Abstract
Vertebrate immune system molecules that bind directly to parasites are commonly subject to strong directional natural selection, probably because they are engaged in an evolutionary arms race with parasites. We have investigated whether similar patterns of evolution are seen in components of the Drosophila immune system that bind parasite-derived molecules. In insects, TEPs (thioester-containing proteins) function as opsonins, binding to parasites and promoting their phagocytosis or encapsulation. The Drosophila melanogaster genome encodes four TEPs, three of which are upregulated after an immune challenge. We report that two of these three Drosophila genes evolve rapidly under positive selection and that, in both TepI and TepII, the “bait-like region” (also known as the variable region) shows the strongest signature of positive selection. This region may be the site of proteolytic cleavage that leads to the activation of the molecule. It is possible that the proteolytic activation of TEPs is a target of host-parasite coevolution, with parasites evolving to prevent proteolysis, which in turn favors mutations in the bait-like region that restore the response. We also sequenced three gram-negative binding proteins (GNBPs) and two immune-induced peptides with strong homology to the GNBPs. In contrast to the Tep genes, the GNBP genes are highly conserved. We discuss the reasons why different components of the immune system have such different patterns of evolution.
Keywords: Drosophila, Gram-negative binding protein, Thioester-containing protein, Immunity
Introduction
Vertebrate immune system receptors such as MHC molecules and immunoglobulins are very specific in the parasite-derived epitopes that they bind to. Furthermore, different alleles and copies of the MHC and immunoglobulin genes encode proteins that bind to a diverse range of different epitopes. This diversity of recognition molecules has arisen through positive Darwinian selection, as the regions of these genes that determine the binding specificity of the receptors have an excess of nonsynonymous relative to synonymous mutations (Hughes and Nei 1988; Tanaka and Nei 1989). This is believed to result from either overdominant or frequency-dependent selection enhancing the diversity of receptor specificities in the population.
The innate immune system is another important component of vertebrate immunity and is the only immune response available to invertebrates. Innate immunity relies entirely on germline-encoded receptors, unlike the adaptive immune system, which generates an enormous repertoire of receptors through somatic mutation and recombination. Despite this limitation, the innate immune response can recognise and eliminate a broad array of pathogens and parasites. This is thought to be the result of pathogens being recognized by highly conserved “pathogen-associated molecular patterns” and then eliminated using effector molecules that act on other highly conserved targets (Medzhitov and Janeway 1997). For this reason, innate immune system receptors are believed to recognise a far lower diversity of molecules than MHC molecules or immunoglobulins.
It is therefore of interest to compare the molecular evolution of innate immune system molecules that bind directly to pathogens with the patterns observed in MHC and immunoglobulin molecules. It is possible that similar selective forces act on both classes of molecules, and positive selection may act to diversify or change the binding specificity of innate immune system receptors. However, if innate immune system molecules recognize highly conserved pathogen targets, there may be little or no selection to either change or diversify their specificities.
Previous studies on the molecular evolution of two families of Drosophila immune receptors have produced contrasting results. The first of these protein families are the peptidoglycan recognition proteins (PGRPs), some of which bind to pathogens and initiate the production of antimicrobial peptides. These proteins were found to evolve slowly under predominantly purifying selection (Jiggins and Hurst 2003). In contrast, several scavenger receptors, which bind to pathogens and play a role in their phagocytosis, evolve rapidly under positive selection (Lazzaro 2005). In this study we investigated two further classes of immune receptors that are believed to bind directly to pathogens, the thioester-containing proteins (TEPs) and the gram-negative bacteria-binding proteins (GNBPs). This will provide a fairly complete picture of the molecular evolution of Drosophila proteins thought to bind to the surface pathogens and elicit an immune response.
The first family of genes we studied are the TEPs (Blandin and Levashina 2004). In vertebrates, this family includes two key components of the immune system, the α2-macroglobulins and complement factors C3, C4, and C5. The α2-macroglobulins are protease inhibitors. They are cleaved by proteases released by pathogens, resulting in a conformational change in the α2-macroglobulin that entraps the protease, inhibiting its action and ultimately leading to its endocytosis. The complement factors C3 and C4 are also activated by proteolytic cleavage to expose their reactive thioester bond. In this case, however, the activating proteases are the host-derived convertase complex. The larger cleavage product then acts as an opsonin, covalently binding to the pathogen and promoting phagocytosis. The proteolytic cleavage also produces a smaller fragment (anaphylatoxin) whose functions include the attraction of marcrophages.
TEPs are also an important component of insect immune systems (Blandin and Levashina 2004). The most detailed functional studies of insect TEPs have been on aTepI in Anopheles mosquitoes. Following infection with bacteria, the aTepI gene is upregulated and its protein product proteolytically cleaved (Blandin and Levashina 2004). It then binds to the bacteria through the thioester bond and functions as an opsonin, promoting the phagocytosis of the pathogen. aTepI also binds to Plasmodium parasites, and knocking down the expression of the gene by RNAi prevents melanization of the parasites (Blandin et al. 2004). Intriguingly, the sequence of the 280-amino acid-long C3d domain of aTepI is very variable, and it has been postulated that this may be responsible for variation in the ability of mosquitoes to transmit malaria (Blandin et al. 2004).
The genome of Drosophila melanogaster contains four Tep genes, three of which (TepI, TepII, and TepIV) are strongly upregulated following immune challenge (Lagueux et al. 2000). The function of these genes has been investigated using RNAi in cell culture, and it was found that TepII and TepIII are required for the efficient phagocytosis of E. coli and Staphylococcus aureus, respectively (Stroschein-Stevenson et al. 2006). A gene called Mcr, which is related to the Tep genes but lacks the thioester motif, was required for phagocytosis of the fungal pathogen Candida albicans (Stroschein-Stevenson et al. 2006). Therefore, different members of this protein family target different pathogens and promote their phagocytosis. None of the Drosophila Tep genes have 1:1 orthologues in the Anopheles genome, and the three immune-upregulated genes have probably arisen by gene duplication in the Drosophila lineage (Christophides et al. 2002).
The hypervariable or bait-like region lies near the center of the Tep coding sequence in D. melanogaster. The corresponding region in vertebrate TEPs encodes the bait region of α2-macroglobulins and the anaphylatoxin fragment of complement protein C3 (Lagueux et al. 2000). In α2-macroglobulins, interspecific sequence variation in this region causes changes in the range of proteases that cleave the α2-macroglobulin (Sottrup-Jensen et al. 1989). In D. melanogaster, the amino acid sequence of this region is poorly conserved in comparisons both with TEPs in other animals and between paralogous Tep genes in the genome (Lagueux et al. 2000). Furthermore, alternative splicing of the TepII transcript can result in proteins with five different bait-like regions (Lagueux et al. 2000), suggesting that sequence variation in this region is functionally important. In the crustacean Daphnia, this region of a Tep gene evolves rapidly under positive selection (Little et al. 2004). Therefore, the bait-like region is a candidate target of host-parasite coevolution.
