Abstract
Saccharomyces cerevisiae telomerase-negative cells undergo homologous recombination on subtelomeric or TG1–3 telomeric sequences, thus allowing Type I or Type II post-senescence survival, respectively. Here, we find that the DNA damage sensors, Mec1, Mec3 and Rad24 control Type II recombination, while the Rad9 adaptor protein and the Rad53 and Chk1 effector kinases have no effect on survivor type selection. Therefore, the Mec1 and Mec3 checkpoint complexes control telomeric recombination independently of their roles in generating and amplifying the Mec1-Rad53-Chk1 kinase cascade. rfa1-t11 mutant cells, bearing a mutation in Replication Protein A (RPA) conferring a defect in recruiting Mec1-Ddc2, were also deficient in both types of telomeric recombination. Importantly, expression of an Rfa1-t11-Ddc2 hybrid fusion protein restored checkpoint-dependent arrest, but did not rescue defective telomeric recombination. Therefore, the Rfa1-t11-associated defect in telomeric recombination is not solely due to its failure to recruit Mec1. We have also isolated novel alleles of RFA1 that were deficient in Type I but not in Type II recombination and proficient in checkpoint control. Therefore, the checkpoint and recombination functions of RPA can be genetically separated, as can the RPA-mediated control of the two types of telomeric recombination.
INTRODUCTION
Telomeres, the ends of eukaryotic linear chromosomes, are required for overall genome stability. Telomeric DNA is made of tandem arrays of TG-rich nucleotide repeats of variable length depending on the organisms (∼300 bp in the yeast Saccharomyces cerevisiae, 5–15 kb in humans) and kept at a constant value in a given organism. Telomeric proteins, organized around telomeric DNA, provide both telomere end protection and processes for access of telomerase during telomere replication (1,2). Telomere length homeostasis is crucial for two main reasons. First, removal of the terminal primer at the end of lagging-strand during DNA synthesis leaves a small gap that cannot be filled in, a phenomenon known as the ‘end-replication problem’ [e.g. see Ref. (3)]. Telomere replication, by a specialized reverse transcriptase, telomerase (4), solves this problem by maintaining telomeres at a constant length. Second, maintenance of telomeric tracts of a minimal size is needed for recruitment by telomeric DNA sequences of specialized telomeric proteins. The major function of telomere end protection proteins is to prevent inappropriate access of DNA repair or modification proteins, such as exonucleases. The processes underlying telomere replication and telomere end protection are intimately linked (5). For instance, in yeast and humans, both telomere erosion and telomere uncapping can trigger telomeric senescence (6,7). Classically, inactivation of telomerase or of the yeast EST proteins trigger telomeric senescence during which telomeres shorten critically so as to lead to cell death after ∼75–100 generations (8,9). Telomeric senescence activates the DNA damage checkpoint that ultimately leads to a cell cycle arrest of the senescing cells at the G2/M transition (10).
In recent years, increasing interest has been focused on a telomerase-independent mechanism for maintaining telomeric repeats that relies on homologous recombination between the repeated telomeric DNA sequences (11,12). This pathway, first discovered in S. cerevisiae (13), allows senescing cells deficient in telomerase function, such as est or tlc1 mutants (8,9), to maintain indefinitely functional telomeres, and, hence, viability, in the absence of telomerase. These mechanisms are receiving much attention at the moment due to the recent finding that they are also operating in some human tumor cells, in which they are known as the alternative lengthening of telomeres (ALT) pathway (14). In S. cerevisiae, the rare cells escaping telomeric senescence, termed post-senescence survivors, use two distinct pathways of recombination, based on the initial observation that Rad52, essential for recombination (15), was needed for this telomerase-independent maintenance of telomeres (13). In telomerase-negative, otherwise wild-type, cells, a first class of survivors (of type I), relying on Rad51, were found to amplify the subtelomeric Y′ elements and had very short terminal tracts of TG1–3 DNA, while Rad50-dependent type II survivors amplified the terminal TG1–3 sequences with no evidence for rearrangement of Y′ elements (11,13,16). In the yeast Kluveromyces lactis, small rolling circles of single- or double-stranded telomeric DNA resulting from the resolution of an intratelomeric invasion provide one mechanism that is responsible for the generation of the long type II telomeres (12,17). Very recently, extra-chromosomal circles of DNA have also been identified in both type I and type II post-senescent survivor S. cerevisiae cells (18). In addition to these recombination-dependent pathways of survival to telomeric senescence, S. cerevisiae telomerase-negative cells have also been shown to utilize recombination-independent pathways that involved repair of DNA double-strand break by palindromic DNA structures (19).
A number of factors have been found to affect the selection of the survivor type in S. cerevisiae. In most of these cases, factors affecting telomere end protection or the processing of damaged telomeric DNA were implicated (11,20,21). Interestingly, the checkpoint proteins of the phosphatidyl-inositol 3-kinase-like family, Mec1 and Tel1 (homologues to the human ATR and ATM, respectively), have been recently found to be required for type II recombination in S. cerevisiae (22). Here, we focused our attention on the role of Mec1, as well as that of other checkpoint proteins, in survivor type selection. We also document the role of Replication Protein A (RPA) in telomeric recombination by analyzing a particular allele of its larger subunit, rfa1-t11, previously reported to be defective in recruiting Mec1/ATR and Ddc2/ATRIP at sites of damaged DNA (23), as well as in recombination (24). Finally, we have isolated separation-of-function rfa1 mutants that exhibit checkpoint proficiency but telomeric recombination defects.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains used in this study were in the BF264-15D genetic background used in our laboratory (25). The rfa1::KanMX4/RFA1, chk1::KanMX4, ddc1::KanMX4, rad51::KanMX4 and rad59::KanMX4 strains were purchased at Euroscarf (Frankfurt, Germany). The rfa1-t11 allele, from the Kolodner lab (24), was integrated at the RFA1 locus after cutting with NheI. In all experiments rfa1-t11 was integrated at the RFA1 locus, after cutting with NheI. The mec1::TRP1 sml1::KanMX4 and rad53::TRP1 sml1::KanMX4 mutant strains are from the Hartwell and Emili laboratories (26,27). The tlc1::LEU2 was from the Gottschling laboratory (28). The rad50::hisG-URA3-hisG strain was from the Haber laboratory (29). The rad9::LEU2 and cdc13-1 strains are from the Hartwell laboratory (30). The clb2::LEU2 strain was from the Reed laboratory (31). The mec3::TRP1 strain was from the Lucchini laboratory (32). The rad17::URA3 and rad24::URA3 strains were from the Friedberg laboratory (33). Yeast cells were grown in yeast extract-peptone-dextrose (YEPD) medium at 30°C unless otherwise indicated. The integrative TRP1-based pRS304 RAD53-2 HA construct (containing the last 344 nt, out of 2463 nt total of RAD53), a generous gift from Andrew Emili (26,27), was used as such and also subcloned into YIp211 (integrative URA3 plasmid) and integrated at the RAD53 locus after cutting with HpaI. The integrative YIp-352-mec1-kd (URA3) construct, containing the last ∼2.3 kb, out of ∼7.1 kb total, of MEC1 in which two substitutions in the kinase domain [D-A at position 2224 and N-K at position 2229 had been introduced (34,35), a kind gift from Akira Matsuura (35)], was integrated at the MEC1 locus after cutting with NruI. Actual introduction of the mutations was verified by sequencing the sequence of interest at the MEC1 locus. CLB2, CLB2-2 HA and RAD52 were expressed either from an integrative, centromeric (CEN) or episomal (2 μ) plasmid under the control of their own respective promoters.
Analysis of telomere organization and structure
Genomic DNAs were prepared, separated in a 0.9% agarose gel (in TBE) run in TBE buffer overnight and, after denaturation, transferred onto nitrocellulose membrane and immobilized by baking at 80°C for 1 h, as described previously (25). The membrane was then pre-hybridized in 6× SSC, 0.1% SDS, 1% non-fat milk and hybridized with a 270 bp TG1–3 32P-labeled telomeric probe. Following digestion with XhoI, to cut within the Y′ regions of chromosomes (36), telomere tracts of wild-type cells appear as a broad-band of ∼1.2–1.3 kb, which represents the average length of most chromosomes, those containing Y′ subtelomeric regions. From non-Y′ chromosomes, XhoI cutting typically generates fragments migrating at ∼2.1, 2.3, 3.3 and 3.9 kb in Southerns. In senescing cells, the disappearance of the non-Y′ fragments attests to the fact that survivors have arisen by homologous recombination [see, for instance, Ref. (16)]. In some experiments, genomic DNA was digested with a mixture of four restriction enzymes (AluI, HaeIII, HinfI and MspI) using a 4 bp recognition sequence and the Southern revealed with a TG1–3 probe. Since these enzymes cut within telomeric Y′ sequences but not within the TG1–3 sequences, they are currently used to confirm identification of type II telomeres (16). In some cases, a 32P-labeled Y′ probe was used. Results were analyzed using a Storm PhosphorImager (Amersham Biosciences) and ImageQuant.
Analysis of the kinetics of senescence/survival and of growth rates
Telomerase-negative cells undergo senescence in 75–100 generations (9). In all kinetics, senescence of tlc1Δ haploid mutant cells, and of their derivatives, was initiated after the tlc1Δ/TLC1+ diploids or their derivatives were induced to sporulate. In the ‘liquid assays’, selected spores of the desired genotype were individually propagated for over 100 generations as exponentially growing liquid cultures that were, under standard conditions, diluted to 105 cells/ml every day. In some experiments, wherever indicated, liquid cultures of senescing cells were diluted to 106 or 107 cells/ml every day, as described previously (31). Under such conditions, telomeric senescence took place in the liquid culture, thus resulting in a mixture of both possible survivor types and allowing the conversion from types I to types II that normally occurs in telomerase-negative cells, due to a selective growth advantage of types II (16). In ‘streak assays’, which allowed the detection of both type I and type II survivors, re-streaking of single colonies on a YEPD agar-based plate was repeated every 48 h (typically, cells underwent ∼25 generations per streakout or passage at 29°C) to allow loss of viability and appearance of survivors (16,37). The occurrence of senescence was determined from analyzing the progression with time of the growth rate or from the appearance of post-senescence survivors on the plates, as well as from the analysis of telomere structure by Southern blotting with a telomeric probe, as described above.
