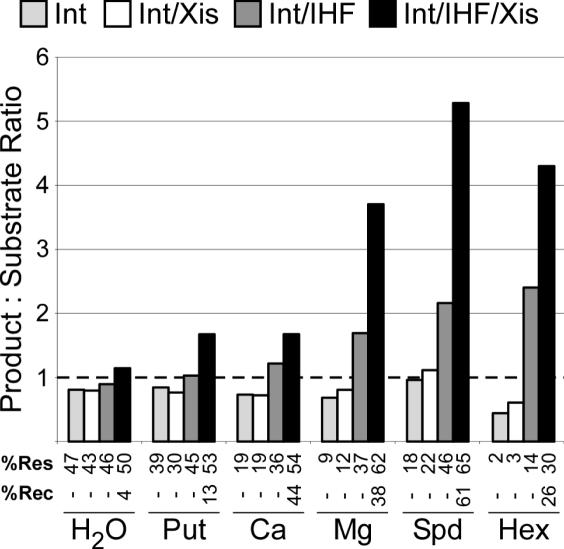
The role of other polycations on directionality
To address the effect of other common cations on directionality, excisive recombination and HJ resolution assays were performed, as above, in the presence of either water (H2O), putrescine (Put), CaCl2 (Ca), MgCl2 (Mg), hexamminecobalt chloride (Hex), spermidine (Spd), spermine or ZnCl2. ZnCl2 and spermine were not permissive for either recombination or resolution (<3% junction resolution and <6% recombination) and they were omitted from further study. Each cation tested affected resolution directionality ranging from a slightly reverse bias (higher chance of resolving to substrates than products) to over a 5× preference towards products (Figure 5), depending which proteins were present. In every case, resolution by Int alone showed no bias or a slightly reverse bias, which was unaffected by the addition of Xis. In the presence of every polycation except Put, Int and IHF together increased the forward bias of resolution modestly. Regardless of cation, Int, IHF and Xis together provided the largest forward resolution bias. Except for zinc and spermine, all the cations tested increased the amount of recombination compared to the recombination achieved in the absence of any di- or trivalent cation (Figure 5). The amount of resolution in the absence of cations or in the presence of putrescine were largely unaffected by either IHF or Xis. Conversely, the remaining cations supported only low resolution by Int or Int and Xis, while adding IHF or IHF and Xis together to Int provided maximal resolution (Figure 5). Of all the cations tested, spermidine resulted in the combination of highest recombination efficiency, and highest junction resolution efficiency and directionality.
Figure 5.
Effect of several polycations and protein combinations on Int-mediated HJ resolution directionality and extent. HJ were resolved with the indicated proteins in the presence of the indicated polycation. The dotted line indicates a product:substrate ratio of 1, i.e. no resolution bias. % Res, resolution. %Rec, recombination. In this experiment, recombination was only measured in the presence of spermidine. The average of two independent experiments is shown.
Since these di- and trivalent cations influence directionality, we tested their effect on the related tyrosine recombinase Cre, which catalyzes bidirectional recombination (the products are identical to the substrates). Different cations influence the extent of recombination with a different pattern than is observed in λ Int-mediated recombination: the divalent cations Mg2+ and Ca2+ supported the most recombination (21 and 20%, respectively), while cations with increasing numbers of positive charges or size supported less recombination (5% for Hex, and 11% each for both Put and Spd, compared to 4% recombination in the absence of di- or trivalent cations). While we were able to trap and isolate Cre-generated HJs (in our hands, Cre aborts recombination more frequently at the HJ stage than Int in vitro), they did not resolve efficiently. Minimal resolution occurred in the absence of polycations or in the presence of putrescine or Mg2+, and there was no significant difference in the resolution bias in any of these conditions (data not shown). Cre binds to its recombination substrates with higher affinity than the catalytic domain of Int and Cre-loxP complexes are much more stable than Int-attB complexes (the equivalent accessory site-independent complexes); this higher affinity of Cre for its core-binding sites may make the complexes less sensitive to their environment.
