Abstract
CD4+ T lymphocyte responses are thought to play a major role in control of the hepatitis C virus (HCV). Few, however, have been mapped down to the level of peptide and HLA restriction. Furthermore, the ability of such T cells to respond to viruses which differ in genotype has not been addressed in detail. In most cases of persistent infection with HCV, CD4 proliferative responses are weak or absent. From a large cohort of persistently infected patients, we identified an individual with unusually robust and persistent responses in the face of chronic infection. We firstly mapped two peptide epitopes to regions of the nonstructural protein NS4 (aa1686–1705 and aa 1746–1765). However, in contrast to the genotype 1a derived antigens used for mapping, the infecting virus was identified as genotype 3a. Strikingly, the patient's CD4 response to these epitopes were specific only for the genotype 1a sequence, and did not recognize genotype 3a synthetic peptides. Serologic assays indicated that prior exposure to HCV of genotype 1 had occurred. This patient therefore maintains strong CD4 proliferative responses which are genotype specific and not cross-reactive. The apparent ‘misdirection’ of these nonprotective responses has important implications for the role of natural and vaccine induced CD4 responses in the face of variable viruses.
Keywords: immune escape, HCV genotype, NS4, T helper cell, MHC Class II
INTRODUCTION
HCV is an RNA virus which persists in the majority of individuals infected in the face of a host immune response [1]. Data from multiple groups have shown that CD4 lymphocytes play a major role in clearance of the virus [2–5] although how this occurs and what other factors are involved in elimination of the virus are still not clear. Early interactions between host and virus are likely to play a crucial role in the variable outcome of infection with HCV [6]. Evidence from animal models suggests that failure of CD4 cells at such a stage can lead to subsequent failure of both CD8 and B cell responses to control virus [7,8]. In patients with persistent HCV infection, it is unusual to detect robust CD4+ T cell responses in peripheral blood [3,9,10], and this failure of CD4 responses may therefore play a central role in allowing the virus to persist.
HCV is highly variable, both between genotypes and, in certain genes such as envelope, within an individual over time [11]. Studies of other viral infections such as HIV and HBV have shown that variation within CD4 epitopes can affect T cell recognition and potentially influence antiviral control [12,13]. Since in HCV infection, most of those with persistent viremia have very weak T cell responses, this issue is difficult to address in general. Indeed, very few epitopes (approximately 10 to date) have been mapped down to the peptide level and HLA restrictions performed [5,14–16]. In this study, we have analysed in detail the HCV specific T cell responses of an individual who, surprisingly, maintained strong CD4 helper responses in the face of persistent infection. We firstly identified two epitopes in the NS4 gene and then observed that both these peptide-specific CD4+ T cell populations were directed against sequences which were no longer circulating in his blood. These CD4+ T cells therefore lack crossreactivity in vitro and, apparently, cross-protection in vivo. The issue of crossreactivity of T cell responses between genotypes is an important consideration in the future design of vaccines against HCV.
MATERIALS AND METHODS
Subject
Subject (OX-78) was part of the OXHEP study in which CD4 responses have been analysed in 67 patients as previously reported [17]. This study had been approved by the local Ethics Committee (COREC 98·137) and the informed consent of the patients obtained. OX-78 was a 37-year-old-male who had been persistently infected with HCV for at least eight years, the source of which was not known. He presented with abnormal liver function and chronic active hepatitis with fibrosis was observed on biopsy (Knodell score 4/6). At the time of study, he had failed treatment with IFN-α alone firstly and then in combination therapy with ribavirin. Blood was taken on seven occasions over a period of two years for CD4 lymphocyte studies and for viral sequencing.
The complete HLA type of subject OX-78 was determined by PCR-SSCP [18] to be A*02/*68; B*51/*5108; Cw*15/*1602; DRB1*16/*0301; DRB3*0202; DQB1*0201/*0501.
Sequencing of autologous virus
Bulk sequencing of the NS4 region of autologous virus from serum and sequencing of 4–5 clones from two time points was performed as previously described [19]. RT-PCR was performed using the protocol as described and products cleaned and cloned using Invitrogen TOPO cloning kit.
HCV antigens
Recombinant proteins covering HCV core (C22), NS3 (C33c), NS4 (C100), NS5 and NS3 + NS4 (C200) plus yeast and E. coli control proteins were kindly provided by Chiron (Emeryville, CA, USA). They were used at a final concentration of 1 µg/ml in proliferation assays.