Construction of fusion proteins
Plasmids expressing the Rfa1-t11-Rad52, Rfa1-t11-Ddc2 and Rfa1-t11-HA in-frame fusion proteins were constructed by cloning in a single-copy, centromeric, plasmid the entire ORF of the first part of the hybrid protein plus upstream promoter sequences, in front of the ORF, (which included its natural stop codon) of the second part of the hybrid protein, as described previously (38). The codon for the first amino acid of the second protein in the hybrid construct was directly following the codon for the last amino acid of the first protein, in reading frame with it.
Construction of checkpoint-proficient, recombination-deficient alleles of RFA1
The entire RFA1 ORF plus 267 bp upstream of the ATG and 306 bp downstream of the stop codon were amplified by PCR under the following mutagenic conditions. The concentration of dNTPs was either kept as in standard conditions (200 μM each) or one of the dNTP concentration changed to 500 μM, those of the other three were kept to 200 μM and, in both cases, the concentration of MgCl2 was changed from 1.5 to 3.0 or 4.0 mM. Standard Taq polymerase and PCR buffer (Invitrogen) were used. Following a 30-cycle amplification, the PCR products were cleaned and, according to the gap repair method, transformed directly into rfa1::Kan-MX4 YCp33-RFA1-URA3 (including promoter sequences, ORF and post-stop sequences) strains, together with another centromeric LEU2 plasmid (YCp111) made linear by digestion with NdeI and carrying RFA1 flanking regions at each extremity: region −267 to +166 (position of an endogenous NdeI site) at one end, and region +2000 to +2169 (position of another endogenous NdeI site) (the TAA stop codon starts at position +1861) at the other end. Thus, sequences upstream and downstream of RFA1 were available for homologous recombination, while all of RFA1 ORF (with the exception of the first 166 bp) were available for PCR-based mutagenesis. Cells were plated onto leucine-lacking medium at 25°C. Around 10 000 transformants grew up which were transferred to leucine-lacking liquid medium and then plated on 5-FOA-containing medium at 25°C in order to force the loss of the YCp33-RFA1-URA3 plasmid. Growing colonies were then re-streaked on YEPD medium and then analyzed for possible defects in telomeric recombination, as explained above, after introduction, by crossing, of a telomerase-negative (tlc1Δ) mutation. Selected mutants were again crossed to a cdc13-1 strain that also contained RAD53-2 HA integrated at the RAD53 genomic locus and checkpoint activation assessed by analyzing Rad53 phosphorylation following immunoprecipitation and western blotting using anti-HA antibodies, as explained below.
Immunoprecipitation and immunoblotting
Proteins were immunoprecipitated from yeast cell extracts and separated by electrophoresis, as described previously (39). All strains analyzed harbored a chromosomal copy of RAD53 tagged in 3′ with 2 HA epitope, the corresponding integrative plasmid being a generous gift from Andrew Emili (26,27). Mouse monoclonal anti-HA raw ascites fluid (BabCo, 16B12, Eurogentec) and mouse anti-HA monoclonal 12CA5 antibody (Roche) were used for immunoprecipitation and western blotting, respectively. Monoclonal mouse anti-actin antibody (clone C4) from MP Biomedicals was purchased at ICN Pharmaceutical. Blotted proteins were detected by chemifluorescence via fluorescein-labeled anti-mouse secondary antibodies and a tertiary anti-fluorescein alkaline phosphatase conjugate (ECF western blotting kit, Amersham Biosciences) and signals analyzed using a Storm PhosphorImager (Amersham Biosciences) and ImageQuant.
RESULTS
Checkpoint proteins affect recombination in survivors from telomerase-negative cells
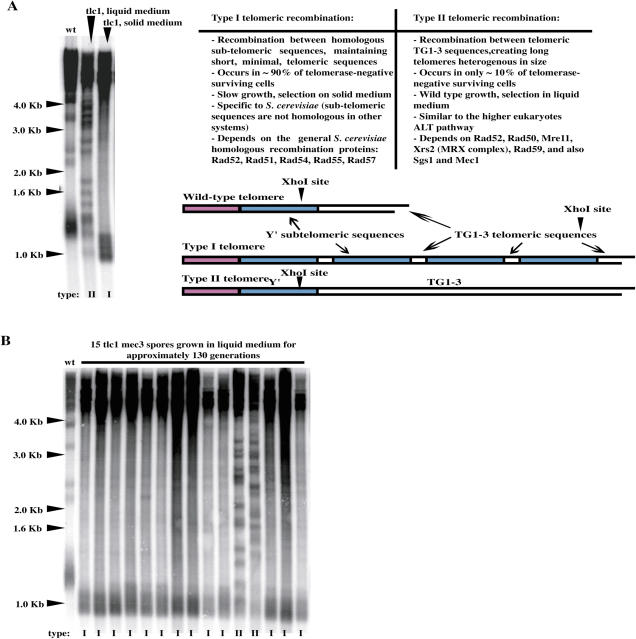
A very small percentage of S. cerevisiae telomerase-negative cells (here we used tlc1Δ cells, deleted for the RNA subunit of telomerase, throughout) survive telomere erosion by performing recombination between telomeric sequences (13). Two types of recombination can be generated (Figure 1A). Type I involves recombination between the homologous subtelomeric sequences Y′ and, in telomerase-negative, otherwise wild-type, cells, is Rad51-dependent, while type II involves recombination between the telomeric TG1–3 sequences and is Rad50- and Rad59-dependent (11). It is currently agreed, as previously explained in detail (16,22), that the type I telomere pattern is most easily visualized by XhoI digestion Southern blot analysis. XhoI cleaves 0.9 kb from the 3′ end of the Y′ element yielding a ∼1.3 kb terminal fragment corresponding to the distal part of the Y′ element plus the ∼0.3–0.4 kb of terminal TG1–3 tracts (Figure 1A). In type I survivors, the TG1–3 terminal tracts have become eroded and the XhoI-restricted terminal fragment is now ∼0.9–1.0 kb long (Figure 1A). In contrast to type I survivors, type II survivors exhibit many XhoI fragments of different sizes [Figure 1A; (16,22)]. The terminal part of the Y′ elements contain short TG1–3 tracts, allowing type I survivors to be also conveniently detectable with a TG1–3 probe [Figure 1A; (16)]. Although type II survivors grow better than type I survivors, ∼90% of tlc1Δ surviving cells display type I telomeres when grown on solid (agar-based) media [Figure 1A; (16)]. On the other hand, and most importantly for the experiments described below, when tlc1Δ surviving cells are grown in liquid medium, 100% type II cells are typically recovered (Figure 1A), because, due to their faster growth, which is similar to that of wild-type cells, they quickly overtake the population of type I survivors (16).
Figure 1.
(A) Survivors from telomerase-negative (tlc1) cells show two distinct patterns. Genomic DNAs from wild-type cells (no recombination, lane 1), type II (lane 2) and type I (lane 3) senescing tlc1Δ cells were digested with XhoI and the Southern blot revealed with a TG1–3 32P-labeled probe. In both types of survivors, the disappearance of the non-Y′ fragments (migrating at ∼2.1, 2.3, 3.3 and 3.9 kb) attests to the occurrence of homologous recombination. In type II survivors, XhoI cutting reveals amplification of very long and heterogeneous TG1–3 sequences (lane 2), located more distal than the single XhoI site that is present at the distal part of Y′ sequences (16). In contrast, in type I survivors telomere erosion is evident after XhoI cutting (lane 3) in comparison with the mean size of the terminal telomere tracts in wild type corresponding to the broad band at ∼1.2 kb (lane 1), the amplified Y′ sequences being located more proximal than the XhoI site, as schematically represented. (B) Mec3 inhibits the generation of type II survivors in liquid assays. Individual spores from tlc1Δ mec3Δ strains were grown in liquid YEPD for ∼130 generations, at 29°C, with a daily dilution of 105 cells/ml (see Materials and Methods). The conversion from types I to types II that is readily seen in liquid culture of telomerase-negative, otherwise wild-type, cells (16) was largely inhibited in the absence of Mec3. Thus, the samples shown in lanes 12 and 13 were the only two ones, among the 15 ones shown here, to exhibit many XhoI fragments of different sizes, which entitle them as type II survivors, those amplifying the TG1–3 tracts. tlc1Δ MEC3+ cells generated 100% type II survivors (data not shown), like shown for wild-type cells in (A) above. Analysis of telomere structure, as described above in (A), allowed to classify these 15 tlc1Δ mec3Δ spores according to the type of recombination accomplished, I or II, as labeled below each lane.
Telomerase-negative cells also bearing a null mutation in the MEC3 checkpoint gene had difficulties in generating type II cells, even in liquid medium. Thus, when grown in liquid rich medium for about 130 generations (crisis occurs ∼80–100 generations after inactivation of telomerase), only ∼20% of tlc1Δ mec3Δ cells displayed type II telomeres versus 100% of type II for the tlc1Δ MEC3+ control (Table 1; Figure 1B). We verified that in the mec3Δ background type II survivors also grew better than type I survivors (data not shown). The Mec3 complex (Ddc1 and Rad17 are the other two components) represents, together with the Mec1–Ddc2 complex, DNA damage sensors (40,41). The Mec1 and Mec3 complexes are recruited independently to DNA double-strand breaks (42,43). Interestingly, telomerase-negative cells bearing a mec1Δ mutation have been previously shown to be deficient in type II recombination (22). To better document the relationships between telomeric recombination and the DNA damage checkpoint, we next constructed additional checkpoint-negative telomerase-negative double mutants and analyzed their pattern of telomeric recombination after extended propagation in liquid cultures. Among these, tlc1Δ rad17Δ, tlc1Δ rad24Δ and tlc1Δ mec1Δ sml1Δ, similarly to tlc1Δ mec3Δ, had difficulties in generating type II survivors after crisis. On the other hand, tlc1Δ rad9Δ, tlc1Δ rad53Δ sml1Δ and tlc1Δ chk1Δ were fully proficient in type II recombination (Table 1). It should be noted that Rad24 is required for the loading of the Mec3–Ddc1–Rad17 complex (44,45). The Rad9 checkpoint adaptor and the Rad53 and Chk1 effector kinases are subsequently activated after the Mec1 and Mec3 complexes have bound damaged DNA (40,41). It appears, therefore, that among the different classes of checkpoint proteins, only the DNA damage sensors have an effect on the type of telomeric recombination. In agreement with the finding above that in mutants of the Mec3 complex type II recombination, although diminished, was still possible, we recovered 100% type II survivors after RAD51 had been deleted in the tlc1Δ rad17Δ mutant (Table 3).