The interactions between sequence and polycations and their effects on directionality
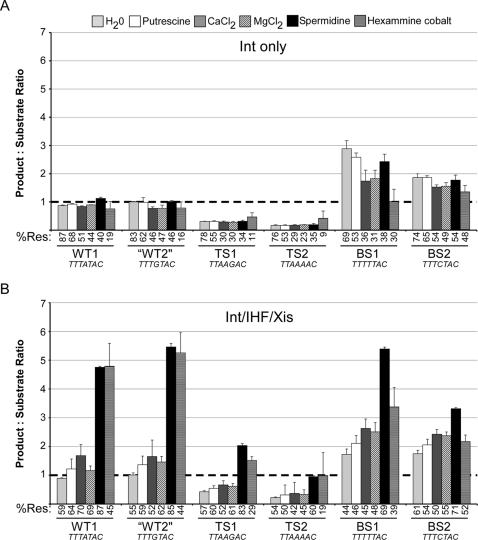
Azaro and Landy (21) investigated the role of the sequence of the central three bases of the seven base overlap regions on the isomerization of the HJ. They determined that skewing the ratio of purines in this region favored one of the two junction isomers and thus controlled directionality. When the three central bases were all purines, the top strand (TS) isomer was favored and resolution occurred mostly towards recombination substrates rather than products; when these bases were pyrimidines, the BS isomer was favored and resolution occurred towards recombination products. These previous experiments were done in the absence of any polycations, without IHF or Xis, with junctions that contained only the core Int sites and with a catalytic fragment of Int (21).
We used our assay conditions to reinvestigate the role of both sequence and polycations on directionality. The overlap sequences used are the same as those used by the Landy lab (21) and are shown in Figure 6. In this experiment, the substrates were made by primer extension and their arms were appreciably shorter than the junctions used elsewhere in this paper; this may account for the small differences seen in the extent of resolution and directionality in Figure 6 versus other Figures.
Figure 6.
Effect of overlap sequence, polycations and proteins on excision HJ resolution. (A) Effect on HJ resolution by Int alone. Bars indicate bias of directionality while the numbers below indicate overall % resolution obtained (% Res), regardless of the direction of resolution. The sequence of the overlap region is given below the name of the junction. (B) Effect on HJ resolution by Int together with IHF and Xis. Data is formatted exactly as in part A. All reactions were performed as described in Materials and Methods and incubated at 37°C. The average of three independent experiments is shown. The dotted line denotes a product:substrate ratio of 1 (no resolution bias).
We compared junction resolution performed by Int alone with resolution performed in the presence of IHF and Xis. In the absence of cations, our results (Figure 6A) agree with those obtained previously (21)—junctions with the wild-type overlap sequence (WT1) or a sequence with similar purine content in the TS (‘WT2’; we used similar names for the junctions as the Landy lab) resolve without bias. A preponderance of purines in the TS favors exchange of the top strands, and lack of purines in the TS favors exchange of the bottom strands. Moreover, adding various polycations in the absence of accessory factors had a negligible effect on directionality. Interestingly, Hex supported significantly lower resolution efficiency in the case of WT and TS overlap sequences than the other cations. Junctions with similar overlap sequences give similar results (Figure 6A and B). Thus in the absence of accessory factors, sequence is the dominant effector of directionality.
In the presence of Int, IHF, and Xis, polycations can have a pronounced effect depending on the sequence of the overlap (Figure 6B). Junctions with wild-type purine content in the overlap sequences (WT1 and WT2) showed a 4.8–5× forward resolution bias towards attP and attB in the presence of either Spd or Hex over their absence, with 85–87% or 44–45% resolution, respectively. Spd showed the largest bias in directionality and the most resolution. The TS junction isomers still exhibited their sequence-imparted bias towards recombination substrates, but Spd and Hex both countered this bias. The BS isomers resolved mostly as in the ‘Int only’ conditions, except with added spermidine, where the bias increased to as much as 5×. Although in most of these cases Hex attains parity with Spd with respect to directionality, it always supported less resolution overall.