Twenty-mer peptides overlapping by 10 amino acids covering all of HCV core (amino acids 1–190) were obtained from Research Genetics (Huntsville, Alabama, USA) and those covering all of NS3 (aa 1007–1616) and NS4 (aa1617–2015) from Chiron Mimotopes. These were combined into pools of five peptides each for initial identification of specific epitopes in proliferation assays and were used at a final concentration of 1 µm for each peptide. Three known CD4 epitopes within NS3 were tested separately as individual peptides (NS3A 1247-1266; NS3B 1347-1366; NS3C 1457-1476) [15]. Three known HLA-A2 CD8 epitopes described in previous studies were also tested in an Elispot assay – NS31073-1081; NS31406-1415; NS52594-2602 [6].
Proliferation assays
PBMC were separated from whole blood by centrifugation on a density gradient (Lymphoprep, Nycomed, Oslo, Norway) and were washed and cultured in RPMI 1640 cell culture medium containing glutamine, penicillin/streptomycin and 10% heat inactivated human A positive serum (RH10). PBMC were cultured with and without antigen in RH10 at 2 × 105 cells per well in round bottomed 96 well plates in a total volume of 200 µl. Triplicate wells were set up for each of the antigens which included control antigens PHA and tetanus toxoid (2 µg/ml) plus yeast and E. coli control proteins. After five days, cultures were pulsed with 1 µCi of 3H-thymidine per well for 16 h before harvesting and counting in a Topcount scintillation counter (Packard, Canberra, Australia). Results are expressed as counts per minute (cpm) or as stimulation indices (SI = cpm in the presence of antigen divided by cpm without antigen). An SI > 3 was regarded as significant.
CD4 T cell lines
CD4 positive HCV specific T cell lines were isolated from fresh PBMC from which CD8 positive T cells had been depleted by magnetic bead separation (Dynal (UK) Ltd, Wirral, Merseyside, UK). The remaining cells were cultured with the appropriate antigen in RH10 for five days and the lymphoblasts separated by density gradient centrifugation (Percoll, Pharmacia, Uppsete, Sweden). The resulting cells were then expanded in culture medium containing 10% IL-2 (Lymphocult-T, Biotest, Dreieich, Germany). Long-term cultures were maintained on a regime of restimulation in the presence of appropriate peptide and antigen presenting cells (APC, irradiated autologous or HLA Class II matched PBMC) followed by expansion in 10% IL-2 (Lymphocult-T).
Proliferation assays using peptide specific CD4 T cell lines were carried out using 2·5 × 104 T cells and 1 × 105 APC in the presence of appropriate antigens. They were incubated for 3 days before labelling and harvesting.
HLA Class II restriction
This was identified in each case by assaying the CD4 lines for proliferation using HLA matched and mismatched irradiated PBMC as antigen presenting cells.
IFN-γ Elispot assay
Fresh PBMC were plated in 96 well nitrocellulose plates (Millipore, Watford, Herts, UK) that had been coated with 0·5 µg/ml of anti-human IFN-γ mAb. Antigens were added at the same concentrations as those used in proliferation in duplicate wells and then 3 × 105 PBMC added, in a final volume of 200 µl RH10. The plates were incubated overnight at 37°C and developed as previously described [20]. The number of specific T cells was enumerated by the use of an Elispot plate reader (Autoimmune Diagnostika (AD, Strassbourg, Germany), Germany using software version v.2·1).
Cytokine production
Production of cytokines (IL-2, IL-4) by the CD4 cells on stimulation with the specific peptides was assessed by ELISA assays (R and D Systems, Abingdon, Oxon, UK) carried out on culture supernatants harvested after 48 h in culture.
Phenotype
T cell lines were stained with anti-CD4-FITC, CD8-PerCP, CD3-PE, CD45RO- FITC and CD38-APC (Becton Dickinson, Biosciences, San Diego, CA, USA) and analysed on a FACScalibur™ flow cytometer using CellQuest software (Becton Dickinson)
ELISA for serotype antibodies
An ELISA assay for detection of genotype specific anti-HCV was carried out on patients' sera. Microtitre immunoplates were coated with 2·5 µg/ml of NS4-8 or NS4-14 peptides, genotype 1a and 3a, or with recombinant NS4 at 1 µg/ml in PBS for 1 h at 37°C. Unbound antigen was removed and wells blocked with 5% w/v nonfat milk for 30 min at room temperature (RT). The wells were then washed three times with PBS/0·5% Tween 20 and 100 µl of serum dilutions (1:50–1:500) added. Plates were incubated for 90 min at RT and then washed as before. 100 µl of goat-anti-human IgG conjugated to HRP (Dako, Glostrup, Denmark) at 1/1000 in PBS was added and the plates incubated for one hour at RT. The final immune complexes were detected by addition of 100 µl TMB substrate solution (Sigma, Poole, Dorset, UK) and colourimetric determination of the absorbance at 450 nm.