The second family of proteins that we investigated is the GNBPs. These proteins have sequence similarities to bacterial glucanases, and probably represent a case of either horizontal gene transfer or convergent evolution (Ferrandon et al. 2004; Lee et al. 1996). Although they do not show enzymatic activity, various GNBPs are able to bind to fungal β-1,3-glucans, bacterial lipopolysaccharides, and/or bacterial lipoteichoic acid (Dimopoulos et al. 1997; Kim et al. 2000). Two Drosophila GNBPs have been shown to function as pattern recognition molecules. GNBP1 together with another pattern-recognition molecule, called PGRP-SA, is required to activate the Toll pathway in response to infection by gram-positive bacteria (Gobert et al. 2003). The Toll pathway leads to the production of antimicrobial peptides, and loss-of-function mutants in GNBP1 are very susceptible to infection by gram-positive bacteria. It has also been reported that loss-of-function mutants in another gene, GNBP3, are sensitive to fungal infection, although the primary data to support this have yet to be published (Ferrandon et al. 2004).
The Drosophila genome contains three full-length GNBP genes (GNBP1, GNBP2, and GNBP3), none of which is upregulated following infection (De Gregorio et al. 2001; Irving et al. 2001). There are also two genes (CG13422 and CG12780) that are very similar to the N-terminal part of the GNBPs and are upregulated following bacterial infection (De Gregorio et al. 2001; Irving et al. 2001). One of these (CG13422) is also upregulated following fungal infection (De Gregorio et al. 2001).
In this study we have tested whether natural selection drives causes rapid evolution of these proteins. Polymorphism data from Drosophila simulans for GNBP1 and part of TepI have previously been collected by Schlenke and Begun (2003). Neither of these genes showed individual departures from neutrality (Schlenke 2003, Supplementary Material). In this paper we present a larger and more comprehensive dataset on these two gene families from the closely related species D. melanogaster.
Methods
Isofemale D. melanogaster lines that had originally been collected in the Netherlands or Gabon by Peter Andolfatto and Bill Ballard were supplied by Penny Haddrill. The appropriate chromosomes were made isogenic using standard crosses to balancer stocks (SM1 and TM6). Analysis of this preliminary dataset showed that the bait-like region of TepII evolved extremely rapidly but did not show unequivocal evidence of positive selection. Therefore, we increased our dataset for this gene by sequencing eight alleles of from Kenyan isofemale lines of D. simulans that had been inbred by sib mating for nine generations.
Population genetic analyses can be confounded by the presence of chromosomal inversions in the population because they suppress recombination. Therefore, inversions may introduce strong haplotype structure into the dataset. Furthermore, it is well-known that genes within inversions are often under strong selection and so selection, even on loci far from the gene of interest, may alter patterns of polymorphism in the target gene (Powell 1997). For this reason we only used inversion-free chromosomes for sequencing. These were identified by crossing the isogenic chromosomes to an inversion-free stock and checking the salivary gland chromosomes for the presence of inversion loops.
All of the Tep genes are on chromosome arm 2L. In the sample from Gabon, 8 of 21 2L chromosome arms had inversions, and 10 inversion-free lines were retained for sequencing. GNBP1, GNBP2, and GNBP3 are all on chromosome arm 3L. Unfortunately, we were unable to obtain a sufficient number of homozygous lines for this chromosome and therefore sequenced the GNBP genes from the Netherlands lines. In the Netherlands lines, all 16 of the 3L chromosome arms inspected were inversion-free, and 12 of these were retained for sequencing. The two shorter GNBPs (CG13422 and CG12780) are on chromosome arm 2R. Of 14 Netherlands 2R chromosome arms inspected, 13 were inversion-free. D. melanogaster is thought to have originated in Africa and passed through a population bottleneck during the colonization of Europe (David and Capy 1988). This should not affect patterns of interspecific divergence. The out-of-Africa range expansion had little effect on autosomal genetic diversity (Andolfatto 2001), but care should nonetheless be taken when comparing the European GNBP and African Tep polymorphism data.
We sequenced 12 alleles of all five GNBP genes and 10 alleles of the three Tep genes that are upregulated following infection (TepI, TepII, and TepIV). We sequenced the entire coding region and introns of the five GNBP genes. The Tep genes are longer than the GNBP genes, so we only sequenced the regions shown in Fig. 1. Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ973199-AJ973208, AJ973615-AJ973634, and AM050187-AM050254. We also analyzed sequences of the Tep genes obtained by Blast searching the annotated genome sequences of several Drosophila species (Smith 2004; Wilson 2004). The genome assemblies used were D. melanogaster Flybase release 4.0, D. pseudobscura Flybase release1.0, D. simulans Langley Group assembly 29/9/2004, D. yakuba Langley Group assembly 22/5/2004, D. mojavensis Agencourt Bioscience Corporation assembly 6/12/2004, D. virilis Agencourt assembly 29/10/2004, D. erecta Agencourt assembly 28/10/2004, and D. annanasae Agencourt assembly 6/12/2004. We checked that all the sequences of a given gene were reciprocal best tBLASTn hits (Altschul et al. 1997). We also aligned all the inferred amino acid sequences of the Tep genes (excluding the most variable regions) and reconstructed a neighbor-joining tree (data not shown). In all cases, the sequences of each Tep gene from different species formed a monophyletic group. Furthermore, the relationships within each of these clades were the same as the known phylogeny of different species of flies. All analyses were based on ClustalW alignments of the nucleotide sequence that were corrected by eye to account for the amino acid sequence.
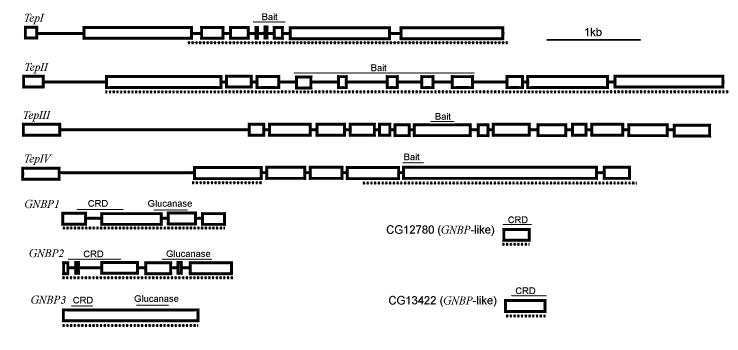
Fig. 1.