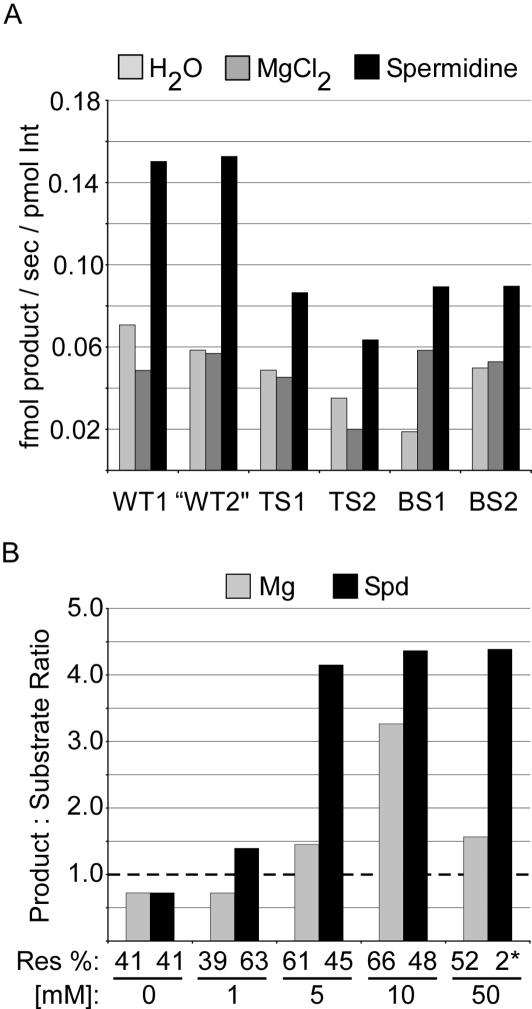
In order to further characterize the effect of Mg2+ and spermidine, we compared the kinetics of HJ resolution at 37°C in their presence (Figure 7A). In all cases, Int, IHF and Xis together resolved junctions faster with Spd than with Mg2+ or without polycations. The greatest effect was seen with junctions that had a more balanced purine:pyrimidine content in the middle 3 nt of the overlap (WT1 and WT2)—they were resolved more than twice as fast as in the absence of polycation. Spd also increased the resolution rate of TS and BS junction substrates, although not as much.
Figure 7.
Resolution of junctions in the presence of Mg2+ or spermidine compared to no polycations. (A) The effect of core sequence on the rates of HJ resolution with or without Mg2+ or Spd. Core sequences are shown in Figure 5B. (B) Effect of Mg2+ or Spd concentrations on the resolution and directionality bias of wild-type excision HJ. The dotted line denotes lack of resolution bias.
The previous experiments were done in the presence of 5 mM polycation, which is near physiological levels for both Mg2+ and spermidine. To determine how much the concentration of cations influenced resolution, we tested the effect of Mg2+ or Spd (Figure 7B) in the range of 1–50 mM. Mg2+ at 1 mM does not improve directionality or resolution over the absence of cations, while 1 mM Spd supported maximal resolution and a modest increase in directionality. Mg2+ at 10 mM gave the highest resolution bias and the maximal amount of resolution, the closest to approaching the effects of Spd. At 50 mM Mg2+ the bias in directionality decreased significantly while the extent of resolution decreased only moderately. In contrast, 50 mM Spd did not allow resolution, probably due to precipitation of the DNA. Thus, while increasing the concentration of Mg2+ can increase the directionality bias of junction resolution, with few exceptions Spd gives more resolution bias.
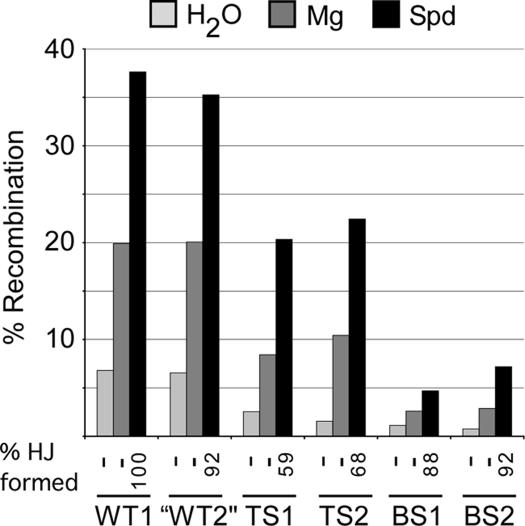
Finally, we tested the effect of Mg2+ and spermidine on the extent of excisive recombination between substrates with different purine:pyrimidine composition in the overlap region to determine whether sequence or polycations have the dominant effect. In the absence of polycations, only minimal recombination occurred between substrates with WT overlap regions (<7%) and even less (1–3%) between substrates with TS or BS overlaps (Figure 8). Adding Mg2+ results in ∼3× increase in recombination extent, while spermidine causes a 6× increase, regardless of sequence. Absolute values showed that the TS isomer is more amenable to recombination than the BS isomer. The low recombination values for the BS isomer may be due to a lower frequency of TS cleavage at the beginning of recombination.