Additionally, serum samples from subject OX-78 were tested with a commercially available and a redeveloped version of an HCV serotyping assay (Abbott Murex, Destford, Kent, UK). Briefly, immunoplates were coated with synthetic peptides representing variable regions within NS4 from HCV types 1, 2, 3, 4, 5 and 6. Serum dilutions were then added in the presence of competing peptide in free solution to block cross-reactivity. Captured antibodies were then detected as described above for the previous ELISA.
RESULTS
Seventeen of the 67 subjects in the OXHEP study showed CD4 proliferative responses [17] seven of whom were PCR +ve, and three of which were consistently detectable at multiple time points. Of these the responses of OX-78 were consistently the greatest and hence this subject was the focus of the detailed mapping studies. In addition, unlike the other PCR +ve, patients with CD4+ T cell responses detectable, this patient was found to be infected with genotype 3a virus (see below) and therefore crossreactivity of such responses in vivo could be addressed.
CD4 epitopes
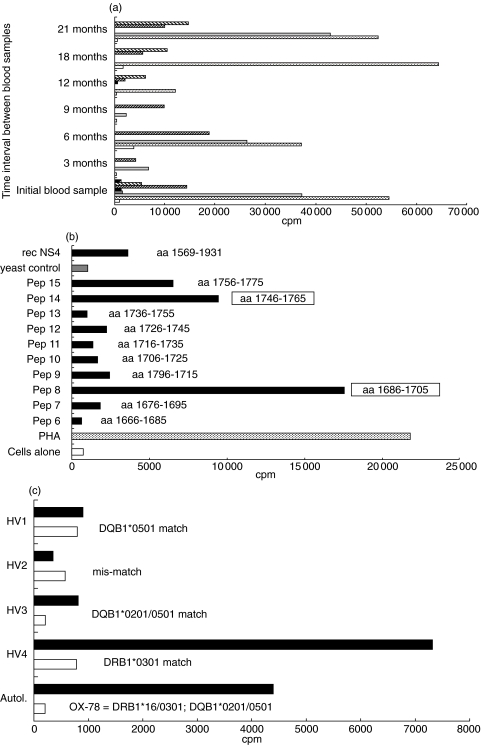
Proliferative responses of fresh PBMC obtained from subject OX78 on seven occasions over a two year period are shown in Fig. 1a. Responses were NS4 specific as proliferation to the other proteins and peptides were negative at all time points tested. Long-term CD4 T cell lines specific for NS4 were isolated from three of these blood samples and their peptide specific epitopes identified by the use of 39 overlapping peptides covering all of NS4. Two HCV CD4 epitopes were identified – NS4-8 (aa1686–1705) and NS4-14 (aa1746–1765) (Fig. 1b).
Fig. 1.
Mapping of sustained HCV specific proliferative responses in a persistently infected individual. (a) Responses of PBMC from OX-78 over time. Fresh peripheral blood mononuclear cells (PBMC) obtained from subject OX-78 were tested for a proliferative response to recombinant HCV antigens (▪ rec NS3,  rec NS4,
rec NS4,  rec NS3/4,
rec NS3/4,  rec NS5,
rec NS5,  rec core) and control antigens, phytohaemagglutinin (
rec core) and control antigens, phytohaemagglutinin ( , PHA) and tetanus toxoid (
, PHA) and tetanus toxoid ( , TT) and cells alone (□). Raw data is displayed and these are equivalent to a positive stimulation index of 6 (min) to 30 (max). (b) Epitope mapping of HCV NS4 specific CD4 T cell lines from subject OX-78. An NS4-specific CD4+ T cell ine derived from subject OX-78 was tested for a proliferative response to 20mer peptides overlapping by 10 amino acids, covering all of NS4 (aa1616–2015). Immunodominant epitopes were identified as NS4-8 (SI = 24) and NS4-14 (SI = 13). (c) HLA Class II restriction of CD4 line specific for peptide NS4-8 (aa1686–1705). The HLA Class II restriction of a CD4 +ve T cell line derived from OX-78 which was specific for peptide NS4-8 was determined using HLA Class II partially matched and mismatched irradiated PBMC from healthy volunteers (HV1, 2, 3, 4) as antigen presenting cells (APC) in a proliferation assay with 1 µm peptide NS4-8 as stimulating antigen (▪). Cells alone (□).