The arrangement of Tep and GNBP introns and exons. The region sequenced is indicated by a dashed line below each gene. The location of the bait-like region, CRD-like domain (carbohydrate recognition domain), and glucanase-homology domain are marked above.
We compared the Ka/Ks ratio of the Tep and GNBP genes with genome-wide estimates of Ka/Ks between D. simulans and D. melanogaster. The genes were selected for analysis if all exons were identifiable as unique best reciprocal BLAST hits between known D. melanogaster genes and the April 2005 release of the D. simulans genome. Those in which the D. simulans data were not valid coding sequence were rejected. Codeml (PAML) (Yang 1997) was used to provide maximum-likelihood estimates of Ka/Ks for all genes (runmode = -2). Because the Ka/Ks ratio has both a higher variance and a higher mean in short genes, we only included genes that had coding sequences of 1 kb or more, resulting in a final set of 4558 genes.
Functionally important regions of the genes are shown in Fig. 1. The location of the bait-like region (also known as the variable region) was taken from Fig. 1 of Lagueux et al. (2000) when it fell at exon boundaries. When this was not the case, the end of the bait-like region was defined as the end of Block D (Lagueux et al. 2000). The starts of the bait-like regions within exons were identified by aligning the amino acid sequence of the four genes. The C3d-like domain was predicted by aligning the amino acid sequences of the Drosophila genes with the C3d-like region of aTepI of Anopheles (Blandin et al. 2004).
The full-length GNBP transcripts encode a signal peptide followed by a carbohydrate-recognition domain, a link region, the glucanase-homology domain, and, finally, a C-terminal section (Fabrick et al. 2004) (Fig. 1). The signal peptides were predicted using the program SignalP 3.0 (Bendtsen et al. 2004). The location of the carbohydrate recognition domain (CRD) was predicted by aligning the amino acid sequence with this domain from Bombyx mori, where it has been identified experimentally (Ochiai and Ashida 2000). The glucanase homology domain was taken from Kim et al. (2000).
We tested for heterogeneity in the polymorphism-to-divergence ratio across the gene sequences using the program DNAslider (McDonald 1996, 1998). This method first classifies variable sites into intraspecific polymorphisms or interspecific fixed differences. Following the recommendation of McDonald (1998), we used the three different statistics (the Kolmogorov-Smirnov statistic, the number of runs, and the mean sliding G) for detecting heterogeneity, as each is most powerful in different situations. The significance of these statistics was assessed by generating 1000 replicate datasets by coalescent simulation. These simulations were conditioned on the recombination rate R. In D. simulans and D. melanogaster there is no recombination in males, and therefore for autosomal genes R=2Nc (where N is the effective population size and c the crossing-over rate/bp/generation in females). We assumed that in D. simulans N = 2 × 106 and in D. melanogaster N = 106 (Andolfatto and Przeworski 2000). We used the estimate of c in D. melanogaster obtained by Marais et al. (2003) using the polynomial method of Hey and Kliman (2002). We conservatively assumed the same value of c for D. simulans. The resulting estimates of R in D. melanogaster were 0.02826 for TepI, 0.09006 for TepII, 0.00204 for TepIV, 0.07066 for GNBP3, 0.01266 for GNBP2, 0.01266 for GNBP1, 0.02472 for CG12780, and 0.07726 for CG13422.
The statistical significance of the rate of nonsynonymous substitutions being higher than the rate of synonymous substitutions (Ka>Ks) was estimated by simulating 50,000 replicate datasets based on the Comeron (1995) model of nucleotide substitution using the program K estimator. The maximum likelihood analysis of Ka/Ks ratios was performed using the program PAML (Yang 1997). This analysis was based on published phylogenies of these species (((((melanogaster,simulans),(yakuba,erecta)),ananassae),pseudoobscura),(mojavensis,virilis)) (Ko et al. 2003; Powell 1997). Most population genetic analyzes were implemented with the program DNAsp (Rozas and Rozas 1999). The null distributions of statistics describing the frequency distribution of mutations were obtained from 1000 coalescent simulations conditioned on θ and the recombination rates described above.
Results
Tep Interspecific Divergence
TepI is one of the most rapidly evolving genes in the Drosophila genome. The Ka/Ks ratio between the D. melanogaster and the D. simulans TepI sequences was 0.71, which is among the highest 0.5% of Drosophila genes over 1 kb long. TepII has a Ka/Ks ratio of 0.23, which is in the top 13th percentile of our genomic sample. TepIV evolves slightly slower (Ka/Ks=0.17) and falls in the highest 21st percentile. TepIII evolves slower than the genome average (Ka/Ks=0.06; 60th percentile).
TepI also evolves rapidly compared to other immune-related genes. In a sample of 64 immunity genes of all lengths, only an antimicrobial peptide expressed in the male reproductive tract, called andropin, has a higher Ka/Ks ratio. In this list, the Ka/Ks ratios of TepII, TepI, and TepIII had the 13th, 24th, and 49th highest Ka/Ks ratios, respectively.
The differences in the Ka/Ks ratios both among the four Tep genes and between the Tep genes and the rest of the genome are largely accounted for by differences in the nonsynonymous substitution rate (Ka; Table 1). In contrast, the divergence at synonymous sites (Ks) is less heterogeneous across genes and gene regions (Table 1). Furthermore, these estimates of Ks are similar to the genome average in our comparison of D. simulans and D. melanogaster (unweighted mean in our dataset of 4558 genes, Ks=0.12). Therefore, the high nonsynonymous substitution rate is not due to increased mutation rates in the Tep genes but results from either positive selection or low selective constraints on the protein sequence.
Table 1.