Figure 8.
Effect of various core sequences on the extent of recombination between attL and attR obtained in the absence of polycations or in the presence of Mg2+ or Spd. In these reactions, both substrates have the same overlap sequence, as indicated (see Figure 5B for the sequences of the overlap regions).
DISCUSSION
Spermidine has long been known to stimulate Int-mediated strand exchange and inhibit Int's topoisomerase activity (1,3,31). While spermidine might be predicted to promote synapsis by countering the negative charges in the partner DNA's phosphate backbone, synapsis does not require nor is it stimulated by spermidine (30,32). To test the effect of spermidine on Int's catalytic activities without the involvement of synapsis, we investigated Int's catalytic activities in the context of the full-length HJ intermediates, containing the arms of the att sites in addition to the core DNA binding sites. Spermidine somewhat inhibited resolution of synthetic HJs containing only the Int core-binding sites [(39) and data presented herein]. More striking, however, is that spermidine affects the bias of HJ resolution: in the absence of spermidine, resolution by Int is essentially equal towards either the attP and attB products or the attL and attR substrates and is independent of Xis and/or IHF. In the presence of spermidine, resolution favors products ∼4×, and the full effect is dependent on both Xis and IHF. This bias is also seen on supercoiled excisive HJ, indicating that this effect is physiologically relevant. In the absence of accessory factors, Int by itself resolves HJ better in the absence of spermidine than in its presence. Thus, while on its own spermidine inhibits Int catalysis, in the complete recombination reaction it reduces the reversal of recombination to substrates at the HJ stage. Evidently overall recombination efficiency has been sacrificed to some extent in order to promote a greater fraction of recombination substrates proceeding to completion.
Spermidine affects the overall conformation of HJ recombination intermediates and alters specific contacts between Int and the core-binding site. The functional significance of these alterations becomes apparent during Holliday junction resolution: the repositioning of the Int core-binding domain by IHF and Xis in the presence of spermidine results in greater catalysis at the locus of BS exchange.
Putrescine, not spermidine, is the most abundant polycation (20 mM) in E.coli. However, while putrescine allows similar resolution levels to spermidine, it promotes a significantly lower directionality bias (less than a 2× preference towards products in the presence of IHF and Xis); this may be the reason for the lesser extent of overall recombination in the presence of putrescine. The key difference between putrescine and spermidine is the difference between 2 and 3 positive charges, respectively. Ca2+ was midway between putrescine and Mg2+ with respect to recombination: like putrescine, it promoted essentially no bias in HJ resolution towards products, and promoted less recombination than Mg2+, spermidine or hexammine cobalt. The latter polycation, with 3 positive charges, has properties most similar to spermidine, although it does not support as high an extent of junction resolution or bias towards products. Thus, while the number of charges matters, their geometry is also very important and more optimal in spermidine than in hexammine cobalt.
The sequence of the overlap region has been shown to affect the directionality of resolution by biasing the isomeric conformation adopted by the junction. In the absence of polycations and accessory factors, our results were very similar to those obtained previously (21)—the purine:pyrimidine content of the overlap region has the dominant effect on HJ resolution. Without accessory factors, we observe the same polycation-dependent inhibition of resolution, ranging from 18 to 82% reduction in the case of wild-type overlap sequences and 9–90% reduction among all sequences tested. The largest inhibitory effects cannot be offset by a favorable bias in directionality; thus, it is possible that one factor which led to the recruitment of accessory proteins in Int-mediated recombination was to help Int overcome the inhibitory effect of polycations. In the presence of accessory factors, spermidine and hexammine cobalt impose a strong ∼5× resolution bias in the case of junctions without a sequence-imposed bias. Spermidine, and to a lesser extent hexammine cobalt, even counteract the sequence-imposed bias in the case of junctions that favor TS exchange, although they cannot overcome it completely. In the case of junctions already biased towards BS exchange, spermidine is the most effective at reinforcing this bias. Thus spermidine and similar polyvalent cations are dominant in imposing a resolution bias towards completion of recombination in physiological conditions.
