Abstract
The cyclin D1 gene is overexpressed in human breast cancers and is required for oncogene-induced tumorigenesis. Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor selectively activated by ligands of the thiazolidinedione class. PPARγ induces hepatic steatosis, and liganded PPARγ promotes adipocyte differentiation. Herein, cyclin D1 inhibited ligand-induced PPARγ function, transactivation, expression, and promoter activity. PPARγ transactivation induced by the ligand BRL49653 was inhibited by cyclin D1 through a pRB- and cdk-independent mechanism, requiring a region predicted to form an helix-loop-helix (HLH) structure. The cyclin D1 HLH region was also required for repression of the PPARγ ligand-binding domain linked to a heterologous DNA binding domain. Adipocyte differentiation by PPARγ-specific ligands (BRL49653, troglitazone) was enhanced in cyclin D1−/− fibroblasts and reversed by retroviral expression of cyclin D1. Homozygous deletion of the cyclin D1 gene, enhanced expression by PPARγ ligands of PPARγ and PPARγ-responsive genes, and cyclin D1−/− mice exhibit hepatic steatosis. Finally, reduction of cyclin D1 abundance in vivo using ponasterone-inducible cyclin D1 antisense transgenic mice, increased expression of PPARγ in vivo. The inhibition of PPARγ function by cyclin D1 is a new mechanism of signal transduction cross talk between PPARγ ligands and mitogenic signals that induce cyclin D1.
The cyclin-dependent kinase holoenzymes are a family of serine/threonine kinases that play a pivotal role in controlling progression through the cell cycle (38, 47). Dysregulation of the cell cycle control apparatus is an almost uniform aberration in tumorigenesis (48). The cyclins encode regulatory subunits of the kinases which phosphorylate specific proteins, including the retinoblastoma (pRB) protein, to promote transition through specific cell cycle checkpoints (47, 57). Cyclin D1 plays a pivotal role in G1/S phase cell cycle progression in fibroblasts and is rate limiting in growth factor- or estrogen-induced mammary epithelial cell proliferation (29, 67). Cyclin D1 overexpression is found in >30% of human breast cancers, correlating with poor prognosis (23). Several different oncogenic signals induce cyclin D1 expression, including mutations of the Ras and Wnt/APC/β-catenin pathway (2, 49). Mammary-targeted expression of cyclin D1 is sufficient for the induction of mammary adenocarcinoma, and cyclin D1−/− mice are resistant to ErbB2-induced tumorigenesis (53, 64).
In addition to binding cyclin-dependent kinases 4 and 6 (cdk4 and cdk6) and pRB, cyclin D1 forms physical associations with P/CAF (p300/CBP-associated factor), Myb, MyoD, and the cyclin D1 myb-like binding protein (DMP1) (16, 20, 31, 39). Binding of cyclin D1 to the estrogen receptor alpha (ERα) enhances ligand-independent reporter gene activity, and liganded androgen receptor reporter gene activity is inhibited by cyclin D1 (33, 39, 68). The in vivo or genetic evidence indicating a requirement for cyclin D1 in nuclear receptor function remained to be determined.
The peroxisome proliferator-activator receptors, including PPARα, PPARγ, and PPARδ, are ligand-activated nuclear receptors (42). Their modular structure resembles those of other nuclear hormone receptors with N-terminal AF-1, a DNA binding domain, and a carboxyl-terminal ligand-binding domain (LBD). PPARγ was cloned as a transcription factor involved in fat cell differentiation and is required for the induction of adipocyte differentiation (41, 51). Adenoviral delivery of PPARγ to the livers of mice induces hepatic steatosis, consistent with an important role for PPARγ in hepatocellular lipid biosynthesis (65). The PPARγ ligands include eicosanoids, such as 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), and synthetic ligands of the thiazolidinedione (TZD) class. PPARγ agonists inhibit the growth of human colorectal cancer cells (45) and promote fibroblast and breast epithelial cell differentiation (14, 32).