Estimated number of nonsynonymous (Ka) and synonymous (Ks) substitutions per site between the D. melanogaster and D. simulans Tep and GNBP genes
| Gene | Region | No. codons | K a | K s | Ka/Ks | p (Ka/Ks > 1) |
|---|---|---|---|---|---|---|
| TepI | Bait | 62 | 0.160 | 0.035 | 4.57 | <0.001 |
| Nonbait | 1299 | 0.073 | 0.109 | 0.67 | ||
| TepII | Bait | 263 | 0.086 | 0.085 | 1.01 | n.s. |
| Nonbait | 1346 | 0.028 | 0.130 | 0.22 | ||
| TepIII | Bait | 72 | 0.000 | 0.110 | 0.00 | |
| Nonbait | 1360 | 0.007 | 0.118 | 0.06 | ||
| TepIV | Bait | 52 | 0.045 | 0.148 | 0.30 | |
| Nonbait | 1406 | 0.016 | 0.080 | 0.20 | ||
| CG12780 | CRD | 300 | 0.032 | 0.145 | 0.22 | |
| CG13422 | CRD | 387 | 0.015 | 0.155 | 0.10 | |
| Other | 69 | 0.087 | 0.048 | 1.81 | n.s. | |
| GNBP1 | CRD | 312 | 0.004 | 0.100 | 0.04 | |
| Glucanase | 351 | 0.008 | 0.127 | 0.06 | ||
| Other | 807 | 0.012 | 0.131 | 0.09 | ||
| GNBP2 | CRD | 288 | 0.015 | 0.194 | 0.08 | |
| Glucanase | 363 | 0.003 | 0.184 | 0.02 | ||
| Other | 732 | 0.021 | 0.129 | 0.16 | ||
| GNBP3 | CRD | 312 | 0.009 | 0.041 | 0.22 | |
| Glucanase | 351 | 0.004 | 0.129 | 0.03 | ||
| Other | 807 | 0.016 | 0.114 | 0.14 |
Note. CRD, carbohydrate recognition domain; glucanase, glucanase homology domain. The “nonbait” region includes the entire protein outside the bait region. The genetic distances and probability that Ka/Ks > 1 were estimated following Comeron (1995).
As discussed in the Introduction, we had an a priori prediction that the bait-like region might be the target of antagonistic coevolution with parasites. In all three immune-upregulated genes, this region has a higher nonsynonymous substitution rate than the rest of the gene, while in TepIII it is highly conserved (Table 1). In TepI, Ka is significantly higher than Ks in the bait-like region (p<0.001; Table 1), and this result holds after correction for multiple tests (p<0.008). This provides strong evidence that positive selection has acted on this region during the divergence of these species. The TepII bait-like region has Ka/Ks=1, which suggests either that this region evolves neutrally or that some sites are under positive selection.
It is possible that positive selection is acting on parts of the gene other than the bait-like region, so we also conducted a sliding-window analysis of the Ka/Ks ratio along the length of the genes (Fig. 2). This confirms that the bait-like regions of TepI and TepII have the highest Ka/Ks ratios. In the Anopheles gambiae aTepI gene, the C3d-like domain is very polymorphic (Blandin et al. 2004). This domain surrounds the thioester active site, and it has been suggested that these polymorphisms may have important effects on the binding properties of the protein. We identified the homologous region in the Drosophila genes by aligning the protein sequences with the Anopheles aTepI sequence (Blandin et al. 2004; Levashina et al. 2001). In none of the Drosophila genes did this region have an accelerated rate of protein evolution, suggesting that this is not a target of positive selection (Fig. 2).
Fig. 2.
Sliding window analysis of the Ka/Ks ratio along the entire coding sequence of the four Tep genes in D. simulans and D. melanogaster. The bait-like and C3d-like regions are marked. Window size = 100 bp; step size = 1 bp; Ka/Ks ratio estimated following Nei and Gojobori (1986).
The second approach that we used to estimate the Ka/Ks ratio was to align sequences from multiple species and fit a maximum likelihood model of codon substitution along the phylogenetic tree of those species (Nielsen and Yang 1998). This analysis used unpublished data from the genome projects and there may be some errors in the sequences. We have assumed that any such errors will be equally likely at synonymous and nonsynonymous sites and will, therefore, not result in Ka being significantly higher than Ks. Different species were included for the different genes either because it was impossible to align the most variable regions of the sequences from distant relatives or because homologues could not be identified reliably in some species (i.e., there were no reciprocal best blast hits). To test whether the protein sequence of the different species has diverged under positive selection, we compared models of codon substitution where a proportion of sites was allowed to have Ka/Ks >1 (Model M8) with models where all codons had Ka/Ks ≤ 1 (Model M8A) (Swanson et al. 2003). In model M8A, the codons fell into either one of eight Ka/Ks categories that followed a beta distribution bounded between 0 and 1 or a ninth category where Ka/Ks=1. Model M8 is identical except that the ninth category is free to vary above 1 (i.e., positive selection is allowed). In both TepI and TepII, model M8A was the better fit to the data (Table 2). Therefore, this analysis suggests that both of these genes are subject to positive selection.
Table 2.
Maximum likelihood test for codons with Ka/Ks> 1
| Gene | Speciesa | Model | No. codons | Ka/Ksb | Proportion of sitesb | Likelihood | 2Δl | p |
|---|---|---|---|---|---|---|---|---|
| TepI | msye | M8A | 1,361 | 1 | 0.46 | -10,946.50 | ||
| M8 | 1,361 | 188.81 | 0.005 | -10,942.62 | 7.76 | <0.01 | ||
| TepII c | msye | M8A | 1,529 | 1 | 0.23 | -10,106.94 | ||
| M8 | 1,529 | 2.05 | 0.09 | -10,101.92 | 10.04 | <0.005 | ||
| TepIII | msyeapvo | M8A | 1,432 | 1 | 0.01 | -16,464.20 | ||
| M8 | 1,432 | 1.00 | 0.01 | -16,464.20 | 0 | 1 | ||
| TepIV | msyeapv | M8A | 1,458 | 1 | 0.04 | -16,166.77 | ||
| M8 | 1,458 | 1.18 | 0.03 | -16,166.78 | 0 | 1 |
m, melanogaster; s, simulans; y, yakuba; e, erecta; a, annanasae; p, pseudoobscura; v, virilise; o, mojavensi.;
Refers to the Ka/Ks category of sites in addition to β distribution (see text).
Excludes exon 9, which is too divergent to align.
Tep McDonald-Kreitman Test
An alternative approach to test whether the high Ka of TepI and TepII is the result of positive selection is to compare polymorphism and divergence at synonymous and nonsynonymous sites. Under the neutral model, the ratio of synonymous:nonsynonymous polymorphic sites will be the same as the ratio of synonymous:nonsynonymous interspecific differences. The McDonald-Kreitman (1991) test simply compares these two ratios in a 2×2 contingency table. These ratios are significantly different in TepI (Table 3). This is the result of the large number of nonsynonymous substitutions that have occurred since D. simulans diverged from D. melanogaster (Table 1), as the synonymous divergence (Table 1) and hS/hR ratio (see below) of TepI are similar to those of the other two genes. This suggests that the significant McDonald-Kreitman test is caused by positive selection favoring changes to the TEPI amino acid sequence during the divergence of the two species. Interestingly, this test is significant even when the bait-like region is excluded from the analysis (Table 3), indicating that positive selection is not solely confined to this region of TepI. Using these data, it is possible to estimate that 71% of the amino acid substitutions that have occurred were fixed by positive selection (Smith and Eyre-Walker 2002). This equates to positive selection fixing 121 nonsynonymous substitutions in the ∼2.5 million years since these species diverged (Powell 1997). The McDonald-Kreitman test was not significant for TepII or TepIV.