How does spermidine mediate its effects on junctions to bias their resolution? Interestingly, the addition of spermidine to junctions increases their resolution rate over the resolution rate in the absence of polycations (Figure 7A). This rate increase is largest in the case of those junctions whose isomeric preference is not biased by the composition of the overlap sequence, but is also seen in the case of junctions whose overlap sequence is biased towards either one of the two isomers. One possibility is that spermidine increases the rate of catalysis, perhaps by direct interactions with Int's active site or with residues involved in positioning the active site residues with respect to the scissile phosphate. If that were true, spermidine would not be expected to counter the sequence-imposed bias in resolution; if spermidine simply increased the catalytic rate, it would presumably do so regardless of which isomer was being processed, and thus it would increase the extent of resolution but would not affect the bias of resolution. A second possibility is that spermidine increases the rate of junction isomerization from the isomer favoring TS exchange to the one favoring BS exchange by stabilizing the second isomer. This would effectively increase the rate of catalysis by affecting the position of the DNA with respect to the second pair of active sites. The results of our assays are inconsistent with spermidine simply increasing an unbiased rate of isomerization, since faster toggling between the two possible isomers would also be expected to increase the extent of resolution but would not affect the resolution bias. Because spermidine counters the sequence-imposed resolution bias, it is most likely that spermidine together with the accessory factors increases the amount of time spent by junctions in the BS conformation. In this way, spermidine also increases the catalysis rate on junctions already favoring BS cleavage. This stabilization of the ‘BS’ junction isomer depends on the presence of accessory factors—they may contribute to the rate of isomerization, or may simply help position Int's catalytic domain such that its active sites can be more easily activated once isomerization occurs.
Our data suggests that spermidine (and the polyvalent cations acting similarly) affects the structure of the junction intermediate generated during Int-mediated recombination in a manner that favors one isomer over the other. Polyvalent cations like spermidine and hexammine cobalt bind at the entrance of the major grooves of DNA and attract the phosphates of the DNA backbone towards the cation, resulting in a localized DNA bend (40). Such spermidine-induced bends have been proposed to help the cooperative interactions between two Cre monomers bound to the same loxP site (41). In support of the role of such cations, spermidine and hexammine cobalt (but not Mg2+) appear to compete directly with uranyl ions for a high affinity site at or near the center of Holliday junctions (42). Spermidine may also alter the position of the Int core-binding domains, either directly or indirectly via the accessory factors, by affecting hydrogen bonding networks between protein(s) and DNA. The two possible actions of spermidine, inducing a bend in the core DNA and affecting protein–DNA interactions, are not mutually exclusive and may in fact be related.
Recent crystal structures of an Int tetramer with a Holliday junction have revealed a surprising arrangement of protein domains within the complex (18). The Int arm-binding domain of each Int protomer contacts the core-binding domain of its neighbor. The tetramer of arm-binding domains appears to form a very tight-knit and stable complex, while the core-binding and catalytic domains appear to be much more flexible, and change their position in the three different crystal forms presented. Within the strand exchange crystal, both isomer forms were detected; however, in the same crystal, the arm-binding domains appeared skewed towards the BS exchange isomer. This arrangement suggested that the arm-binding domains influence the directionality of the complex and propel it towards the reaction products by favoring the structural arrangement representing the end of the reaction (18).
The core DNAs are bent in all of the three complexes crystallized, with the bend position changing depending on the crystal form. This movement of the bend within the core region presumably reflects the isomerization process and accompanies the movement of the core and catalytic domains of the protein. It is quite possible that the crystal structures may not accurately reflect the position of Int protomers with respect to DNA and with each other within complexes that include the accessory factors, and that spermidine affects features of the complex invisible in the present structures. Even if the structures do accurately reflect the complete nucleoprotein complexes, our experiments suggest a modification of the model describing how Int performs catalysis in the context of the 2-fold symmetric junctions. This stage of recombination is crucial to the efficiency of the reaction, since the symmetry of the junction intermediate could, in principle, reduce the efficiency of the overall reaction by permitting its reversal. We propose that in the presence of spermidine, the accessory factors may trigger the conversion of the first junction isomer, once it is formed after the first strand exchange, to the second isomer, thereby poising the second pair of scissile phosphates appropriately with respect to the second pair of Int active sites. Spermidine may influence primarily the bend within the core, in which case it is reasonable to assume that the Int core-binding and catalytic domains are the ones that induce, via the bends introduced by the accessory factors, the rearrangement of the arm DNA and consequently influence the position of the tetramer of arm-binding domains. We prefer this first model, although it differs from that proposed by Biswas and colleagues, because of the clear influence of the DNA core sequence on the directionality of the reaction [(21) and the data presented here]. Alternatively, the relevant spermidine interaction may be with a bend closer to the arm sites, first influencing the position of the tetramer of arm-binding domains, which subsequently rearrange the core and catalytic domains with help from the accessory factors. The crystals of the Int tetramer with DNA contain neither the polycations that affect directionality most nor the accessory factors and their binding sites. Nevertheless, our modifications of the models proposed by Biswas et al. (21) account for important features of the recombination reaction that are not yet captured by crystallographic studies.