Table 1.
Survival type frequencies in checkpoint-defective telomerase-negative double mutantsa
| Relevant genotype | % of type I/% of type II |
|---|---|
| tlc1 | 0/100; n = 22b |
| tlc1 rad9 | 0/100; n = 18 |
| tlc1 mec3 | 60/40; n = 25 |
| tlc1 chk1 | 0/100; n = 16 |
| tlc1 rad17 | 80/20; n = 24 |
| tlc1 rad24 | 75/25; n = 18 |
| tlc1 sml1 | 0/100; n = 12 |
| tlc1 rad53 sml1 | 0/100; n = 14 |
| tlc1 mec1 sml1 | 50/50; n = 20 |
aAfter ∼130 generations in liquid cultures at 29°C.
bn = number of survivor populations tested.
Mec1 is, together with Tel1 (homologues of ATR and ATM in humans, respectively), a protein kinase of the phosphatidyl-inositol 3-kinase-like family (46). In budding yeast, phosphorylation of target checkpoint proteins by Mec1 is necessary for initiating the DNA damage response (26,34,47–49). To know whether Mec1 protein kinase activity was required for the control of type II telomeric recombination described above, we used a kinase-dead mec1 (mec1-kd) mutant (34,35). After verifying by sequencing actual introduction of the mec1-kd mutation at the MEC1 locus in our wild-type background, as well as associated-hypersensitivity to the genotoxic agent methyl methane sulfonate (MMS) and concomitant failure to activate endogenous Rad53-HA (data not shown), we constructed the tlc1Δ mec1-kd sml1Δ mutant strain and analyzed its survivor type during post-senescence. Only around one-third of the tlc1Δ mec1-kd sml1Δ senescing cells generated type II survivors in liquid cultures, in contrast with the usual 100% type II in the tlc1Δ MEC1+ control (data not shown). We conclude that, in this assay, the mec1-kd mutation behaves like a null mec1 mutation and, therefore, the control of telomeric recombination necessitates Mec1 protein kinase activity.
Overexpression of CLB2 rescues the defect in type II recombination conferred by a mutation in the Mec3 complex
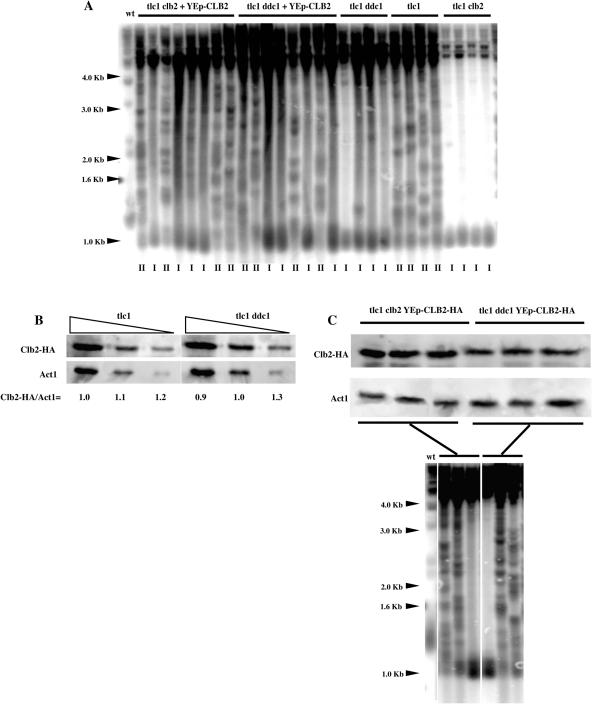
A null mutation in CLB2, coding for the major S. cerevisiae B-type cyclin, as well as temperature-sensitive mutations in CDC28 (Cdk1), were recently found to confer a defect in type II telomeric recombination (31), similar to those found here for mec1Δ and mec3Δ mutants. We reasoned that both mechanisms might be related and set out to establish a functional link between the two events. To this end, we overexpressed CLB2 from an episomal plasmid (under the control of its native promoter) in tlc1Δ ddc1Δ double mutants. As expected, because Ddc1 is a component of the Mec3 complex, tlc1Δ ddc1Δ mutants were deficient in type II recombination (Table 2; Figure 2A). We noted that in these experiments with CLB2 overexpression, some of the samples did not exhibit as clear-cut type I characteristics as usual (Figure 2A). This may have been due to the fact that these cells were cultivated on selective medium to maintain the plasmid within the cells, thereby generating poorer growth and less efficient recombination. However, under identical conditions, overexpression of CLB2 could rescue the defect conferred by ddc1Δ in telomerase-negative cells to the same extent as it did in rescuing the defect of CLB2-deleted telomerase-negative cells (Table 2; Figure 2A). We therefore conclude that the defect in type II recombination conferred by a mutation in the Mec3 complex might be functionally related to that conferred by a deficit in Cdc28-Clb2.
Table 2.
Effect of CLB2 overexpression on survival type frequenciesa
| Relevant genotype | % of type I/% of type II |
|---|---|
| tlc1 | 0/100; n = 22b |
| tlc1 ddc1 | 80/20; n = 18 |
| tlc1 clb2 | 100/0; n = 26 |
| tlc1 ddc1 + YEp-CLB2 | 50/50; n = 16 |
| tlc1 clb2 + YEp-CLB2 | 50/50; n = 8 |
aAfter ∼130 generations in liquid cultures at 29°C.
bn = number of survivor populations tested.
Figure 2.
Overexpression of CLB2, the major B-type mitotic cyclin gene, rescues type II telomeric recombination in tlc1Δ ddc1Δ mutant cells. (A) Telomerase-negative cells (tlc1) bearing a null mutation in either CLB2 (clb2) or DDC1 (ddc1), as indicated, and also overexpressing or not CLB2 (2 μ plasmid, under endogenous promoter) were propagated in liquid cultures for ∼130 generations, from the time the strains were created, at 29°C. As usual, this procedure resulted in the generation of 100% type II survivors in telomerase-negative cells (tlc1) due to the growth advantage of types II over types I in liquid cultures (survivor type has been labeled below each lane). tlc1Δ clb2Δ mutants were defective in generating type II survivors, as reported previously (31), as were tlc1Δ ddc1Δ mutants. Overexpression of CLB2 partially corrected that defect in both tlc1Δ clb2Δ and tlc1Δ ddc1Δ. It is important to note that tlc1Δ clb2Δ mutants were somewhat sick and grew poorly, hence the difficulty in obtaining optimal amounts of DNA for nice signals. XhoI cutting and a TG1–3 32P-labeled probe were used. (B) Endogenous Clb2 levels remain unchanged in the absence of DDC1. Cell extracts from pre-senescent telomerase-negative cells bearing or not a mutation in DDC1, both harvesting CLB2-HA integrated at CLB2 locus, were immuno-precipitated with anti-HA antibody and processed for western blotting with anti-HA antibody. Various dilutions of samples were loaded, as indicated, in order to better appreciate possible variations in Clb2 levels. Quantification was done using ImageQuant. (C) Overproduced Clb2 levels are slightly decreased when DDC1 has been deleted. Immuno-precipated CLB2-HA, expressed from a multi-copy (episomal) plasmid under the control of native promoter in pre-senescent tlc1Δ clb2Δ and tlc1Δ ddc1Δ mutants to serve as a control for the experiment shown above in (A), was detected as described above. The fate of these six individual clones after post-senescence recombination had occurred was followed by Southern (XhoI cutting, TG1–3 32P-labeled probe). For each strain, two type II and one type I survivors were recorded, as shown in the lower panel.
Possible cell cycle alterations in Clb2 levels could be invoked to explain the type II recombination defects in telomerase-negative cells bearing a mutation in the Mec3–Ddc1–Rad17 complex. To evaluate this possibilty, we measured Clb2 levels by immunoprecipitation of endogenous CLB2-HA in the tlc1Δ ddc1Δ and tlc1Δ mutants. Clb2 levels did not vary in the absence of Ddc1 (Figure 2B). We also assessed CLB2-HA levels following overexpression from a multi-copy plasmid, the same conditions as those used for determining the survivor type in these mutants (Figure 2A). Similar levels of overproduced Clb2 from one isolate to the other in both strains (Figure 2C) indicated that, under these experimental conditions, comparing the survivor type in various mutants, as shown in Figure 2A, is technically relevant. This was expected as in the multi-copy plasmid, CLB2-HA was under the control of its native promoter, like in the integrative plasmid. In addition, we observed that Clb2 levels were slightly decreased in the absence of Ddc1 (Figure 2B). This may reflect a slight effect of the ddc1Δ mutation on endogenous Clb2 levels, but detectable only at higher levels of expression for reasons of increased sensitivity. Alternatively, this may reflect the exsitence of a mechanism of control of CLB2 expression in this checkpoint-deficient mutant.
We next evaluated the possibility that the type II defect observed in telomerase-negative cells bearing a mutation in the Mec3–Ddc1–Rad17 complex might in fact reflect alterations in the type I recombination machinery. For instance, it was possible that the intracellular levels of Rad51, (which controls type I recombination) were increased in these mutants and that this could affect telomeric recombination. To test this hypothesis, we overexpressed RAD51 (from a multi-copy, episomal, plasmid) in various strains and analyzed the pattern of telomeric recombination. This had no visible effect as tlc1Δ single mutants overexpressing RAD51 still continued to produce 100% type II survivors, while overexpression of RAD51 in tlc1Δ rad17Δ double mutants did not alter their rate of type II defect (data not shown). It is therefore unlikely that the type II defect conferred by a mutation in the Mec3–Ddc1–Rad17 complex is due to this complex somehow inhibiting Rad51 at the telomeres.