Table 3.
McDonald-Kreitman test on the Tep genes
| Fixed differences |
Polymorphism, melanogaster |
Polymorphism, simulans |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Region | Silent | Replace | Silent | Replace | p a | Silent | Replace | p a |
| TepI | Coding (bait) | 2 | 18 | 0 | 0 | - | |||
| Coding (nonbait) | 70 | 153 | 10 | 7 | 0.03 | ||||
| All coding | 72 | 171 | 10 | 7 | 0.03 | ||||
| Intron + coding | 129 | 171 | 18 | 7 | 0.006 | ||||
| TepII | Coding (bait exon 5) | 3 | 3 | 0 | 0 | n.s. | 2 | 2 | n.s. |
| Coding (bait exon 6) | 0 | 2 | 2 | 0 | n.s. | 1 | 1 | n.s. | |
| Coding (bait exon 7) | 3 | 9 | 1 | 0 | n.s. | 0 | 4 | n.s. | |
| Coding (bait exon 8) | 2 | 5 | 0 | 0 | n.s. | 0 | 2 | n.s. | |
| Coding (bait exon 9) | 9 | 26 | 1 | 3 | n.s. | 0 | 6 | n.s. | |
| Coding (bait all exons) | 17 | 45 | 4 | 3 | n.s. | 3 | 15 | n.s. | |
| Coding (nonbait) | 105 | 61 | 48 | 26 | n.s. | 57 | 37 | n.s. | |
| All coding | 122 | 106 | 52 | 29 | n.s. | 60 | 52 | n.s. | |
| Intron + coding | 265 | 106 | 102 | 29 | n.s. | 129 | 52 | n.s. | |
| TepIV | Coding (bait) | 4 | 6 | 1 | 1 | n.s. | |||
| Coding (nonbait) | 59 | 29 | 14 | 14 | n.s. | ||||
| All coding | 63 | 35 | 15 | 15 | n.s. | ||||
| Intron + coding | 76 | 35 | 20 | 15 | n.s. | ||||
Two-tailed Fisher exact test. Dataset includes a single additional allele from the genome sequences of D. simulans and D. melanogaster.
Tep Polymorphism
The above analyses provide strong evidence that positive selection has acted on TepI. They also suggest that TepII may have evolved under positive selection. If this has been the result of directional selection causing recurrent selective sweeps, then the genetic diversity of these genes may have been reduced. Alternatively, some models of host-parasite coevolution predict that frequency dependent or diversifying selection may act on immune system molecules (Barrett 1988), which may result in increased genetic diversity at linked sites.
The synonymous site heterozygosity (Table 4) is similar to that of other genes in these species. For example, six other genes sampled from the same population had mean heterozygosities of πs=0.013 in D. melanogaster and πs=0.033 in D. simulans (Dro1-6 [Jiggins and Kim 2005]). These estimates are similar to those reported for larger datasets from different populations by both Moriyama and Powell (1996) and Andolfatto (2001). The three genes have fairly high levels of nonsynonymous polymorphism relative to synonymous polymorphism (Table 4). This is most marked in D. simulans, where πs/πa = 0.67 in the bait region and πs/πa = 4.42 in the rest of the gene. This compares to a mean πs/πa = 10.2 across six other genes in this population (Dro1-6 [Jiggins and Kim 2005]; similar estimates were obtained by Andolfatto (2001).
Table 4.
Nucleotide diversity of the Tep and GNBP genes
| Gene | Species | Region | Length (bp) | na | Sb | π × 102 | πa×102 | πs×102 | πs/πa | θd×102 |
|---|---|---|---|---|---|---|---|---|---|---|
| TepI | mele | Coding (bait) | 186 | 10 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Coding (not bait) | 2671 | 10 | 17 | 0.21 | 0.11 | 0.53 | 4.82 | 0.23 | ||
| Intron | 487 | 10 | 10 | 0.50 | 0.73 | |||||
| TepII | mele | Coding (bait) | 786 | 10 | 7 | 0.22 | 0.15 | 0.43 | 2.87 | 0.32 |
| Coding (not bait) | 4050 | 10 | 74 | 0.69 | 0.33 | 1.86 | 5.64 | 0.66 | ||
| Intron | 2045 | 10 | 58 | 1.00 | 1.00 | |||||
| TepII | simf | Coding (bait) | 753 | 8 | 18 | 0.82 | 0.89 | 0.60 | 0.67 | 0.92 |
| Coding (not bait) | 3958 | 8 | 94 | 0.91 | 0.50 | 2.21 | 4.42 | 0.92 | ||
| Intron | 1762 | 8 | 75 | 1.56 | 1.64 | |||||
| TepIV | mele | Coding (bait) | 186 | 10 | 2 | 0.23 | 0.16 | 0.47 | 2.94 | 0.41 |
| Coding (not bait) | 3288 | 10 | 28 | 0.31 | 0.19 | 0.67 | 3.53 | 0.30 | ||
| Intron | 288 | 10 | 6 | 0.82 | 0.74 | |||||
| GNBP1 | mele | Coding | 1476 | 12 | 17 | 0.42 | 0.13 | 1.36 | 10.46 | 0.38 |
| Intron | 292 | 12 | 6 | 0.62 | 0.68 | |||||
| GNBP2 | mele | Coding | 1383 | 12 | 16 | 0.44 | 0.04 | 1.86 | 46.50 | 0.38 |
| Intron | 503 | 12 | 11 | 1.02 | 0.84 | |||||
| GNBP3 | mele | Coding | 1470 | 12 | 24 | 0.55 | 0.18 | 1.73 | 9.61 | 0.54 |
| CG12780 | mele | Coding | 300 | 12 | 4 | 0.37 | 0.19 | 0.92 | 4.84 | 0.44 |
| CG13422 | mele | Coding | 456 | 12 | 6 | 0.58 | 0.15 | 1.91 | 12.73 | 0.44 |
Analysis excludes positions with alignment gaps.
Number of alleles.
Number of segregating sites.
Watterson (1975) estimate of 4Neµ.
D. melanogaster.
D. simulans.
The neutral model of molecular evolution predicts that polymorphism and divergence will be positively correlated across different genes or regions of genes. We did not formally compare levels of polymorphism and divergence among the three Tep genes, as they are found in regions of the genome with different rates of recombination (this can affect the nucleotide diversity [Begun and Aquadro 1992]). However, we did look for variation within each of the genes, and in TepII the polymorphism-to-divergence ratio was significantly heterogeneous using both the D. simulans and the D. melanogaster datasets (D. melanogaster, mean G=15.4 [p<0.001], number of runs=183 [p=0.006], K-S=0.03 [p=0.03]; D. simulans, mean G=6.49 [p<0.03], number of runs=235 [p=0.22], K-S=0.04 [p=0.001]). There was no significant heterogeneity in either TepI or TepIV.