, TT) and cells alone (□). Raw data is displayed and these are equivalent to a positive stimulation index of 6 (min) to 30 (max). (b) Epitope mapping of HCV NS4 specific CD4 T cell lines from subject OX-78. An NS4-specific CD4+ T cell ine derived from subject OX-78 was tested for a proliferative response to 20mer peptides overlapping by 10 amino acids, covering all of NS4 (aa1616–2015). Immunodominant epitopes were identified as NS4-8 (SI = 24) and NS4-14 (SI = 13). (c) HLA Class II restriction of CD4 line specific for peptide NS4-8 (aa1686–1705). The HLA Class II restriction of a CD4 +ve T cell line derived from OX-78 which was specific for peptide NS4-8 was determined using HLA Class II partially matched and mismatched irradiated PBMC from healthy volunteers (HV1, 2, 3, 4) as antigen presenting cells (APC) in a proliferation assay with 1 µm peptide NS4-8 as stimulating antigen (▪). Cells alone (□).
Subsequently, two separate CD4 lines were raised against NS4-8 and NS4-14. One of the CD4 lines specific for NS4-8 was cloned and most of the assays described here were performed on both the polyclonal line and clones. The response to epitope NS4-8 was found to be clearly restricted by HLA DRB1*0301 (Fig. 1c) and the response to NS4-14 by HLA DRB1*16 or DRB3*0202.
All the long-term cultured T cell lines and clones described in this study were confirmed to be CD3 +ve (>95%), CD4 +ve (>90%), CD8-ve (<10%) and CD45RO +ve (>90%), indicating a memory phenotype. Both CD4 T cell lines were shown to produce IL-2 on stimulation with peptide (range 36–110 pg/ml) and not IL-4 (<2 pg/ml), indicating a Th1 type of response.
Recognition of viral variants
Sequencing of autologous virus from serum samples obtained from this patient at two time points (initial sample and 12 month sample) revealed that the individual was persistently infected with HCV of genotype 3a (Table 1). This was consistent with conventional genotyping data using a PCR-RFLP for 5′UTR. Bulk sequencing confirmed by clonal analysis revealed that the viral sequence was identical to the consensus sequence for NS4 genotype 3a, and quite distinct from genotype 1a, the sequence used in the peptides and antigens (Table 1). PCR specific for genotype 1 using primers in the NS3 region [21] were negative (data not shown).
Table 1.
Sequences of HCV CD4 epitopes from patient OX-78
| Epitope | Position | Genotype | Amino acid sequence | HLA Class II restriction |
|---|---|---|---|---|
| NS4-8 | 1686–1705 | 1a | VVLSGKPAIIPDREVLYREF | DRB1*0301 |
| 3a | IELGGKPALVPDKEVLYQQY | |||
| Autologous | IELGGKPALVPDKEVLYQQY | |||
| NS4-14 | 1746–1765 | 1a | IAPAVQTNWQKLETFWAKHM | DRB1*16 or |
| 3a | IEPIVATNWQKLEAFWHKHM | DRB3*0202 | ||
| Autologous | IEPIVATNWQKLEAFWHKHM |
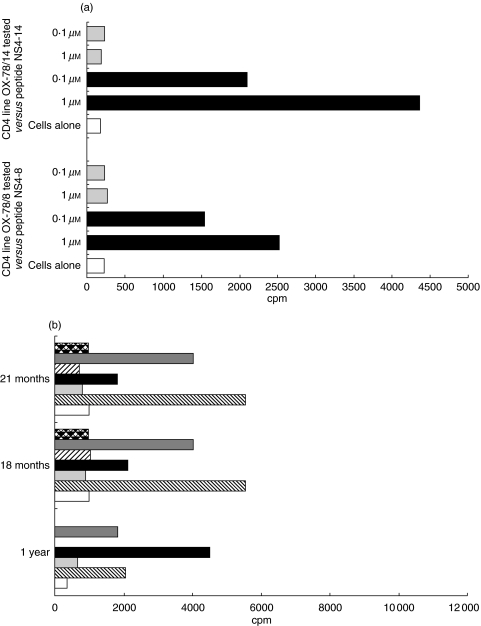
Peptides corresponding to the patient's autologous (and consensus) genotype 3a sequence for NS4-8 and to the consensus genotype NS4-14 sequence were synthesized for use in both proliferation and Elispot assays with the CD4 T cell lines. Proliferative responses of the NS4-8 and NS4-14 specific CD4 lines were completely specific for the genotype 1a peptide and did not recognize the 3a synthetic peptides (Fig. 2a).
Fig. 2.