RPA controls both types of telomeric recombination
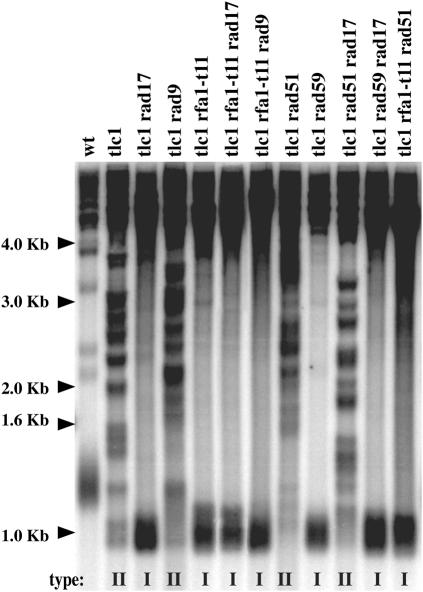
RPA, a single-stranded DNA-binding protein essential for DNA replication, has been implicated in a number of DNA metabolism reactions (50,51). RPA is generally considered to be the initial sensor of DNA damage (23,52), but, because some studies have found that Rad51 (53) or Mre11 (54) were recruited first at DNA double-strand breaks, the issue is still the matter of debate (50). RPA has been evolutionary conserved and, in humans, consists, of three subunits, RPA70, RPA32 and RPA14, homologues in S. cerevisiae to Rfa1, Rfa2 and Rfa3, respectively. In yeast and humans, RPA binds single-stranded DNA generated after resection of DNA double-strand breaks, then Ddc2/ATRIP, thereby loading the Mec1–ATR complex and activating the checkpoint (23). Numerous mutants of RFA1, mainly, have been isolated based on their sensitivity to genotoxic stress, defects in various types of recombination or in DNA replication. Because RPA has been recently shown to recruit Ddc2-Mec1 at sites of DNA damage, a function lost in the rfa1-t11 mutant (23), we chose to use the rfa1-t11 (K45E) mutant to further document the mec1Δ-induced telomeric recombination defect described above. Moreover, rfa1-t11-induced defects in recombination and adaptation to DNA damage have been described (15,24,50,55). As detailed in Table 3 (see also Figure 3), telomerase-negative cells bearing the rfa1-t11 mutation were severely affected in telomeric recombination events. Incidentally, survivors (all of type I) from tlc1Δ rfa1-t11 mutants appeared when the liquid cultures were propagated using a lower dilution [107 cells/ml every 24 h; Table 3; the rate of dilution has recently been proven to be important in such experiments; Ref. (31)]. However, intriguingly, deletion of RAD51 (essential for type I recombination in telomerase-negative, otherwise wild-type, cells) in tlc1Δ rfa1-t11 mutants still allowed the production of survivors (Table 3). Although we have not directly tested whether Y′ elements were amplified in these tlc1Δ rfa1-t11 mutants, the absence of any amplification of TG1–3 sequences (Figure 3, last lane), together with the existence of only two types of telomeric sequences being amplified in telomerase-negative cells, namely Y′ and TG1–3 (16), most probably identify these survivors as type I survivors. The complete absence of any survivor in senescing tlc1Δ rfa1-t11 rad59Δ mutants indicated that this rfa1-t11-associated type I recombination (atypically) relied on the Rad59 pathway (Table 3).
Table 3.
Pattern of telomeric recombination in telomerase-negative rfa1-t11 mutantsa
| Relevant genotype | Frequency of recombinationb | Type of recombination |
|---|---|---|
| tlc1 | 1/105 | II |
| tlc1 rad17 | 1/105 | mostly I |
| tlc1 rad9 | 1/105 | II |
| tlc1 rfa1-t11 | 1/107 | I |
| tlc1 rfa1-t11 rad17 | 1/107 | I |
| tlc1 rfa1-t11 rad9 | 1/107 | I |
| tlc1 rad51 | 1/105 | II |
| tlc1 rad59 | 1/105 | I |
| tlc1 rad51 rad17 | 1/105 | II |
| tlc1 rad59 rad17 | 1/105 | I |
| tlc1 rad51 rfa1-t11 | 1/107 | I |
| tlc1 rad59 rfa1-t11 | N/Ac | N/A |
| cdc13-1 tlc1 | 1/105 | II |
| cdc13-1 tlc1 rfa1-t11 | 1/105 | II |
| cdc13-1 tlc1 rad17 | 1/105 | II |
| cdc13-1 tlc1 rfa1-t11 rad17 | 1/105 | I |
aCells were grown in liquid cultures for ∼150 generations, at 29°C (except for the ts cdc13-1 strains grown at 24°C).
bCells were diluted every 24 h at 1 × 105 or 1 × 107 cells/ml; survivors appeared at the indicated value when their frequency of appearance exceeded the rate of dilution.
cN/A: recombination never occurred.
Figure 3.
Effects of the recombination-deficient, checkpoint-deficient rfa1-t11 mutation on telomeric recombination. Analysis of telomere organization and structure by Southern blotting (see Materials and Methods), in the strains (of the indicated relevant genotype), plus a wild-type non-senescing control strain (wt, lane 1). Mutant cells were propagated in liquid cultures, at 29°C, until senescence took place and beyond (a total of ∼130 generations), with a daily dilution of 107 cells/ml. The survivor type, determined according to criteria described in Materials and Methods and in Figure 1A, is indicated under each lane. XhoI cutting and a TG1–3 32P-labeled probe were used.
Targeting Rad52 at Rfa1-t11 binding sites restores type I, but not type II, recombination
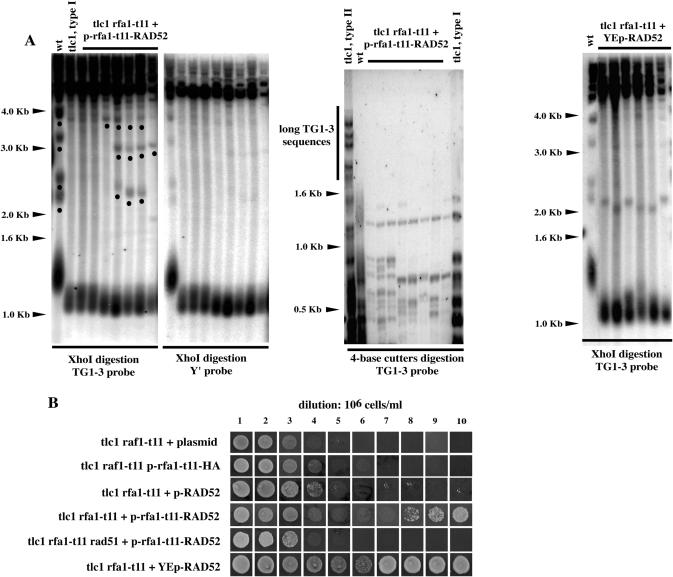
Some aspects of type II recombination are still very mysterious because the Rad51 recombinase, essential for strand invasion, is dispensable during these homologous recombination processes (11,15,17). Interactions between RPA and Rad51 (and Rad52) are relatively well understood (15,50). On the other hand, the putative role of RPA in type II telomeric recombination is totally unknown. As Rad52 participates in -and is essential for- both type I and type II recombination (13,16,56), we set out to determine whether targeting Rad52 at the sites for binding of the recombination-deficient Rfa1-t11 protein would have the same effect on these two types of recombination. Expression of fusion (hybrid) proteins has been shown to represent a valuable tool to decipher telomeric pathways (38,57). Here, we constructed an rfa1-t11-RAD52 chimeric gene and expressed it, together with the appropriate control constructs, in tlc1Δ rfa1-t11 mutants. Survivors from tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 were generated, in the liquid assay, while tlc1Δ rfa1-t11 mutants did not generate survivors at all under identical conditions (Figure 4B). Under the usual criteria, these survivors appeared to be of type I, with the reserve, however, that bands in the 2.0–3.0 kb range were still present in some isolates (Figure 4A, left panel). In both types of survivors, the disappearance of the non-Y′ fragments (migrating at ∼2.1, 2.3, 3.3 and 3.9 kb) attests to the occurrence of homologous recombination (16). Since in tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 cells, some of these bands were still visible, we set out to further characterize these survivors. To this end, the same blot as that shown in Figure 4A (left panel) was reprobed with an Y′ probe. The telomeric fragments migrating at ∼1.1 kb, representative of type I recombination (Figure 1A), were heavily labeled over the background (Figure 4A, left panel). We also digested the genomic DNA from the tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 cells with a mixture of four base cutter restriction enzymes that is sometimes used to identify type II recombination on the TG1–3 substrate [(16); see also Materials and Methods]. This also identified these survivors as type I survivors (Figure 4A, middle panel). Finally, the survivors from the tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 strain depended on Rad51 for their maintenance (Figure 4B), as do normal survivors from telomerase-negative, otherwise wild-type, cells. Therefore the Rfa1-t11-Rad52 fusion protein does rescue telomeric recombination but leaves cells with an rfa1-t11-associated specific defect in type II recombination.
Figure 4.
Targeting Rad52 at the sites of DNA damage restores type I, but not type II, recombination in tlc1Δ rfa1-t11 mutants. (A) Pattern of telomeric recombination in tlc1Δ rfa1-t11 cells carrying the rfa1-t11-RAD52 fusion construct on a centromeric plasmid. Left two panels: Seven different isolates from tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 (lanes 3–9 in both panels) plus a wild-type, non senescing, strain (lane 1 in both panels) and a tlc1Δ strain having recombined with a type I (lane 2 in both panels) are illustrated. Cells were grown in liquid cultures for about 200 generations, at 29°C, after the senescence crisis and processed for Southern blot analysis using either a telomeric TG1–3 (leftmost panel) or Y′ probe (rightmost panel) following DNA digestion with XhoI in both cases. In leftmost panel, non-recombining DNA fragments, that appear to be non-Y′ fragments have been labeled with a black dot on the leftmost panel. These bands were not detected when the blot was re-probed with a probe detecting Y′ sequences (rightmost panel). Middle panel: Genomic DNAs from the same cells as those shown above in the left two panels were digested with a mixture of restriction enzymes (AluI, HaeIII, HinfI and MspI) using a 4 bp recognition sequence and the Southern revealed with a TG1–3 probe. This allowed to discriminate between type I and type II recombination because these enzymes do not cut within TG1–3 sequences (16). The slow migrating bands (labeled with the black dots in the leftmost panel) were no longer visible on the four base cutter blot, indicating that these bands are not the result of some limited type II recombination. DNAs from type II (lane 1) or type I (lane 11) survivors from a tlc1Δ strain are shown for comparison. Right panel: Overexpression of RAD52, from a 2 μ episomal plasmid under the control of native promoter also rescued telomeric recombination in tlc1Δ rfa1-t11 cells. Only type I survivors were recovered. (B) tlc1Δ rfa1-t11 cells expressing the rfa1-t11-RAD52 fusion from a centromeric plasmid (or the rfa1-t11-HA control or plasmid alone), as indicated, or expressing RAD52 from a low-copy (centromeric) plasmid (row 3) or from a multi-copy (episomal) plasmid (row 6), were propagated in liquid cultures for 10 days, at 29°C, with a dilution to 106 cells/ml every 24 h, before re-streaking on plates to assess survival. Both YCp-rfa1-t11-RAD52 and YEp-RAD52 (but not YCp-RAD52) restored telomeric recombination, which, in both cases, was of type I (data not shown). No survivors were generated in the absence of RAD51 (row 5). Therefore, type I recombination in tlc1Δ rfa1-t11 p-rfa1-t11-RAD52 cells is conventional in the sense that it is Rad51-dependent.