Since at least 1.6 million patients take TZDs as antidiabetic agents (42), it is important to understand PPARγ function, including its possible role in cancer. The possibility of inhibiting tumor cellular proliferation using PPARγ ligands as nontoxic therapeutics has provided the impetus to assess their efficacy in animal models. Mutation within the adenomatous polyposis coli (APC) pathway occurs frequently in human colon cancer and is sufficient for the induction of gastrointestinal polyposis in the Min mouse. PPARγ ligands inhibited the growth of implanted colonic tumors with APC mutations in one study (44); however, treatment of Min mice with PPARγ ligands increased polyposis in other studies (27, 43). Together these findings raise the possibility that PPARγ ligand effects in vivo may be governed by specific genetic determinants. Identifying the molecular genetic determinants governing PPARγ regulation of cellular proliferation and tumorigenesis in vivo is therefore of considerable importance. We examined the role of cyclin D1 as a genetic determinant of PPARγ function, since cyclin D1 has been implicated in the genesis of breast and colon cancer.
MATERIALS AND METHODS
Reporter genes, expression vectors, DNA transfection, and luciferase assays.
The acyl-coenzyme A oxidase triple PPARγ response element (PPRE) luciferase reporter gene [(AOX)3LUC], PPARγ-GAL4, pCMX-PPARγ, and UAS5E1B-TATALUC, the adenoviral vectors for ecdysone receptors (Ad-DB-Ecr-IRES-GFP and DB-Ecr), and retroviral vectors pBPSTR1 cyclin D1 sense and MSCV-IRES-GFP (9) were previously described (19, 39, 56). Cyclin D1 mutants were generated by PCR and subcloned into p3xFLAG-CMV-10 (Sigma). Cells were transfected by Superfect Transfection reagent (Qiagen, Valencia, Calif.). The HeLa, cyclin D1−/− mouse embryonic fibroblasts (MEFs,) and 3T3 cells (cyclin D1−/− and cyclin D1+/+) were previously described (1, 3a, 52). The medium was changed after 5 h, cells were treated with ligand or vehicle as indicated (see figure legends), and luciferase activity was determined after 24 h. Luciferase activity was normalized for transfection efficiency with β-galactosidase reporters as an internal control. Luciferase assays were performed at room temperature with an Autolumat LB 953 (EG&G Berthold) (55). The fold effect was determined by comparison to the empty expression vector cassette, and statistical analyses were performed using the Mann Whitney U test.
Viral infection.
The construction and preparation of the pBPSTR1-cyclin D1, ectopic, replication-defective helper virus was previously described (25). Retroviruses were prepared by transient cotransfection with helper virus into 293T cells using calcium phosphate precipitation (http://www.stanford.edu/group/nolan). cyclin D1−/− mouse embryonic fibroblasts (MEFs) were centrifuged for 90 min at 400 × g rpm at room temperature in DMEM medium with 10% fetal bovine serum, half volume of fresh retroviral supernatants, and 8 μg of polybrene/ml and then incubated overnight at 37°C in 5% CO2. The following day the media of the cyclin D1−/− MEFs was changed to DMEM with 10% fetal bovine serum and cultured for differentiation assay. A retroviral expression plasmid encoding green fluorescent protein (GFP) was included for monitoring infection efficiency (9).
Western blots and semiquantitative RT-PCR.
The antibodies were polyclonal cyclin D1 antibody Ab3 for Western blot analysis, anti-PPARγ polyclonal antibody H100, and monoclonal E8 (Santa Cruz Biotechnology), anti-S3/12 (46), and anti-guanine dissociation inhibitor (GDI) (25) as a protein loading control. The membrane was incubated with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) and washed three times with 0.05% Tween 20-phosphate-buffered saline (PBS). Immunoreactive proteins were visualized by the enhanced chemiluminescence system (Amersham, Arlington Heights, Ill.), and their abundance was quantified by phosphorimaging (computing densitometer [Image Quant, version 1.11]; Molecular Dynamics, Sunnyvale, Calif.). Semiquantitative reverse transcription (RT)-PCR was conducted using 4 μg of total liver RNA from three age-matched pairs of wild-type (wt) and cyclin D1−/− mice which was reverse transcribed with the SuperScript II kit (Invitrogen) using Oligo(dT)12-18. The cDNA was diluted threefold with water sequentially three times; 1 μl of each of the dilutions was used in a PCR. Taq polymerase (TaKaRa) was used to amplify PPARγ and β-actin using 35 cycles and 20 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s, respectively. The primers were the following: PPARγ, GTTGACACAGAGATGCCATTC (5′ primer) and GGTTCTTCATGAGGCCTG (3′ primer); β-actin, TGTTACCAACTGGGACGACA (5′ primer) and AAGGAAGGCTGGAAAAGAGC (3′ primer).