CD4+ T lymphocyte responses to genotype-specific peptides. (a) Responses of OX-78 NS4-8 and NS4-14 specific CD4 lines to genotype peptide variants. HCV specific CD4 +ve T cell lines from subject OX-78 were tested for a proliferative response to genotype 3a variants of the NS4-8 and NS4-14 peptide epitopes.  Genotype 3a variant; ▪ genotype 1a varient. (b) Responses of PBMC from OX-78 to variant peptides. Fresh PBMC from subject OX-78 were tested for an ex vivo response to the genotype variants of peptides NS4-8 and NS4-14 used at 1 µm concentration.
Genotype 3a variant; ▪ genotype 1a varient. (b) Responses of PBMC from OX-78 to variant peptides. Fresh PBMC from subject OX-78 were tested for an ex vivo response to the genotype variants of peptides NS4-8 and NS4-14 used at 1 µm concentration.  Pep NS4-14/3a,
Pep NS4-14/3a,  PepNS4-14/1a,
PepNS4-14/1a,  Pep NS4-8/3a, ▪ Pep NS4-8/1a,
Pep NS4-8/3a, ▪ Pep NS4-8/1a,  Yeast control,
Yeast control,  rec NS4, □ cells alone.
rec NS4, □ cells alone.
In order to confirm that this was not a phenomenon induced by selection and long-term culture of the cells and restimulation with the 1a genotype peptide, PBMC from fresh blood samples taken from the patient were assayed by both proliferation and Elispot assays against the 1a and 3a variant peptides for both the NS4-8 and NS4-14 specific cell lines. There was no proliferative response to the 3a variant of either NS4-8 or NS4-14 (Fig. 2b) although responses to the 1a variant were detectable. Similarly, in an IFN-γ Elispot assay, specific responses of 28 spots per million PBMC were detectable against 1a peptides with no responses detected against 3a peptides. Interestingly, although at the first time point tested, the dominant proliferative response was to peptide NS4-8, in the subsequent blood samples this had almost disappeared and the response to peptide NS4-14 dominated (Fig. 2b).
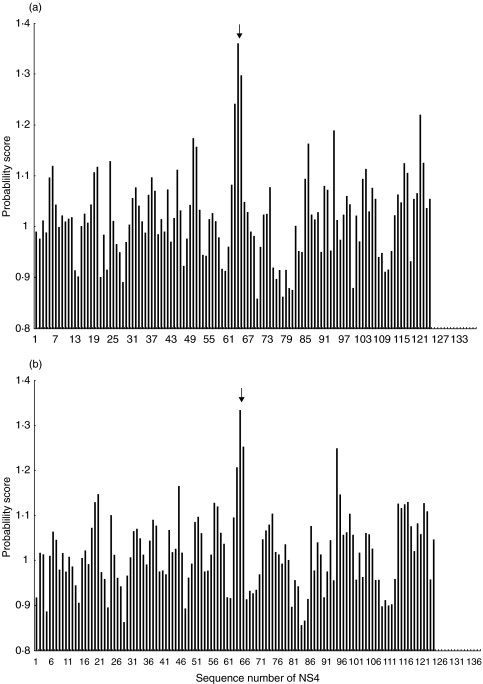
We tested whether the genotype 3a peptides could act as altered peptide ligands for genotype 1a specific cells by coincubating the APC with the 3a peptide after which a standard proliferation assay was performed using the 1a peptide. In addition to the fact that they could not induce proliferation or cytokine secretion, the 3a peptides were unable to block proliferation of these responses when pulsed onto targets either prior to or after incubation with genotype 1a peptide (data not shown). We conclude that they are not partially stimulating responses nor acting as TCR antagonists. A validated epitope prediction program for HLA-DR3 [22,23] was also used to assess whether the NS4-8/3a peptide was capable of binding to the relevant HLA Class II molecule, DRB1*0301. The greater the score achieved the more likely the peptide is to represent an epitope in vivo. This showed that the region of NS4-8 had the highest probability of being processed and presented (PPP) score of the entire protein in both the type 1a and 3a sequence (Fig. 3). This data strongly suggests that the lack of recognition of the 3a peptides is not due to poor binding or presentation by APC.
Fig. 3.
Probability of being processed and presented (PPP) score for peptides from NS4 scanned with the HLA-DR3 probability matrix. (a) Predicted binding of peptide NS4-8/genotype 1a to HLA-DR3. Graph showing the PPP scores for iterative 15mers from NS4 scanned with the HLA-DR3 probability matrix. The T cell epitope NS4-8 is marked with an arrow. (b) Predicted binding of peptide NS4-8/genotype 3a to HLA DR3. As above for 3a genotype
To try to confirm whether or not this individual had previously been exposed to both genotype 1a virus, analysis of B cell responses was undertaken. Firstly, an ELISA for serotype specific antibodies, using peptides NS4-8 and NS4-14 derived from genotype 1a and 3a, was performed. This revealed the presence of anti-NS4-14 peptide antibodies to peptides of both 1a and 3a genotype in the patients serum but only monospecific antibodies in sera from five patients infected with genotype 1a or 3a, respectively. A second confirming result was also obtained for subject OX-78 using a commercial ELISA although in this instance only antibodies to HCV NS4 of serotype 1 were detected (see methods). The fact that genotype 3 antibodies were not detected in the second ELISA is possibly due to the use of peptides from different regions of NS4 regions in the two assays. However, both results strongly suggests that exposure of B cells to genotype 1 specific antigens has previously occurred in patient OX-78.