Selection and demography can also alter the frequency of segregating sites within a population. We calculated Tajima’s (1989) D, which reflects the frequency distribution of polymorphisms in the population, and Fay and Wu’s (2000) H, which is a measure of the frequency of derived polymorphisms (Table 5). Of these statistics, only Fay and Wu’s H for TepI was marginally significantly different from the null distribution generated by coalescent simulations conditioned on θ.
Table 5.
Summary statistics of the frequency spectrum of segregating sites of the Tep and GNBP genes for D. melanogaster (mel) and D. simulans (sim)
| No. sites |
Tajima’s D |
Fay & Wu’s H |
||||
|---|---|---|---|---|---|---|
| Gene | mel | sim | mel | sim | mel | sim |
| TepI | 3347 | -0.818 | -5.600 a | |||
| TepII | 6807 | 6473 | 0.011 | -0.213 | -2.667 | 4.21 |
| TepIV | 3750 | 0.028 | 0.800 | |||
| GNBP1 | 1768 | 0.25 | 1.48 | |||
| GNBP2 | 1886 | 0.81 | -1.97 | |||
| GNBP3 | 1470 | 0.08 | -2.73 | |||
| CG12780 | 300 | -0.54 | 0.09 | |||
| CG13422 | 456 | 1.25 | -0.55 | |||
p<0.05. Analysis includes coding sequence and introns but excludes positions with alignment gaps.
GNBP Interspecific Divergence
The ratio of nonsynonymous-to-synonymous substitutions (Ka/Ks) between the D. melanogaster and the D. simulans GNBP genes is similar to the genome average. In our sample of 4558 genes over 1 kb long, the GNBP1, GNBP2, and GNBP3 Ka/Ks ratios fall in the 48th, 53rd, and 61st percentiles, respectively. In our sample of 64 immunity genes, the Ka/Ks ratios of CG12780, CG13422, GNBP1, GNBP2, and GNBP3 had the 16th, 28th, 37th, 41st, and 47th highest Ka/Ks ratios, respectively.
The highest amino acid divergence in the GNBP proteins is seen in the signal peptides (data not shown), which are cleaved from the mature proteins and probably never interact directly with pathogens. Therefore, this divergence is unlikely to be driven by parasite-induced positive selection. The carbohydrate-recognition and glucanase-homology domains are candidate sites of host-parasite coevolution, as they have been found to bind directly to various parasite-associated polysaccharides in the GNBPs of other insects (Fabrick et al. 2004). However, both these domains are highly conserved (Table 1). Furthermore, sliding-window analyzes along the gene did not reveal any high peaks of Ka/Ks (data not shown). We also repeated the maximum likelihood test for positive selection described above for the Tep genes. In none of the five GNBP genes did allowing a class of positively selected sites increase the likelihood of the model (data not shown).
GNBP Polymorphism
The nucleotide diversity of the GNBP genes is typical of that observed for D. melanogaster genes (Table 4). The level of polymorphism at nonsynonymous sites relative to synonymous sites is lower than was the case for the Tep genes and more typical of other D. melanogaster genes (Table 4). Summary statistics based on the frequency spectrum of polymorphisms are close to the neutral expectation, and none differed significantly from null distributions generated by coalescent simulations conditioned on θ (Table 5).
The McDonald-Kreitman test also failed to give any evidence of positive selection acting on the GNBP genes (Table 6). We first conducted the test on each gene separately, and in all cases the synonymous:nonsynonymous ratio was the same for fixed differences between species and for polymorphic sites within species. We repeated the test separately for each of the different functional domains, summing the data across all five genes (Table 6). Again, the ratio did not differ significantly between intraspecific polymorphisms and interspecific divergence. There was significant heterogeneity in the polymorphism to divergence ratio within GNBP2 (mean G=6.2, p=0.01; number of runs=35, p=0.01; K-S=0.05, p=0.03). This test was not significant for any of the other four genes.
Table 6.
McDonald-Kreitman test on GNBP genes
| Fixed differences |
Polymorphic sites |
|||||
|---|---|---|---|---|---|---|
| Gene | Region | Synonymous | Nonsynonymous | Synonymous | Nonsynonymous | p |
| GNBP1 | All | 43 | 10 | 12 | 5 | n.s. |
| GNBP2 | All | 47 | 15 | 16 | 2 | n.s. |
| GNBP3 | All | 34 | 11 | 16 | 8 | n.s. |
| CG12780 | All | 9 | 7 | 3 | 1 | n.s. |
| CG13422 | All | 12 | 6 | 6 | 2 | n.s. |
| All | Signal | 2 | 13 | 0 | 3 | n.s. |
| All | CRD | 42 | 16 | 19 | 3 | n.s. |
| All | Link | 48 | 11 | 17 | 4 | n.s. |
| All | Glucanase | 39 | 4 | 7 | 2 | n.s. |
| All | end | 14 | 5 | 10 | 6 | n.s. |
| All | All | 145 | 49 | 53 | 18 | n.s. |
Note. Polymorphism in D. melanogaster; divergence from D. simulans.
Discussion
The Evolution of Tep and GNBP Genes
The Ka/Ks ratio of TepI is among the highest 0.5% of genes in the Drosophila genome. Furthermore, there is strong evidence that this rapid evolution was driven by positive selection. First, the bait-like region has a Ka/Ks ratio that is significantly >1. Second, a maximum likelihood analysis identified a proportion of codons as being positively selected. Finally, the McDonald-Kreitman test suggested that over 100 amino acid substitutions have been fixed by selection during the divergence of D. simulans and D. melanogaster. Therefore, it is likely that TEPI is continually adapting to novel parasite challenges.
Some immune-related genes in other species are highly polymorphic due to balancing selection maintaining variation. This is clearly not the case in TepI, which has slightly lower synonymous site diversity than is normal for other genes. However, TepI does not have a skewed frequency spectrum of polymorphisms or very low genetic diversity, as might be expected given that repeated selective sweeps have occurred in this gene. It is possible that the last selective sweep occurred sufficiently far in the past that the patterns of polymorphism have recovered to near the neutral equilibrium or that the small number of alleles means that analyses of the frequency spectrum of polymorphisms have little power (Ramos-Onsins and Rozas 2002; Simonsen et al. 1995). It is also possible that diversifying selection within or between populations is acting on TepI, which may have different effects on patterns of polymorphism to a simple selective sweep.