Induction of differentiation.
Primary MEFs were isolated from 14-day-postcoitus mouse embryos (1). 3T3-L1 and MEF cells were maintained at confluence for 1 day before being switched to basal differentiation medium (DMEM supplemented with 10% charcoal-stripped serum and 10 mg of insulin/liter). Differentiation was induced by serum supplemented with 0.2 mM methylisobutylxanthine (Sigma), 5 μM dexamethasone (Sigma), and insulin (Sigma) for 3 days. Subsequently, cells were maintained in basal differentiation medium supplemented with BRL49653 (0.2 μM) (P. G. Treagust [Smithkline Beecham, West Sussex, United Kingdom]) or troglitazone (5 μM) (Sanky Co., Ltd., Tokyo, Japan) or vehicle as indicated. Retroviral infection was conducted as previously described (25) (http://www.stanford.edu/group/nolan). For Oil Red-O staining, cells were fixed in 10% formaldehyde in PBS for 1 h and rinsed with water and ethanol. Cells were stained with Oil Red-O solution (six parts saturated Oil Red-O dye in isopropanol plus four parts water) at 37°C for 15 min, washed with 70% ethanol, and then rinsed with ddH2O. Cells were inspected by microscopy, and staining was quantified after incubation with 4% NP-40 in isopropanol and measuring of absorbance at 520 nm.
Ponasterone-inducible cyclin D1 antisense-IRES-GFP transgenic mice.
The cDNA for the mouse cyclin D1 gene (a gift from V. Fantl) was subjected to internal deletion of bases 294 to 640 by PstI/StuI digestion followed by blunt-end ligation. An EcoRI fragment containing the modified murine cyclin D1 cDNA was inserted in the antisense orientation into the transgene shuttle vector, EGRE3ΔMMTV-BGH PolyA (3), modified to contain the IRES-GFP component from IRES2-EGFP (Clontech). The purified DNA was microinjected at the AECOM transgenic facility.
Immunostaining of human breast tumor samples.
The studies were approved by the Albert Einstein College of Medicine Institutional Review Board. Material from 36 benign breast disease specimens was selected randomly from the Kaiser Permanent Northwest's Benign Breast Disease Registry using breast tissue obtained over 24 years. The tissues were evaluated by two pathologists using a standard template to identify histological features of benign breast disease, including proliferative and nonproliferative fibrocystic change, epithelial hyperplasia with and without atypia, and papillomas. Materials from 33 consecutive patients with infiltrating ductal or lobular carcinoma were obtained from the files of the Einstein Division of the Montefiore Medical Center. The benign and malignant material was formalin fixed and paraffin embedded. The primary antibodies were to cyclin D1 (monoclonal antibody [MAb] DCS-6, and polyclonal (Ab-3,), and to PPARγ (MAb E-8, Santa Cruz). The secondary antibodies were goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase. For immunoperoxidase staining of paraffin-embedded tissue sections, the Santa Cruz Biotechnology ABC Staining Systems were used. Briefly, specimens were incubated for 1 h in 2% normal blocking serum derived from the same species in which the secondary antibody was raised in PBS. Sections were incubated with the primary antibody for 30 min at room temperature, washed with three changes of PBS for 5 min each, incubated for 30 min at 1 μg/ml diluted in PBS with 2% normal blocking serum, and then washed with three changes of PBS for 5 min each. Incubation was for 30 min with avidin biotin enzyme reagent. More than 300 cells were counted and scored for percent immunopositivity as previously described (26).
Morphological analysis of liver tissues.