At the CD8 T cell level, we found the patient to be negative by Elispot for responses to three HLA-A2 restricted epitopes in NS3 and NS5 (see methods). This result was obtained using peptides derived from genotype 1 and 3a in each case [24]. A similar result was obtained using HLA Class I peptide tetramers to the epitopes (data not shown) [25]. These negative results are typical for patients with persistent HCV infection [25].
DISCUSSION
We describe an unusual but informative patient who maintained a strong proliferative response directed against HCV, in the face of ongoing viremia. Although the responses were robust, clearly virus specific and Class II restricted, they were genotype 1 specific and did not recognize genotype 3 peptides – i.e. those present in the circulating virus. Although this patient is unusual with regards to the maintenance of strong T cell responses, the detailed analysis of these responses raises issues which are generally relevant: the ability of T cells to control new or different genotypes of virus and the mechanism for loss of T cell responsiveness in the presence of circulating virus.
Both epitopes when originally mapped, had not been previously described, but one of them (aa 1686–1705) has been recently reported by other workers [16], who identified it in the liver of a single chronically infected patient. The simultaneous evaluation of this epitope by our group adds weight to its validity, and the second epitope – to the best of our knowledge – has not previously been described. Both epitopes appear to elicit robust responses when derived from genotype 1, and are predicted to bind and be presented when derived from genotype 3a sequence. Since in chronically infected patients, where genotype can be assessed accurately, CD4 responses are rarely detected, it has not been possible to date to identify another patient of the same HLA type with a response specifically to the genotype 3a peptide.
The likely cause for the phenomenon observed is that this patient was previously infected with, or perhaps initially coinfected with genotype 1 virus, which he has subsequently cleared below the level of detection. The evidence for genotype 1 infection is not only derived from cellular immune responses but also serological assays. The CD4+ T cell responses have been maintained in the absence of the circulating genotype 1a virus – just as in many patients in whom virus has been cleared, CD4 responses are preserved. Interestingly, after infection with a new variant, such responses were not protective but they were maintained, as has been reported for CD8 + cells in HIV [26,27].
We cannot exclude the possibility that genotype 1a virus persists in subject OX78 at very low levels thereby maintaining the immune response to it. However, a study of HCV infection acquired through blood transfusion, in which the parental genomes of the infecting viruses were clearly defined, suggests that superinfection with different HCV strains can lead to eradication or suppression of the original strain [28]. Similarly, the possibility of a second infection with HCV of a different genotype has been previously described in an intravenous drug user in which antibodies were detected only to the initially infecting genotype 1 virus but not to a genotype 3 virus with which he was apparently subsequently infected [29]. Among multitransfused haemophiliacs, genotype changes over time are common in those with chronic HCV [30] although the frequency of superinfection among HCV positive individuals in general is not know. For intravenous drug users and multiply transfused recipients, the apparent lack of protection conferred by a primary HCV infection against subsequent infection with viruses of different genotypes has important implications.
These results indicate that the loss of CD4+ T cell responses in normal infection requires recognition of the virus through the TCR, and is not a consequence of persistent viremia per se. It is possible that the responses we observed may persist to some extent through immunological ‘ignorance’ i.e. the genotype specific CD4 cells fail to recognize the presence of the circulating genotype 3a virus. In the majority of chronically infected patients, only low levels of HCV specific CD4 cells are detectable and the virus is able to avoid the effector arm of the immune response. The longterm nature of this response and even the slight modulation in immunodominance observable during the course of the study, in which the response to NS4-8 becomes weaker in the face of increased responsiveness to NS4-14 suggests that it is being actively maintained.
Clearly such responses are not protective against genotype 3a, even though they may well have been in concert with other mediators against type 1a. The use of genotype 1a sequences to generate vaccine-induced responses may well lead to strain specific T cells which are not protective. It is possible, and has been proposed for HIV that such ‘established’ responses might interfere with the production of naturally protective responses [31].