To try to confirm these findings above, we also performed experiments with RAD52 expressed from a plasmid. Interestingly, overexpression of RAD52 from a multi-copy (episomal) plasmid also rescued the recombination defect in tlc1Δ rfa1-t11 cells, only type I but not type II, like the Rfa1-t11-Rad52 fusion protein (Figure 4A and B). Importantly, slight overexpression of RAD52 from a centromeric plasmid (the same sort as that used for the rfa1-t11-RAD52 fusion experiments described above) did not rescue the recombination defect in tlc1Δ rfa1-t11 cells (Figure 4B). Therefore, in the experiments with the rfa1-t11-RAD52 fusion, the effect of Rad52 on telomeric recombination is really due to targeting to rfa1-t11's binding sites and not merely to moderate increase in its intracellular amounts. In summary, targeting of Rad52 to Rfa1-t11's binding sites with two different strategies produce the same phenotype, namely selective rescue of telomeric recombination.
An Rfa1–t11–Ddc2 fusion protein restores the checkpoint function of rfa1-t11 cells but not its recombination function
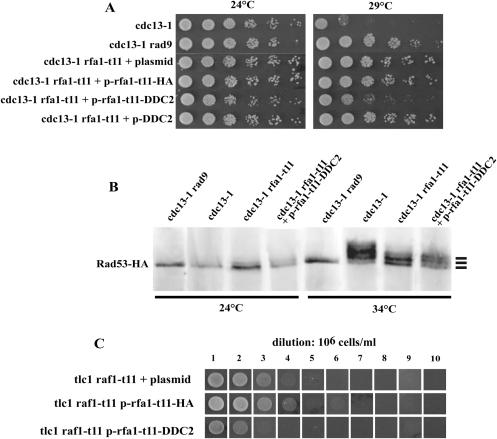
RPA and Rad52 are likely to interact functionally and physically, thereby modulating the interactions between RPA and Rad51 during repair of double-strand breaks by homologous recombination (15,50), as well as during type I telomeric recombination, as seen above. Since in wild-type cells, RPA recruits Mec1-Ddc2 at the sites of DNA double-strand breaks, an event that is defective in rfa1-t11 mutant cells (23), and since mec1Δ cells are defective in type II recombination [(22); present data], we next wanted to determine whether failure of the Rfa1-Ddc2 interaction in rfa1-t11 was responsible for the type II recombination defect in this mutant. Because we were confident that fusion proteins implicating Rfa1-t11 could be reliable, as seen above, we decided to use an Rfa1–t11–Ddc2 fusion protein to target Mec1-Ddc2 at the sites of telomeric damage. First, we determined whether the hybrid protein could rescue the checkpoint defect of the rfa1-t11 mutation in the cdc13-1 background (58). We did not expect to rescue the lethality conferred by the absence of DDC2 (59–61) with this chimeric protein, but only aimed at trying to restore Rfa1-t11's defect in Ddc2 binding without interfering with other Ddc2 functions. Consequently, these experiments were performed in a DDC2+ background. cdc13-1 rfa1-t11 cells expressing the rfa1-t11-DDC2 fusion construct clearly grew less at 29°C than when expressing the plasmid alone, but still grew better than cdc13-1 cells, thereby suggesting partial re-establishment of a functional DNA damage checkpoint (Figure 5A).
Figure 5.
Restoration of rfa1-t11 checkpoint defect by targeting of Ddc2 does not correct the rfa1-t11-induced defect in telomeric recombination. (A) Growth of cdc13-1 rfa1-t11 strains transformed each with a centromeric plasmid expressing the indicated fusion construct at 29°C. Growth of checkpoint-proficient cdc13-1 (lane 1) and checkpoint-deficient cdc13-1 rad9Δ (lane 2) mutants is shown for comparison. Rescue of the checkpoint defect of cdc13-1 rfa1-t11 by plasmid bearing rfa1-t11-DDC2 is suggested by re-acquisition of the cdc13-1-induced growth arrest at 29°C. See text for explanations. (B) Activation of Rad53 assessed by visualizing electrophoretic mobility alterations after western-blot analysis of immunoprecipitated cell extracts using anti-HA antibodies. Strains of the indicated relevant genotype were grown at the temperature of 24 or 34°C (permissive or fully restrictive for growth of cdc13-1 cells, respectively). The non-phosphorylated Rad53 band, as well as the first two bands of phosphorylated Rad53, representing the first two levels of activation of Rad53, are indicated by bars in the right margin. Additional bands representing higher levels of activation of Rad53 could also be detected in the cdc13-1 control (lane 2 of 34°C). All strains carried endogenous RAD53-HA integrated at the RAD53 locus. See text for explanations. (C) The rfa1-t11-DDC2 fusion construct (expressed from a centromeric plasmid) did not restore defective recombination in tlc1Δ rfa1-t11 mutant cells. Cells were propagated in liquid cultures for 10 days, at 29°C, with a dilution of the cultures to 106 cells/ml every 24 h. tlc1Δ rfa1-t11expressing p-rfa1-t11-HA were used as a control.
Improved growth of cdc13-1 cells can result not only from checkpoint inactivation, but also from diminished amounts of damaged DNA, such as in exo1Δ mutants, for instance (62). Since RPA is likely to readily affect processing of damaged DNA and to control exonucleases, we set out to assess checkpoint activation in a different manner. The phosphorylation state of Rad53, an event that reflects its activation (63,64), was visualized by western blotting using anti-HA antibodies in strains in which a RAD53-HA fusion gene had been introduced at the RAD53 locus. As expected, Rad53 was massively phosphorylated in cdc13-1 mutant cells at 34°C, but not at 24°C, and this signal was absent in cdc13-1 rad9Δ cells at either temperature (Figure 5B). In cdc13-1 rfa1-t11 cells, Rad53 phosphorylation was intermediate between these two fully active and inactive states (Figure 5B), in agreement with the previous finding that the rfa1-t11 allele is not completely defective in checkpoint activation (58). Importantly, Rad53 exhibited a higher degree of phosphorylation in cdc13-1 rfa1-t11 cells bearing the rfa1-t11-DDC2 construct than in cdc13-1 rfa1-t11 cells expressing the control fusions or plasmid alone (Figure 5B). Indeed, an additional Rad53 shifted band was present in the former sample compared with the control samples (Figure 5B). Increased Rad53 activation in cells expressing the Rfa1–t11–Ddc2 fusion was also evident from the observation of a decrease in the intensity of the Rad53 lower band (Figure 5B). Altogether, these data indicated that the Rfa1–t11–Ddc2 fusion protein is capable of restoring, at least partially, the DNA damage checkpoint.
We next asked whether the rfa1-11-DDC2 construct could cure the rfa1-t11-associated telomeric recombination defects. tlc1Δ rfa1-t11 cells bearing the rfa1-t11-DDC2 construct were unable to generate any survivors at all (Figure 5C), as observed in tlc1Δ rfa1-t11 cells under identical conditions (Table 3; Figure 4B). Thus, partial rescue of Rfa1-t11's checkpoint defect by the Rfa1–t11–Ddc2 fusion protein does not result in concomitant rescue of telomeric recombination. At this point, we hypothesized that the interaction between RPA and Mec1-Ddc2 might be important for operating some processes only of type II recombination. We therefore set out to provide rfa1-t11 mutants with a functional Rad52 machinery and determine whether the rfa1-t11-DDC2 fusion could now make type II recombination work. tlc1Δ rfa1-t11 cells co-expressing RAD52 and the rfa1-t11-DDC2 fusion could generate type I survivors, as observed above with the rfa1-t11-RAD52 construct alone, but were still unable to produce type II survivors (data not shown). These data suggest that the telomeric recombination defect conferred by rfa1-t11 does not result from improper recruitment of Ddc2-Mec1 at damaged telomeres. Moreover, a functional interaction between RPA and the Mec1-Ddc2 checkpoint complex does not appear to play any role in promoting the functioning of the Rad51-dispensable type II recombination, even when functional Rad52 is present.
RPA might compete with Cdc13 for the occupancy of the telomeric TG1-3 substrate
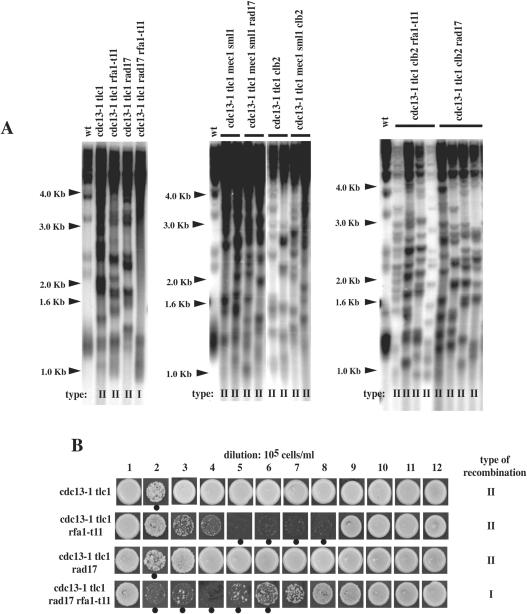
cdc13-1, a temperature-sensitive mutation in CDC13, coding for a single-stranded telomeric DNA-binding protein exhibiting affinity exclusively for TG1–3 sequences (65), generates large stretches of single-stranded telomeric DNA at semi-permissive and restrictive temperatures (30). We reasoned that making more telomeric TG1–3 substrate available within the cell might rescue the type II recombination defect conferred by the rfa1-t11 mutation. Although a null mutation in RAD17 (coding for a member of the Mec3 complex) affects type II recombination in a tlc1Δ background, as seen above, it had no apparent effect on type II recombination in the cdc13-1 tlc1Δ background (Table 3; Figure 6), in agreement with previous results (6). We next conducted similar experiments using the rfa1-t11 mutation. In the cdc13-1 tlc1Δ rfa1-t11 strain, survivors were generated at the 105 cells/ml dilution (Table 3). By Southern analysis, telomeres of the cdc13-1 tlc1Δ rfa1-t11 strain were of type II (Figure 6A). Therefore, presumably, the increase in single-stranded telomeric DNA provoked by the cdc13-1 mutation compensates for the defects conferred by the mec3Δ, rad17Δ and rfa1-t11 mutations in type II recombination by providing more TG1–3 substrate normally masked by Cdc13, as previously proposed (6). Interestingly, the cdc13-1 tlc1Δ rfa1-t11 rad17Δ survivors displayed type I telomeres (Figure 6). The most likely explanation is that the rfa1-t11 and rad17Δ mutation impinge on type II recombination in an additive manner, acting in parallel pathways, such that type I recombination remains the only possible mechanism.