The extremely rapid evolution seen in TepI is not found in the other Tep genes. However, there is evidence that less intense positive selection is acting on TepII, although the data are less clear-cut than for TepI. In TepII, the maximum likelihood analysis strongly indicates that a proportion of codons is positively selected. Additionally, in the bait-like region Ka/Ks = 1, indicating either an absence of selective constraints or positive selection. However, despite its high nonsynonymous divergence, the McDonald-Kreitman test was not significant for this gene. This may result from a lack of statistical power or from diversifying selection increasing the level of nonsynonymous polymorphism. We found no evidence that TEPIV or TEPIII evolves under positive selection. Once functions of the different TEP molecules have been characterized, it may be possible to interpret the reasons that the different genes show different patterns of evolution.
We also conducted a similar set of analyses on the Drosophila GNBPs. These genes showed little or no evidence of adaptive evolution. The only significant deviation from the neutral model was a heterogeneous polymorphism:divergence ratio in GNBP2. Although this may result from positive selection, variation in the mutation rate, the recombination rate, the strength of selection on synonymous sites (e.g.,near splice sites), or the strength of background selection all could generate similar patterns (McDonald 1996, 1998). An unfortunate aspect of our data is that the GNBP and TEP datasets come from different populations (this was largely due to them being collected at different times), which makes it difficult to compare patterns of polymorphism directly. However, the differences we see are due to the higher interspecific divergence of the TEPs compared to the GNBPs. Only about 0.2% of the divergence between D. simulans and D. melanogaster has occurred since the split of European and African populations (David and Capy 1988; Lachaise et al. 1988). Therefore, it is unlikely that the differences we see between GNBP and TEP evolution are the result of population specific effects and it is safe to conclude that the two gene families show different patterns of molecular evolution.
Why Does Natural Selection Act Differently on Different Immune Genes?
Evolutionary analyses such as this on immunity genes in a variety of animals have produced diverse results. Some genes contain ancient balanced polymorphisms, others show evidence of recurrent selective sweeps, and many simply evolve slowly under predominantly purifying selection. It is therefore of interest why different genes evolve in such different ways.
First, it is of interest whether receptor molecules in the innate immune system evolve differently than those in the acquired immune system do. None of the genes in this study showed any evidence of selection maintaining multiple alleles for long periods of evolutionary time, which is in agreement with studies of other Drosophila immune genes (Begun and Whitley 2000; Clark and Wang 1997; Jiggins and Hurst 2003; Jiggins and Kim 2005; Lazzaro and Clark 2003; Schlenke and Begun 2003). Although the ancient polymorphisms found in MHC genes are well known, this pattern has not been replicated in other components of the vertebrate immune system. Therefore, it is possible that ancient balanced polymorphisms are a peculiarity of MHC evolution, rather than a general difference between innate and acquired immunity. Therefore, models of host-parasite coevolution that predict the maintenance of host alleles over long periods of evolutionary time are unlikely to be generally applicable in animals.
It is still possible that diversifying selection may act on some immune genes but not maintain polymorphisms for long periods. Consistent with this, unusually high nonsynonymous heterozygosities have been reported for some Drosophila scavenger receptors (Lazzaro 2005). Some of our Tep datasets also had fairly high levels of nonsynonymous relative to synonymous polymorphism. However, this may simply reflect low selective constraints.
In TepI and TepII, natural selection has recurrently fixed advantageous nonsynonymous mutations. This pattern has been repeatedly observed both in other innate immune system genes (Schlenke and Begun 2003) and in acquired immune system genes. This is consistent with the predominant mode of host-parasite coevolution being a simple arms race between hosts and parasites, where novel adaptations and counter adaptations arise and are fixed within populations. Furthermore, this form of coevolution does not appear to be restricted to highly specific acquired immune responses.
Second, another pattern to explain is why some Drosophila immunity genes evolve under positive selection while others do not. This study combined with previous work means that the molecular evolution of four different classes of molecules that bind to the surface of pathogens and illicit an immune response have been studied. The evolution of the TEPI and TEPII resembles patterns observed in Drosophila class C scavenger receptors (SR-Cs), which also evolve rapidly under positive selection (Lazzaro 2005). SR-Cs, like TEPs, bind to pathogens and are involved in their phagocytosis. The evolution of GNBPs resembles that of another class of recognition protein, the PGRPs (peptidoglycan recognition proteins), which evolve slowly under purifying selection (Jiggins and Hurst 2003). Some PGRPs have similar functions to GNBP1 and GNBP3, in that they bind to parasite polysaccharides and activate pathways that lead to the production of antimicrobial peptides (Leclerc and Reichhart 2004).
Why do the different groups of proteins have such different modes of evolution even though they all bind to the surface of the pathogens? Little et al. (2004) have suggested that the likelihood of host-parasite arms races will depend on the types of host and parasite molecules interacting. They propose that parasite polysaccharides have far less potential to evolve to evade the immune system than parasite proteins. Therefore, host-parasite coevolution is most likely to occur when host and parasite proteins interact (TEPs) than when host proteins interact with pathogen polysaccharides or glycoproteins (e.g., GNBPs and PGRPs). Scavenger receptors interact with a very broad range of ligands, including modified proteins (Krieger et al. 1993). A second hypothesis is that positive selection results from pathogens targeting particular molecules to suppress the host immune response (Begun and Whitley 2000). In this case, the positively selected molecules may have some unknown vulnerability to pathogen suppression. Finally, it may be that the positively selected molecules interact with specialist parasites, but PGRPs and GNBPs do not. Coevolutionary arms races will be much more likely between hosts and specialist parasites than between hosts and opportunistic infections. There are no known specialist bacterial or fungal pathogens of D. melanogaster that could coevolve with GNBPs or PGRPs. However, important targets of the cellular immune system are parasitoid wasps, and some of these are specialists on just a few Drosophila species. In particular, TepI is massively upregulated when flies are attacked by parasitoids and may be important in antiparasitoid defenses (Wertheim et al. 2005).
Finally, it is interesting to compare patterns of evolution seen in the same gene families across different arthropod taxa. A Tep gene from the crustacean Daphnia also evolves rapidly under positive selection (Little et al. 2004). This suggests that TEPs may be key sites of host-parasite coevolution in arthropods. In both Drosophila and Daphnia Tep genes, the bait-like region is a particular hotspot of positive selection. Unfortunately it is unknown whether insect TEPs are activated by parasite-derived proteases (as for α2-macroglobulin) or by host-derived proteases (as for complement proteins C3, C4, and C5). If the former is true, one hypothesis is that parasite proteases continually evolve new specificities that do not cleave the bait-like region, while the bait-like region changes its sequence to match the specificity of the proteases. If host proteases cleave the bait-like region, then it is possible that positive selection results from parasite adaptations to block the activation of the TEP proteins. Similar explanations have been proposed to account for the rapid evolution of other Drosophila immune system molecules (Begun and Whitley 2000).