Acknowledgments
We thank Dr Jeffrey Gardner for discussion. K.K. was the recipient of a scholarship from the Achievement Rewards for College Scientists program. This work was funded by NIGMS grant 2R01 52847 to A.M.S. Funding to pay the Open Access publication charges for this article was provided by the SDSU Research Foundation fund 260020.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nash H. Integrative recombination of bacteriophage lambda DNA in vitro. Proc. Natl Acad. Sci. USA. 1975;72:1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azaro M.A., Landy A. λ Integrase and the λ Int family. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148. [Google Scholar]
- 3.Kikuchi Y., Nash H. The bacteriophage λ int gene product. J. Biol. Chem. 1978;253:7149–7157. [PubMed] [Google Scholar]
- 4.Richet E., Abcarian P., Nash H.A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein–DNA complex. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 5.Nash H.A., Robertson C.A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989;8:3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Duyne G.D. A structural view of tyrosine recombinase site-specific recombination. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 93–117. [Google Scholar]
- 7.Guo F., Gopaul D., Van Duyne G. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz C.J., Sadowski P.D. FLP recombinase of the 2 micron circle plasmid of Saccharomyces cerevisiae bends its DNA target. Isolation of FLP mutants defective in DNA bending. J. Mol. Biol. 1989;205:647–658. doi: 10.1016/0022-2836(89)90310-0. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz C.J., Sadowski P.D. FLP protein of 2 micron circle plasmid of yeast induces multiple bends in the FLP recognition target site. J. Mol. Biol. 1990;216:289–298. doi: 10.1016/s0022-2836(05)80320-1. [DOI] [PubMed] [Google Scholar]
- 10.Luetke K.H., Sadowski P.D. The role of DNA bending in Flp-mediated site-specific recombination. J. Mol. Biol. 1995;251:493–506. doi: 10.1006/jmbi.1995.0451. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Tonozuka T., Jayaram M. Mechanism of active site exclusion in a site-specific recombinase: role of the DNA substrate in conferring half-of-the-sites activity. Genes Dev. 1997;11:3061–3071. doi: 10.1101/gad.11.22.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayaram M., Grainge I., Tribble G. Site-specific recombination by the Flp protein of Saccahromyces cerevisiae. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 192–218. [Google Scholar]
- 13.Rice P.A. Theme and variation in tyrosine recombinases: structure of a Flp-DNA complex. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 219–229. [Google Scholar]
- 14.Kitts P.A., Nash H.A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 1988;204:95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- 15.Nunes-Düby S.E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987;50:779–88. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 16.Franz B., Landy A. The Holliday junction intermediates of λ integrative and excisive recombination respond differently to the bending proteins integration host factor and excisionase. EMBO J. 1995;14:397–406. doi: 10.1002/j.1460-2075.1995.tb07014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gopaul D.N., Guo F., Van Duyne G.D. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas T., Aihara H., Radman-Livaja M., Filman D., Landy A., Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Duyne G.D. Lambda integrase: armed for recombination. Curr. Biol. 2005;15:R658–R660. doi: 10.1016/j.cub.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 20.Radman-Livaja M., Biswas T., Ellenberger T., Landy A., Aihara H. DNA arms do the legwork to ensure the directionality of λ site-specific recombination. Curr. Opin. Struc. Biol. 2006;16:42–50. doi: 10.1016/j.sbi.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azaro M.A., Landy A. The isomeric preference of Holliday junctions influences resolution bias by λ integrase. EMBO J. 1997;16:3744–3755. doi: 10.1093/emboj/16.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen S. Introduction to the Polyamines. Englewood Cliffs, NJ: Prentice-Hall, Inc.; 1971. [Google Scholar]