Figure 6.
The cdc13-1 mutation corrects the rad17Δ- and rfa1-t11-induced defects in telomeric recombination. (A) Telomere structure and organization of one or several representative survivors of the indicated relevant genotype. Survivor type after culturing in liquid medium for around 150 generations, as determined by usual criteria (see Materials and Methods and Figure 1A), is indicated for each strain below each lane. In all three panels, lane 1 is from a wild-type (wt) non-senescing control. See text for explanations. (B) Strains of the indicated relevant genotype were grown for 12 days in liquid cultures at 25°C and diluted every 24 h to 105 cells/ml. Every day, cells were spotted on agar plates to assess survival. After post-senescence survival had been attained for all strains, on day 12, cells were prepared for determination of survivor type, indicated for each strain on the right, by Southern (data not shown). In both A and B, XhoI cutting and a TG1–3 32P-labeled probe were used.
We next asked whether Mec1 was acting through the RPA pathway or the Mec3-Ddc1-Rad17 pathway. To answer this question, we set out to analyze the effects of combining the mec1Δ sml1Δ and rad17Δ mutations, as well as the mec1Δ sml1Δ and rfa1-t11 mutations, both in a cdc13-1 tlc1Δ background, and asked which one of these mutants would behave like the cdc13-1 tlc1Δ rfa1-t11 rad17Δ mutant in which cdc13-1-induced type II recombination is no longer possible, as seen above. The cdc13-1 tlc1Δ mec1Δ sml1Δ rad17Δ mutant generated type II survivors, just like the cdc13-1 tlc1Δ mec1Δ sml1Δ control (Figure 6A, middle panel). Unfortunately, we could not analyze the cdc13-1 tlc1Δ mec1Δ sml1Δ rfa1-t11 survivors because this strain was inviable. However, the finding that combining the mec1Δ sml1Δ and rad17Δ mutations now results in type II recombination, unlike that of combining the rad17Δ and rfa1-t11 mutations (Figure 6A, left panel), strongly suggests that Mec1 functions in the same pathway as the Mec3–Ddc1–Rad17 complex, or at least in an overlapping pathway, in these processes.
We attempted to use the same logic to determine in which pathway of telomeric recombination control Clb2 was acting. To this end, various mutant strains combining a clb2Δ mutation and the mec1Δ sml1Δ, rfa1-t11 or rad17Δ mutations, all in the cdc13-1 tlc1Δ background, were constructed. The analysis of the type of telomeric recombination in these mutants, shown in Figure 6A, indicated that they all generated type II survivors. This data suggests that the Clb2 pathway may overlap with both the RPA, Mec1 and Mec3 pathways.
RPA's checkpoint and recombination functions are genetically separable
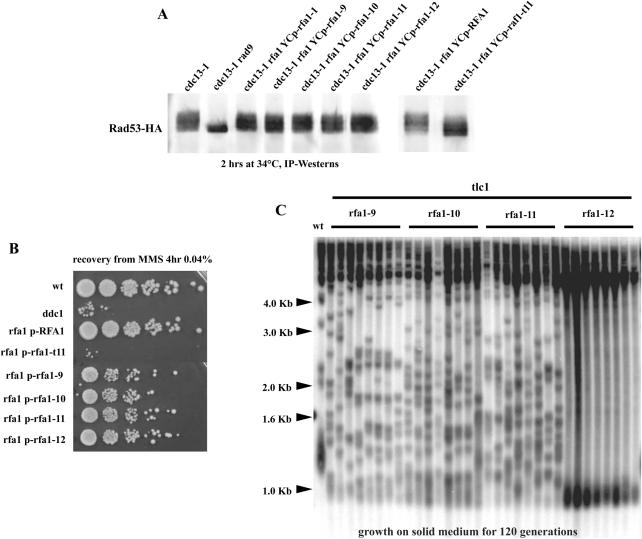
The finding above that Rfa1-t11 with a partially restored checkpoint function still conferred complete defect in telomeric recombination suggested that these two functions of RPA might be distinct. To address this point, we set out to isolate rfa1 mutants that, unlike rfa1-t11, which is deficient in both the checkpoint and recombination functions, would be defective in either one (see Materials and Methods). We could isolate four rfa1 mutants, that we call rfa1-1, rfa1-9, rfa1-10 and rfa1-11, that were affected in telomere recombination but remained checkpoint-proficient, on the basis of their ability to activate Rad53 at a level similar to that in cdc13-1 RFA1 cells (Figure 7A and C; rfa1-12, another mutant isolated in the same screen is shown as a checkpoint-proficient, recombination-proficient allele). In agreement with this observation, these rfa1 mutants did not exhibit significant hypersensitivity to MMS (Figure 7B). Interestingly, these mutants were specifically deficient in type I recombination, as they produced 100% type II survivors in the ‘streak assay’ (Figure 7C), a situation in which, in most strain backgrounds, including ours, ∼90% of the survivors are of type I when grown on agar-based medium (6,16).
Figure 7.
Four novel rfa1 mutants, rfa1-1, rfa1-9, rfa1-10 and rfa1-11, that are deficient in telomeric recombination are nevertheless checkpoint-proficient. (A) Activation of Rad53, a protein kinase with a pivotal role in the DNA damage response, was measured in these four mutants, in the cdc13-1 background (lanes 3–6) as well as in negative controls (cdc13-1 rad9Δ, lane 2 and cdc13-1 rfa1-t11, lane 9) and positive controls (checkpoint-proficient cdc13-1 mutants, lane 1 and cdc13-1 rfa1-t11 expressing RFA1, lane 8), as described in the legend to Figure 5B). rfa1-12 (lane 7) is another mutant isolated in the same mutagenic screen. (B) rfa1-9, rfa1-10, rfa1-11 and rfa1-12 mutants are not hypersensitive to a genotoxic stress, unlike rfa1-t11 and ddc1Δ mutants. 10-fold serial dilutions (from left to right in each row) of cultures of the relevant indicated genotype that have been treated (in liquid) by MMS, as indicated, prior to spotting on agar plates. (C) tlc1 rfa1 mutants were continuously grown, at 29°C, on agar-based medium immediately after sporulation of the diploids. Cells were re-streaked every 48 h and allowed to reach senescence, after which cells were processed for genomic DNA preparation and determination of the survivor type (‘streak assay’, see Materials and Methods), an assay in which the vast majority of survivors is of type I (see text for explanations). The rfa1-1, rfa1-9, rfa1-10 and rfa1-11, but not rfa1-12, mutants were deficient in generating type I survivors in this assay. XhoI cutting and a TG1–3 32P-labeled probe were used.
DNA sequencing revealed that rfa1-1 and rfa1-9 contained multiple point mutations (Table 4). We have not been able so far to identify the mutations responsible for the phenotypes of the corresponding mutants.
Table 4.
Sequence analysis of the amino acid changes in the rfa1-1 and rfa1-9 alleles
| Rfa1-1: I89T, T116S, S160P, D240G, T346P, D415N, D465V, T510S, E562G, Y575C |
| Rfa1-9: S108P, H299D, N310S, V317I, N368Y, S487T, Q571R |
DISCUSSION
Telomere length homeostasis is critical to the maintenance of a stable genome (2,3). In the absence of telomerase, telomeres progressively shorten, thus losing their protective proteins, and eventually fuse together and break during chromosome segregation, leading to death by senescence (8). Rare cells survive this telomeric senescence by performing recombination between telomeric or subtelomeric sequences (11). In budding yeast, telomeric recombination can be affected by mutations in DNA polymerases, a helicase, an exonuclease, a linker histone, telomere end protection proteins, telomere length control proteins, checkpoint protein kinases, cyclin-dependent kinase, non homologous end joining genes and by mating type (11,20–22,31,37,66–68). The present data reveal that, upon loss of telomerase function, the DNA damage sensors Mec1, Rad24 and Mec3 can affect telomere recombination in a pathway distinct from that initiating the Mec1-Rad53-Chk1 kinase cascade normally culminating, upon DNA damage, in cell cycle arrest. This study has also provided insights into how the evolutionary conserved DNA repair/replication/checkpoint RPA complex is involved in promoting cell proliferation by controlling telomeric recombination.
Control of telomeric recombination by DNA damage checkpoint proteins
Here, we find that a subset of checkpoint proteins, namely Mec1 [also found in a previous study, Ref. (22)], Mec3-Ddc1-Rad17 and Rad24, are required for optimal operation of type II telomeric recombination. On the other hand, inactivation of, either RAD9, RAD53 or CHK1 had no impact on telomeric recombination. Inactivation of one of the latter genes totally or partially (Rad53 and Chk1 function in parallel pathways both requiring Rad9) prevents activation of the whole DNA damage response. In particular, the kinase cascade that goes from Mec1 to Rad53 and Chk1 is, of course, interrupted (40,41). Therefore, the whole checkpoint response is not required for the correct control of telomeric recombination by checkpoint proteins. Mec1 kinase activity, which is absolutely necessary for the functioning of the downstream checkpoint components (26,48), was indispensable for these events. This observation leaves open the possibility that Mec1 could act in these pathways by phosphorylating a member of the MRX (Mre11–Rad50–Xrs2) complex or Rad59, as proposed previously (22). In the present situation, the Mec1-dependent DNA damage checkpoint response was shown to branch out early during the process of damage recognition, initiating two separate pathways, one that results in the activation of the kinase cascade and cell cycle arrest, the second one that activates type II recombination. This is not unprecedented as, for instance, mec1Δ tel1Δ double mutants underwent telomeric senescence (69), while single or double mutants of the downstream checkpoint components do not. We propose that Mec1 and Mec3, once loaded onto damaged DNA, affect its conformation. They might, for instance, bend it so as to create conformations compatible with access of proteins subsequently needed for type II recombinational events, such as Mre11, Rad50, Xrs2, Rad59 or other as yet unidentified proteins.