The evolution of GNBPs has been examined in both termites and Daphnia (Bulmer and Crozier 2005; Little et al. 2004). The Daphnia GNBP, like the Drosophila proteins, was under predominantly purifying selection. However, in termites two GNBP genes showed some evidence of positive selection. It is possible that living in colonies exposes termites to higher pathogen pressures and more host-specific pathogens, resulting in stronger selection acting on termite immunity genes. Consistent with this, some other components of the immune system are positively selected in termites but not Drosophila (Bulmer and Crozier 2004; Jiggins and Kim 2005).
Acknowledgments
Acknowledgments. This work was funded by a Wellcome Trust Research Career Development Fellowship to F.J. The fly lines were supplied by Penny Haddrill and originally collected by Peter Andolfatto and Bill Ballard. R. K. Wilson gave permission to use unpublished genome sequences and Dan Halligan generated the genome-wide Ka/Ks estimates.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Contrasting patterns of X-linked and auto-somal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Przeworski M. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics. 2000;156:257–268. doi: 10.1093/genetics/156.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JA. Frequency-dependent selection in plant fungal interactions. Philos Trans Roy Soc Lond Ser B Biol Sci. 1988;319:473–483. [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P. Adaptive evolution of Relish, a Drosophila NF-{k}B/I{k}B protein. Genetics. 2000;154:1231–1238. doi: 10.1093/genetics/154.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40:903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- Bulmer MS, Crozier RH. Duplication and diversifying selection among termite antifungal peptides. Mol Biol Evol. 2004;21:2256–2264. doi: 10.1093/molbev/msh236. [DOI] [PubMed] [Google Scholar]
- Bulmer MS, Crozier RH. Variation in positive selection in termite GNBPs and Relish. Mol Biol Evol. 2005;23:317–326. doi: 10.1093/molbev/msj037. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa N, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu JN, Zheng LB, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Clark AG, Wang L. Molecular population genetics of Drosophila immune system genes. Genetics. 1997;147:713–724. doi: 10.1093/genetics/147.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol. 1995;1:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Scie USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrick JA, Baker JE, Kanost MR. Innate immunity in a pyralid moth—functional evaluation of domains from a beta-1,3-glucan recognition protein. J Biol Chem. 2004;279:26605–26611. doi: 10.1074/jbc.M403382200. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Imler J-L, Hoffmann JA. Sensing infection in Drosophila: Toll and beyond. Semin Immunol. 2004;16:43. doi: 10.1016/j.smim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Hey J, Kliman RM. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics. 2002;160:595–608. doi: 10.1093/genetics/160.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class-I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci USA. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Hurst GDD. The evolution of parasite recognition genes in the innate immune system: purifying selection on Drosophila melanogaster peptidoglycan recognition proteins. J Mol Evol. 2003;57:598–605. doi: 10.1007/s00239-003-2506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kim K-W. The evolution of antifungal peptides in Drosophila. Genetics. 2005;171:1847–1859. doi: 10.1534/genetics.105.045435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-S, Ryu J-H, Han S-J, Choi K-H, Nam K-B, Jang I-H, Lemaitre B, Brey PT, Lee W-J. Gram-negative Bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta -1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster Cells. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- Ko WY, David RM, Akashi H. Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol. 2003;57:562–573. doi: 10.1007/s00239-003-2510-x. [DOI] [PubMed] [Google Scholar]
- Krieger M, Acton S, Ashkenas J, Pearson A, Penman M, Resnick D. Molecular flypaper, host defense, and atherosclerosis. Structure, binding properties, and functions of macrophage scavenger receptors. J Biol Chem. 1993;268:4569–4572. [PubMed] [Google Scholar]
- Lachaise D, Cariou ML, David JR, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in Toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci USA. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP. Elevated polymorphism and divergence in the class C scavenger receptors of Drosophila melanogaster and D. simulans. Genetics. 2005;169:2023–2034. doi: 10.1534/genetics.104.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Clark AG. Molecular population genetics of inducible antibacterial peptide genes in Drosophila melanogaster. Mol Biol Evol. 2003;20:914–923. doi: 10.1093/molbev/msg109. [DOI] [PubMed] [Google Scholar]
- Leclerc V, Reichhart JM. The immune response of Drosophila melanogaster. Immunol Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- Lee W-J, Lee J-D, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible Gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 1996;93:7888–7893. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Little TJ, Colbourne JK, Crease TJ. Molecular evolution of Daphnia immunity genes: polymorphism in a gram-negative binding protein gene and an alpha-2-macroglobulin gene. J Mol Evol. 2004;59:498–506. doi: 10.1007/s00239-004-2641-8. [DOI] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Neutral effect of recombination on base composition in Drosophila. Genet Res. 2003;81:79–87. doi: 10.1017/s0016672302006079. [DOI] [PubMed] [Google Scholar]
- McDonald JH. Detecting non-neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol. 1996;13:253–260. doi: 10.1093/oxfordjournals.molbev.a025562. [DOI] [PubMed] [Google Scholar]
- McDonald JH. Improved tests for heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol. 1998;15:377–384. doi: 10.1093/oxfordjournals.molbev.a025934. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A pattern-recognition protein for beta-1,3-glucan—the binding domain and the cDNA cloning of beta-1,3-glucan recognition protein from the silkworm, Bombyx mori. J Biol Chem. 2000;275:4995–5002. doi: 10.1074/jbc.275.7.4995. [DOI] [PubMed] [Google Scholar]
- Powell JR. Progress and prospects in evolutionary biology: The Drosophila model. Oxford: Oxford University Press; 1997. [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. Drosophila ananassae and D. erecta whole-genome shotgun reads. Beverley, MA: Agencourt Bioscience Corp.; 2004. [Google Scholar]
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sottrup-Jensen L, Sand O, Kristensen L, Fey GH. The alpha-macroglobulin bait region. Sequence diversity and localization of cleavage sites for proteinases in five mammalian alpha-macroglobulins. J Biol Chem. 1989;264:15781–15789. [PubMed] [Google Scholar]
- Stroschein-Stevenson SL, Foley E, Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:87–99. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive Adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nei M. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wertheim B, Kraaijeveld AR, Schuster E, Blanc E, Hopkins M, Pletcher SD, Strand MR, Partridge L, Godfray HCJ. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005;6:R94. doi: 10.1186/gb-2005-6-11-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RK. D. yakuba whole-genome shotgun sequence. St. Louis, MO: Washington University Genome Sequencing Center; 2004. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]