The responses are reminiscent of the phenomenon of ‘original antigenic sin’[32] which has been observed for antibody responses after repetitive infection with different strains of influenza. Such phenomena have also been reported for T cell responses after infection with persistent, variable viruses [33]. In these cases the initial infecting virus appears to lay down a pattern of responses which are maintained or reinforced by subsequent reinfection. There may be a number of effects, including potentially, bystander activation or help delivered by T cell responses to other epitopes, which act to maintain the original response. Unlike other situations, partial cross-reactivity of the TCR for the second variant does not appear to explain the maintenance of such responses in this case. It is also possible, as we have shown previously, that interferon-alpha therapy promotes the induction or maintenance of responses, even if the therapy fails to suppress viraemia in the long term [17].
Previous studies have indicated the presence of apparently cross-reactive responses using recombinant proteins [34] in that genotype 2a and 3a infected patients could recognize genotype 1a antigens. Similar observations have been made in analyses of CD8+ T cell responses [35]. These data suggest that it is essential in future studies of HCV and other variable viruses to match accurately the T cell response with the patient's autologous sequences. Responses which appear robust in vitro may have little protective function in vivo as this example clearly illustrates. This case is especially clear-cut due to the readily detectable differences in viral genotype but similar effects could occur, potentially, due to sequence variation within a genotype or variation within as infected individual. Further detailed, longitudinal studies are required to understand how general the phenomena are and how relevant to viral persistence in the face of host T cell responses. In future design of HCV vaccines, a careful search for crossreactive epitopes is essential to avoid generation of nonprotective responses.
Acknowledgments
We gratefully acknowledge the generous provision of HCV recombinant proteins by Dr Michael Houghton (Chiron Corp., Emeryville, CA, USA) and the work of Lara Sanders from Abbott Laboratories in performing the serotyping. GCH, REP and PK are funded by the Wellcome Trust and ML is funded by the EU (QLK2-CT-1999–00356).
References
- 1.Cohen J. The scientific challenge of Hepatitis C Virus. Science. 1999;285:26–30. doi: 10.1126/science.285.5424.26. [DOI] [PubMed] [Google Scholar]
- 2.Diepolder HM, Zachoval R, Hoffmann RM, et al. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–7. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 3.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4 (+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117:933–41. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 4.Naoumov NV. Hepatitis C virus-specific CD4 (+) T cells: do they help or damage? [Editorial] Gastroenterology. 1999;117:1012–4. doi: 10.1016/s0016-5085(99)70361-6. [DOI] [PubMed] [Google Scholar]
- 5.Godkin A, Jeanguet N, Thurz M, Openshaw P, Thomas H. Characterisation of novel HLA-DR11-restricted HCV epitopes reveals both qualitative and quantitative differences in HCV-specific CD4+ T cell responses in chronically infected and non-viremic patients. Eur J Immunology. 2001;31:1438–46. doi: 10.1002/1521-4141(200105)31:5<1438::AID-IMMU1438>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Lechner F, Wong DK, Dunbar PR, et al. Analysis of Successful Immune Responses in Persons Infected with Hepatitis C Virus. J Exp Med. 2000;191:1499–512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planz O, Ehl S, Furrer E, et al. A critical role for neutralising antibody-producing B cells, CD4+ cells and interferons in persistent infections of mice with LCMV. implications for adoptive immunotherapy of virus carriers. Proc Natl Acad Sci USA. 1997;94:6874–9. doi: 10.1073/pnas.94.13.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciurea A, Hunziker L, Klenerman P, Hengartner H, Zinkernagel RM. Impairment of CD4 (+) T cell responses during chronic virus infection prevents neutralizing antibody responses against virus escape mutants. J Exp Med. 2001;193:297–305. doi: 10.1084/jem.193.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari C, Valli A, Galati L, et al. T-cell response to structural and nonstructural hepatitis C virus antigens in persistent and self-limited hepatitis C virus infections. Hepatology. 1994;19:286–95. [PubMed] [Google Scholar]
- 10.Lechmann M, Ihlenfeldt HG, Braunschweiger I, et al. T- and B-cell responses to different hepatitis C virus antigens in patients with chronic hepatitis C infection and in healthy anti-hepatitis C virus-positive blood donors without viremia. Hepatology. 1996;24:790–5. doi: 10.1002/hep.510240406. [DOI] [PubMed] [Google Scholar]