- 23.Tabor C.W., Tabor H. Polyamines in microorganisms. Microbiol. Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniel R.M., Cowan D.A. Biomolecular stability and life at high temperatures. Cell. Mol. Lif Sci. 2000;57:250–264. doi: 10.1007/PL00000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabor H., Tabor C.W., Irrevere F. Quantitative determination of aliphatic diamines and polyamines by an automated liquid chromatography procedure. Anal. Biochem. 1973;55:457–467. doi: 10.1016/0003-2697(73)90136-x. [DOI] [PubMed] [Google Scholar]
- 26.Lusk J.E., Williams R.J.P., Kennedy E.P. Magnesium and the growth of Escherichia coli. J. Biol. Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 27.Tabor H., Tabor C., Cohn M., Hafner E. Streptomycin resistance (rpsL) produces an absolute requirement for polyamines for growth of an Escherichia coli strain unable to synthesize putrescine and spermidine [Δ(speA-speB) ΔspeC] J. Bacteriol. 1981;147:702–704. doi: 10.1128/jb.147.2.702-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hafner E., Tabor C., Tabor H. Mutants of Escherichia coli that do not contain 1,4-diaminobutane (putrescine) or spermidine. J. Biol. Chem. 1979;254:12419–12426. [PubMed] [Google Scholar]
- 29.Gottesman S., Gottesman M. Excision of prophage λ in a cell-free system. Proc. Natl Acad. Sci. USA. 1975;72:2188–2192. doi: 10.1073/pnas.72.6.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segall A., Nash H. Synaptic intermediates in bacteriophage lambda site-specific recombination: integrase can align pairs of attachment sites. EMBO J. 1993;12:4567–4576. doi: 10.1002/j.1460-2075.1993.tb06145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgin A., Nash H. Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage lambda. Curr. Biol. 1995;5:1312–1321. doi: 10.1016/s0960-9822(95)00258-2. [DOI] [PubMed] [Google Scholar]
- 32.Segall A.M. Analysis of higher order intermediates and synapsis in the bent-L pathway of bacteriophage lambda site-specific recombination. J. Biol. Chem. 1998;273:24258–24265. doi: 10.1074/jbc.273.37.24258. [DOI] [PubMed] [Google Scholar]
- 33.Boldt J., Pinilla C., Segall M. Reversible inhibitors of λ integrase-mediated recombination efficiently trap Holliday junction intermediates and form the basis of a novel assay for junction resolution. J. Biol. Chem. 2004;279:3472–3483. doi: 10.1074/jbc.M309361200. [DOI] [PubMed] [Google Scholar]
- 34.Pargellis C.A., Nunes-Düby S.E., de Vargas L.M., Landy A. Suicide recombination substrates yield covalent λ integrase-DNA complexes and lead to identification of the active site tyrosine. J. Biol. Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 35.Maxam A.M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth. Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 36.Duckett D.R., Murchie A.I., Diekmann S., von Kitzing E., Kemper B., Lilley D.M. The structure of the Holliday junction, and its resolution. Cell. 1988;55:19–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Narendra U., Iype L.E., Cox M.M., Rice P.A. Crystal structure of a Flp recombinase-Holliday junction complex. Assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 38.Cassell G., Klemm M., Pinilla C., Segall A.M. Dissection of bacteriophage λ site-specific recombination using synthetic peptide combinatorial libraries. J. Mol. Biol. 2000;299:1199–1202. doi: 10.1006/jmbi.2000.3828. [DOI] [PubMed] [Google Scholar]
- 39.Hsu P.L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage λ. Nature. 1984;311:721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouzina I., Bloomfield V.A. DNA bending by small, mobile multivalent cations. Biophys. J. 1998;74:3152–3164. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rufer A., Neuenschwander P.F., Sauer B. Analysis of Cre-loxP interactions by surface plasmon resonance: influence of spermidine on cooperativity. Anal. Biochem. 2002;308:90–99. doi: 10.1016/s0003-2697(02)00247-6. [DOI] [PubMed] [Google Scholar]
- 42.Mollegaard N.E., Murchie A.I.H., Lilley D.M.J., Nielsen P.E. Uranyl probing of a four-way DNA junction: evidence for specific metal ion binding. EMBO J. 1994;13:1508–1513. doi: 10.1002/j.1460-2075.1994.tb06412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]