Recently, rad24Δ mutants were shown to exhibit a marked delay in the processing and repair of broken ends at a DNA double-strand break, independently of their defect in G2/M arrest (70). The resulting low survival, not observed in rad9Δ, chk1Δ and pds1Δ mutants, was also observed in rad17Δ, mec3Δ, mec1Δ, and noticeably, in rad53Δ dun1Δ mutants (70). On this basis, the mechanisms described here by which Rad24, Mec1 and Mec3, but not Rad53, control telomeric recombination appear to differ from the mechanisms of resection of sequences at a broken end described by Aylon and Kupiec (70). Moreover, in the study by Aylon and Kupiec (70), Rad51-dependent events of recombination were analyzed, whereas in the present study the defects conferred by the rad24Δ, mec3Δ and mec1Δ mutations concerned a pathway of recombination which, in telomerase-negative, otherwise wild-type, cells, is completely Rad51-independent (71). At telomeres, some checkpoint proteins have also been shown to affect the processing of single-stranded DNA that is produced after cdc13-1-induced telomeric damage (72). It has now been established that Mec1, Rad53 and Rad9 inhibit degradation of telomeric DNA by preventing formation of abnormal single-stranded DNA in these cells, while, on the opposite, Mec3, Rad17 and Rad24 promote degradation of telomeric DNA (73). In fact, human Rad1 and Rad9 (homologues to S. cerevisiae Rad17 and Ddc1) have indeed been shown to possess exonuclease activity (74). Therefore, a member of the Mec3 DNA damage sensor complex could potentially be involved in controlling telomeric recombination via processing damaged structures specifically used for Rad50-, Rad59-dependent recombination.
Although Rad9 also accumulates at double-strand breaks in a Mec1-dependent manner (75), the rad9Δ mutation, unlike the mec1Δ mutation, was not found here to affect telomeric recombination. However, it should be noticed that another study found that Rad9 focus formation was only partially affected in mec1Δ mutants (54). In fission yeast, Crb2 (the homologue of S. cerevisiae Rad9) also physically associated with double-strand breaks (76). Recruitment of Crb2 could be accomplished in the absence of Rad1 and Rad3, (respectively, Rad17 and Mec1 in S. cerevisiae) but necessitated their presence to remain attached (76). Therefore, the physical presence of a checkpoint protein at the sites of damaged DNA is not a sufficient criterion to take into account as determining a positive action on type II recombination. The Mec1 and Mec3 DNA damage sensors may therefore be endowed with specific properties that entitle them to affect type II recombination.
In telomerase-negative, otherwise wild-type, cells, type II recombination takes place on TG1–3 repeats and is operated via the MRX complex, Rad59 and Rad52 (11,56,71). Type II recombination is interesting on a mechanistic point of view because it does not require the intervention of the Rad51 recombinase, the only protein capable of strand invasion during homologous recombination. Rolling-circle replication primed by an intrachromosomal telomeric D-loop has been proposed as the basis for type II recombination (11,12,17,71,77). The observation made here that cdc13-1 rescues the rad17Δ-induced defect in type II recombination demonstrates the possibility of efficient recombination on TG1–3 sequences in the absence of the Mec3 complex. This strongly argues that the unusual predominance of type I survivors in tlc1Δ mec3Δ cells is due to the impairment of the Rad50 pathway in the absence of Mec3 rather than to an impairment of recombination per se. Noticeably, type II recombination in tlc1Δ rad17Δ was restored after RAD51 had been deleted, leaving open the possibility that Rad51 might inhibit type II recombination in wild-type cells and that deletion of a member of the Mec3 complex might affect telomeric recombination through changing Rad51 levels. However, control experiments indicated that variations in Rad51 levels, induced by overexpressing RAD51, had no effect on telomeric recombination and were, therefore, probably not responsible for the observed defect in type II recombination in these mutants. Since Rad52 is essential for type II recombination, unlike MRX and Rad59, which can substitute for each other in this process (56,71,78), it has been speculated that Rad52 might perform the annealing activity involved in D-loop formation (78). Clearly, however, Rad52 cannot be the defective target in mec1Δ and mec3Δ mutants, as if this was the case, then these mutants would also be deficient in type I recombination, Rad52 being essential for both types of recombination.
An important step towards elucidating the processes analyzed here was the observation that CLB2 overexpression could rescue the type II recombination defect conferred by a mutation in the Mec3 complex. These data establish that the cell cycle machinery can impinge on recombinational repair, thus confirming several recent data, including the initial one (79) and one on telomeric recombination (31). Since Cdc28-Clb2, homologue to Cdk1-cyclin B, rescued a null mutation in DDC1, it is not likely that it stimulated the recombination function of the Mec3–Ddc1–Rad17 complex by acting upstream of it. However, Cdc28-Clb2 might act by increasing the intensity of the signal contributed by Mec1. Indeed, it should be noted that our data indicate that Mec1 and Mec3 are both required for the control of telomeric recombination and that, consequently, they perform distinct functions in these processes. The possibility of such a scenario is rendered likely by the recent demonstration that ectopic CLB2 overexpression resulted in direct activation of Tel1 (80). Alternatively, larger amounts of Cdc28-Clb2 than usual might activate a component of the recombinational machinery that is normally put in place by the action of the Mec3 complex, thus bypassing the need for Mec3-induced chromatin modifications. In the same vein, it is possible that Cdc28-Clb2 directly affects Rad51, inhibiting it for instance, rather than acting on the Mec3 complex, either upstream or downstream of it. In this scenario, the Mec3 and Cdc28–Clb2 complexes would affect telomeric recombination independently of each other, but modifying the dosage of the other complex could then counterbalance any effect of either one of these complexes on Rad51.
Our data do not eliminate the possibility that increased dosage of CLB2 might indirectly act by affecting the position of telomerase-negative cells in the cell cycle, possibly enriching a stage favoring type II recombination. For instance, the Cdk1-cyclin B complex has been shown to influence recombinational repair during the G2 phase (79). However, it should be noticed that, unlike constitutive CLB2 overexpression that causes strong mitotic delay or even arrest (81), overexpression of CLB2 under the control of its native promoter has no apparent effect on cell cycle progression (N. Grandin and M. Charbonneau, unpublished data). We did not observe significant changes in endogenous Clb2 levels in the absence of the Mec3–Ddc1–Rad17 complex. Even if the slight decrease in overproduced Clb2 levels observed in ddc1Δ tlc1Δ mutants also takes place in cells expressing endogenous levels of Clb2 but remains undectable, it seems unlikely that such a variation could affect telomeric recombination, because it is only in the complete absence of Clb2 that a defect in type II recombination could be observed. Finally, the slight decrease in overproduced Clb2 levels induced by DDC1 deletion might reflect an effect of the inactivation of the checkpoint on cell cycle position resulting in slight changes in CLB2 expression.
Control of telomeric recombination by RPA
To our knowledge, the role of RPA in mediating telomeric recombination had not been previously studied and the present work therefore provides a platform for further studies. We chose to analyze RPA functions during telomeric recombination using the rfa1-t11 mutant allele (encoding Rfa1-K45E) because it has been well documented in other contexts, not only in DNA repair but also in checkpoint pathways (24,50,52). Several conclusions can be made based on the new phenotypes of rfa1-t11 cells observed in the present study. rfa1-t11 resulted in impairment of both type I and type II recombination. Importantly, however, type I recombination in rfa1-t11 mutants (only possible at decreased dilutions during passages) atypically relied on Rad59, thus stressing the plasticity of these recombinational pathways. This also confirms that rfa1-t11 confers complete impairment of the Rad51 machinery (15,50). The Rfa1–t11–Ddc2 fusion protein partially restored the checkpoint but did not facilitate the generation of type II recombination. The recombination defect caused by rfa1-t11 was more severe than that caused by the absence of a DNA damage sensor. The Rfa1–t11–Rad52 fusion protein restored normal, Rad51-dependent, type I recombination in rfa1-t11 cells, but not type II recombination. The cdc13-1 mutation rescued the defects in type II recombination conferred by rfa1-t11. Elimination by rfa1-t11 of cdc13-1-induced type II recombination in cdc13-1 tlc1Δ rad17Δ cells (Figure 6) seems to indicate that rfa1-t11 and rad17Δ have an additive effect on type II recombination. Moreover, based on genetic arguments illustrated in Figure 6, Rad17 and Mec1 appear to function in the same pathway of telomeric recombination control or, at least, in overlapping pathways, while Cdc28-Clb2 might function in a pathway that overlaps with both the Mec1, Mec3 and RPA pathways.
Although expression of fusion (hybrid) proteins may produce anomalous results under certain circumstances, it should be stressed that it has been previously used with success in studies on telomeric processes (38,57). Noticeably, one of the telomeric proteins of the protein fusion pairs used in these previous studies was Cdc13, which, like Rfa1 used here, is a single-strand DNA-binding protein (65). In the present study, overexpression of RAD52 gave similar results, thus comforting the experiments using the rfa1-t11-RAD52 and rfa1-t11-DDC2 chimeric genes.
The production of abnormally high levels of single-stranded DNA at telomeres in cdc13-1 cells (30) was presumably the cause for the rescue of type II recombination in mec3Δ, rad17Δ and rfa1-t11 cells (Table 3; Figure 6). It is possible that increased single-stranded DNA allows more RPA to be recruited at damaged telomeres, thereby counterbalancing the defect caused by the rfa1-t11 mutation. Increased efficiency of type II recombination in the cdc13-1 mutant background has been previous observed (6). We propose two possibilities to explain the rescue of type II recombination by cdc13-1 in rfa1-t11 cells. In the first scenario, Rfa1-t11 would not bind enough TG1–3 sequences, which would result in a failure to recruit the Mre11–Rad50–Xrs2 complex, needed for type II recombination. This would be a specific characteristic of Rfa1-t11 towards this type of DNA because Rfa1-t11 normally binds other types of single-stranded DNA, in vitro (82). In the second scenario, Rfa1-t11 would normally bind the telomeric TG1–3 DNA but would be defective in binding Mre11-Rad50-Xrs2. In both situations, increasing the amount of single-stranded TG1–3 DNA via expression of the cdc13-1 mutation would correct either defect by increasing the amount of Rfa1-t11, which although defective, would eventually become functional due to the increase in its amounts. Somehow unexpectedly, type II survivors were no longer generated when the control of recombination became too defective, due to the simultaneous mutations in Rfa1 and Rad17 in cdc13-1 rad17Δ cells and type I survivors were generated instead. This observation that there is at least one possible condition that allows the generation of type I survivors in a cdc13-1 background again underlines the remarkable plasticity of these recombinational pathways.