- 11.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 12.Harcourt GC, Garrard S, Davenport MP, Edwards A, Phillips RE. HIV-1 variation diminishes CD4 T lymphocyte recognition. J Exp Med. 1998;188:1785–93. doi: 10.1084/jem.188.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg W. Mechanisms of immune escape in viral hepatitis. Gut. 1999;44:759–64. doi: 10.1136/gut.44.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamonaca V, Missale G, Urbani S, et al. Conserved hepatitis C virus sequences are highly immunogenic for CD4 (+) T cells: implications for vaccine development. Hepatology. 1999;30:1088–98. doi: 10.1002/hep.510300435. [DOI] [PubMed] [Google Scholar]
- 15.Diepolder HM, Gerlach JT, Zachoval R, et al. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–9. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penna A, Missale G, Lamonaca V, et al. Intrahepatic and circulating HLA class II-restricted, hepatitis C virus-specific T cells: functional characterization in patients with chronic hepatitis C. Hepatology. 2002;35:1225–36. doi: 10.1053/jhep.2002.33153. [DOI] [PubMed] [Google Scholar]
- 17.Harcourt G, Hellier S, Bunce M, et al. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat. 2001;8:174–9. doi: 10.1046/j.1365-2893.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- 18.Bunce M, O'Neill CM, Barnardo MC, et al. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–67. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 19.Forns X, Maluenda MD, Lopez-Labrador FX, et al. Comparative study of three methods for genotyping hepatitis C virus strains in samples from Spanish patients. J Clin Microbiol. 1996;34:2516–21. doi: 10.1128/jcm.34.10.2516-2521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–65. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang KM, Rehermann B, McHutchison JG, et al. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest. 1997;100:2376–85. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godkin AJ, Davenport MP, Willis A, Jewell DP, Hill AV. Use of complete eluted peptide sequence data from HLA-DR and -DQ molecules to predict T cell epitopes, and the influence of the nonbinding terminal regions of ligands in epitope selection. J Immunol. 1998;161:850–8. [PubMed] [Google Scholar]
- 23.Davenport MP, Ho Shon IA, Hill AV. An empirical method for the prediction of T-cell epitopes. Immunogenetics. 1995;42:392–7. doi: 10.1007/BF00179401. [DOI] [PubMed] [Google Scholar]
- 24.Ward S, Lauer G, Isba R, Walker B, Klenerman P. Cellular immune responses against hepatitis C virus: the evidence base 2002. Clin Exp Immunol. 2002;128:195–203. doi: 10.1046/j.1365-2249.2002.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes E, Harcourt G, Brown D, et al. The dynamics of T-lymphocyte responses during combination therapy for chronic hepatitis C virus infection. Hepatology. 2002;36:743–54. doi: 10.1053/jhep.2002.35344. [DOI] [PubMed] [Google Scholar]
- 26.McAdam S, Klenerman P, Tussey L, et al. Immunogenic HIV variant peptides that bind to HLA-B8 can fail to stimulate cytotoxic T lymphocyte responses. J Immunol. 1995;155:2729–36. [PubMed] [Google Scholar]
- 27.Klenerman P, Meier UC, Phillips RE, McMichael AJ. The effects of natural altered peptide ligands on the whole blood cytotoxic T lymphocyte response to human immunodeficiency virus. European J Immunol. 1995;25:1927–31. doi: 10.1002/eji.1830250720. [DOI] [PubMed] [Google Scholar]
- 28.Laskus T, Wang LF, Radkowski M, et al. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J Virol. 2001;75:2059–66. doi: 10.1128/JVI.75.5.2059-2066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proust B, Dubois F, Bacq Y, et al. Two successive hepatitis C virus infections in an intravenous drug user. J Clin Microbiol. 2000;38:3125–7. doi: 10.1128/jcm.38.8.3125-3127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyster ME, Sherman KE, Goedert JJ, Katsoulidou A, Hatzakis A. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. The Multicenter Hemophilia Cohort Study. J Inf Dis. 1999;179:1062–9. doi: 10.1086/314708. [DOI] [PubMed] [Google Scholar]
- 31.McElrath MJ, Corey L, Greenberg PD, et al. Human immunodeficiency virus type 1 infection despite prior immunization with a recombinant envelope vaccine regimen. Proc Natl Acad Sci USA. 1996;93:3972–7. doi: 10.1073/pnas.93.9.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De St Groth F, Webster R. Disquisitions on original antigenic sin. I. Evidence in man. J Exp Med. 1966;124:331–45. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klenerman P, Zinkernagel RM. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–5. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 34.Missale G, Bertoni R, Lamonaca V, et al. Different clinical behaviours of acute HCV infection are associated with different vigor of the anti-viral T cell response. J Clin Invest. 1996;98:706–14. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauer GM, Ouchi K, Chung RT, et al. Comprehensive analysis of CD8(+)-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol. 2002;76:6104–13. doi: 10.1128/JVI.76.12.6104-6113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]