The failure of the only one documented checkpoint-deficient mutant protein, Rfa1-t11, to recruit Mec1-Ddc2 (23) also confers an intrinsic recombination defect, due to the fact that Mec1 affects type II recombination [(22); present data]. The present finding that expression of an Rfa1-t11-Ddc2 hybrid protein partially re-established checkpoint signaling in cdc13-1 rfa1-t11 cells without concomitant rescue of recombination, in tlc1Δ rfa1-t11, suggested that RPA-t11's defect in type II recombination not only stems from its defect in recruiting Ddc2, but is also due to a defect in a distinct pathway. Interestingly, the experiments in which cells overproduced RAD52 in addition to the Rfa1–t11–Ddc2 fusion suggest that this latter pathway necessitates something else than Rad52 competent in Rad51-mediated events of recombination. The putative formation of D-loops by Rad52, presumably required for Rad50-, Rad59-mediated type II recombination (11,15), might not involve as tight and direct interactions between RPA, Rad52 and MRX as those between RPA, Rad51 and Rad52 during strand invasion. The present data do not preclude the likely possibility that the function of Mec1 in controlling telomeric recombination necessitates its recruitment by RPA. Our data may also suggest that the defect of the DNA damage sensors mutants mec3Δ and rad17Δ in generating type II recombination is not due to the absence in these cells of RPA–Ddc2 interactions. Finally, the present study also provides rfa1 alleles that are deficient in telomere recombination but proficient in checkpoint function and that may be useful in some biochemical or genetic assays. These alleles, rfa1-1, rfa1-9, rfa1-10 and rfa1-11, may confer a specific defect in type I recombination. Alternatively, they may confer a defect in the processing of damaged DNA, so as to alter the nature of the substrate used for telomeric recombination. In this latter hypothesis, these rfa1 mutants would behave like cdc13-1 or yku70Δ mutants, which, presumably because they accumulate abnormal levels of telomeric single-stranded DNA, also abnormally generate exclusively type II survivors during growth on semi-solid medium (37). Recombinational telomere elongation, which at the moment is best understood in yeast, appears to be involved in a subset of human cancers and its study in yeast is therefore important for biomedical research.
Acknowledgments
The authors thank Richard Kolodner, Andrew Emili, Errol Friedberg, Akira Matsuura, Leland Hartwell, Steve Reed, Dan Gottschling, Giovanna Lucchini, Jim Haber and Lorraine Symington for the gifts of strains and plasmids. This work was supported by grants from the ‘Association pour la Recherche contre le Cancer’ and the ‘Comité Départemental de la Savoie de la Ligue Nationale contre le Cancer’. Funding to pay the Open Access publication charges for this article was provided by ‘Association pour la Recherche contre le Cancer’ (ARC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Ann. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 3.Vega L.R., Mateyak M.K., Zakian V.A. Getting to the end: telomerase access in yeast and humans. Nature Rev. Mol. Cell. Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 4.Kelleher C., Teixeira M.T., Förstemann K., Lingner J. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- 5.Chakhparonian M., Wellinger R.J. Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet. 2003;19:439–444. doi: 10.1016/S0168-9525(03)00135-5. [DOI] [PubMed] [Google Scholar]
- 6.Grandin N., Damon C., Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlseder J., Smogorzewska A., de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 8.Lundblad V., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 9.Lendvay T.S., Morris D.K., Sah J., Balasubramanian B., Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna F., Teo S.H., Jackson S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 11.Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya M.K., Lustig A.J. Telomere dymanics in genome instability. Trends Biochem. Sci. 2006;31:114–122. doi: 10.1016/j.tibs.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 14.Neumann A.A., Reddel R.R. Telomere maintenance and cancer—look, no telomerase. Nature Rev. Cancer. 2002;2:879–884. doi: 10.1038/nrc929. [DOI] [PubMed] [Google Scholar]
- 15.Symington L.S. Role of RAD52 epistastis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng S.C., Zakian V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaska L., McEachern M.J., Nosek J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 2004;567:142–146. doi: 10.1016/j.febslet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 18.Larrivée M., Wellinger R.J. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nature Cell. Biol. 2006;8:741–747. doi: 10.1038/ncb1429. [DOI] [PubMed] [Google Scholar]
- 19.Maringele L., Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertuch A.A., Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–1659. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maringele L., Lydall D. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics. 2004;166:1641–1649. doi: 10.1534/genetics.166.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai Y.L., Tseng S.F., Chang S.H., Lin C.C., Teng S.C. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 2002;22:5679–5687. doi: 10.1128/MCB.22.16.5679-5687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 24.Umezu K., Sugawara N., Chen C., Haber J.E., Kolodner R.D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin N., Reed S.I., Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 26.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 27.Emili A., Schieltz D.M., Yates J.R., III, Hartwell L.H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 28.Singer M.S., Gottschling D.E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 29.Moore J.K., Haber J.E. Cell cycle and genetic requirement of two pathways of nonhomologous end-joining repair of double-strand breaks in S. cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garvik B., Carson M., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grandin N., Charbonneau M. Mitotic cyclins regulate telomeric recombination in telomerase-deficient yeast cells. Mol. Cell. Biol. 2003;23:9162–9177. doi: 10.1128/MCB.23.24.9162-9177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longhese M.P., Fraschini R., Plevani P.V., Lucchini G. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siede W., Nusspaumer G., Portillo V., Rodriguez R., Friedberg E.C. Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallory J.C., Petes T.D. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl Acad. Sci. USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takata H., Kanoh Y., Gunge N., Shirahige K., Matsuura A. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell. 2004;14:515–522. doi: 10.1016/s1097-2765(04)00262-x. [DOI] [PubMed] [Google Scholar]
- 36.Louis E.J., Borts R.H. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics. 1995;139:125–136. doi: 10.1093/genetics/139.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grandin N., Charbonneau M. The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1–3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:3721–3734. doi: 10.1128/MCB.23.11.3721-3734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grandin N., Damon C., Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 2000;20:8397–8408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandin N., Bailly A., Charbonneau M. Activation of Mrc1, a mediator of the replication checkpoint, by telomere erosion. Biol. Cell. 2005;97:799–814. doi: 10.1042/BC20040526. [DOI] [PubMed] [Google Scholar]
- 40.Melo J., Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell. Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 41.Rouse J., Jackson S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 42.Kondo T., Wakayama T., Naiki T., Matsumoto K., Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 43.Melo J.A., Cohen J., Toczyski D.P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connell M.J., Walworth N., Carr A.M. The G2-phase DNA damage checkpoint. Trends Cell. Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 45.Zhou B.S., Elledge S.J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 46.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 47.Sun Z., Hsiao J., Fay D.S., Stern D.F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 48.Vialard J.E., Gilbert C.S., Green C.M., Lowndes N.F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paciotti V., Clerici M., Scotti M., Lucchini G., Longhese M.P. Characterization of mec1 kinase-deficient mutants and of new hypomorphic alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Binz S.K., Sheehan A.M, Wold M.S. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 51.Boschkarev A., Boschkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Carr A.M. Beginning at the end. Science. 2003;300:1512–1513. doi: 10.1126/science.1085689. [DOI] [PubMed] [Google Scholar]
- 53.Wolner B., van Komen S., Sung P., Peterson C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 54.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choregraphy of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 56.Le S., Moore J.K., Haber J.E., Greider C.W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans S.K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 58.Kim H.S., Brill S.J. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisisae. Mol. Cell. Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paciotti V., Clerici M., Lucchini G., Longhese M.P. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 60.Rouse J., Jackson S.P. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakayama T., Kondo T., Ando S., Matsumoto K., Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maringele L., Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toh G.W., Lowndes N.F. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 2003;31:242–246. doi: 10.1042/bst0310242. [DOI] [PubMed] [Google Scholar]
- 64.Pellicioli A., Foiani M. Signal transduction: how Rad53 kinase is activated. Curr. Biol. 2005;15:R769–R771. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 65.Lustig A.J. Cdc13 subcomplexes regulate multiple telomere functions. Nature Struct. Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 66.Downs J.A., Kosmidou E., Morgan A., Jackson S.P. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 67.Liti G., Louis E.J. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell. 2003;11:1373–1378. doi: 10.1016/s1097-2765(03)00177-1. [DOI] [PubMed] [Google Scholar]
- 68.Lowell J.E., Roughton A.I., Lundblad V., Pillus L. Telomerase-independent proliferation is influenced by cell type in Saccharomyces cerevisiae. Genetics. 2003;164:909–921. doi: 10.1093/genetics/164.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie K.B., Mallory J.C., Petes T.D. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aylon Y., Kupiec M. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol. 2003;23:6585–6596. doi: 10.1128/MCB.23.18.6585-6596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Q., Iijpma A., Greider C.W. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lydall D., Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 73.Jia X., Weinert T., Lydall D. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;166:753–764. doi: 10.1534/genetics.166.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bessho T., Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J. Biol. Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 75.Naiki T., Wakayama T., Nakada D., Matsumoto K., Sugimoto K. Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol. Cell. Biol. 2004;24:3277–3285. doi: 10.1128/MCB.24.8.3277-3285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du L.L., Nakamura T.M., Moser B.A., Russell P. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 2003;23:6150–6158. doi: 10.1128/MCB.23.17.6150-6158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Natarajan S., McEachern M.J. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsukamoto M., Yamashita K., Miyazaki T., Shinohara M., Shinohara A. The N-terminal domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics. 2003;165:1703–1715. doi: 10.1093/genetics/165.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caspari T., Murray J.M., Carr A.M. Cdc2-cyclin B kinase activity links Crb2 and Rqh1 topoisomerase III. Genes Dev. 2002;16:1195–1208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clerici M., Baldo V., Mantiero D., Lottersberger F., Lucchini G., Longhese M.P. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 2004;24:10126–10144. doi: 10.1128/MCB.24.23.10126-10144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cross F.R., Schroeder L., Kruse M., Chen K.C. Quantitative characterization of a mitotic cyclin threshold regulating exit from mitosis. Mol. Biol. Cell. 2005;16:2129–2138. doi: 10.1091/mbc.E04-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kantake N., Sugiyama T., Kolodner R.D., Kowalczykowski S.C. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 2003;278:23410–23417. doi: 10.1074/jbc.M302995200. [DOI] [PubMed] [Google Scholar]