The genotyping of cyclin D1−/− mice was conducted as previously described (1). Morphological analysis of hepatic cells was determined using the methyl-green pyronin method for overall histologic study, which detects DNA and RNA in the nucleus and cytoplasm (28, 36); Oil Red-O method for staining of neutral lipids normally seen in Ito cells (28, 35); an acid phosphatase lead enzyme method using CMP as the substrate for Kupffer cells and lysosomes in hepatic cells (e.g., hepatocytes) (36); and a diaminobenzidine method at pH 9.7 for the presence of catalase in peroxisomes and microperoxisomes (34, 35). Livers from wt and knockout mice were fixed in a fixative containing cold 4% paraformaldehyde, 2% glutaraldehyde, cacodylate buffer (pH 7.4) for 3 to 5 h (22). Sections of livers were prepared on a freezing microtome after cryoprotection in increasing concentrations of sucrose. The sections were then processed by the above-described methods, after which they were viewed with the light microscope.
Structural modeling of cyclin D1/PPARγ.
The coordinates for the three-dimension model of cyclin D1 were generated based on the sequence alignment between cyclin D1 and cyclin A and the published crystal structures of cyclin A (6). Modeling was performed using the homology-modeling program PROMOD (37). In the homology model of cyclin D1, the region required for PPARγ repression (from 142 to 178) formed a helix-loop-helix. Within this region a cluster of hydrophobic residues (amino acids [aa] 137 to 148, LLXXXLLLVXXL) was identified. Since this sequence forms a helix, some of the hydrophobic residues were exposed to solvent. We therefore modeled this helix binding to the same region (residues 280 to 318 in PPARα) that corepressors and coactivators bind to. By using the published X-ray crystal structures of PPARα-corepressor (62) and PPARγ-coactivator (61) complexes and the homology model of cyclin D1, a three-dimensional model of the complex between PPARγ and cyclin D1 was generated using the molecular modeling program INSIGHT (Accelrys Inc., San Diego, Calif.). The coordinates for the whole complex were subjected to energy minimization using the AMBER force field (8).
RESULTS
Cyclin D1 repression of PPARγ transactivation.
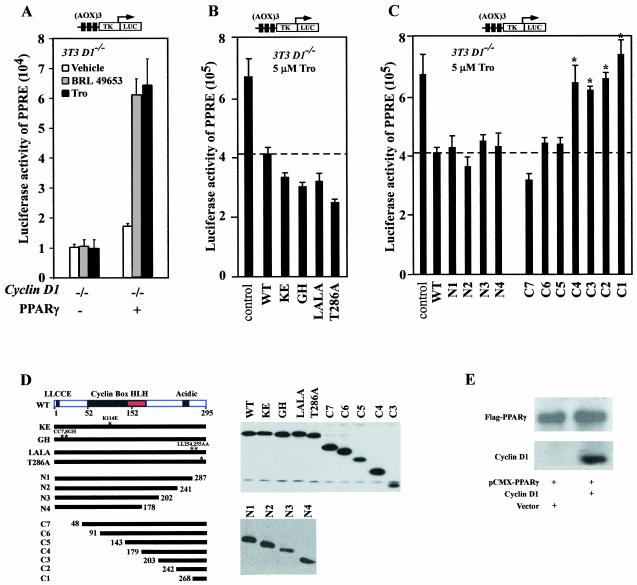
We examined the role of cyclin D1 in regulating PPARγ-dependent gene activity. Cyclin D1-deficient (cyclin D1−/−) 3T3 cells, generated from cyclin D1−/− mice (3a), were transfected with the wt PPARγ and a reporter gene encoding multimeric sequences of a PPARγ-response element from the acyl coenzyme A oxidase gene promoter, (AOX)3LUC. In the presence of a PPARγ expression plasmid, the specific ligands, BRL49653 and Troglitazone, induced (AOX)3LUC three- to fivefold. Coexpression of cyclin D1 wt in cyclin D1−/− 3T3 cells repressed ligand-induced (AOX)3LUC reporter activity by 40% (Fig. 1B). In order to identify the mechanisms by which cyclin D1 repressed liganded PPARγ-dependent gene transcription, point mutations of cyclin D1 were assessed (Fig. 1B to D). Each of the mutants was expressed as well as the wt in cultured cells (Fig. 1D). The cyclin D1 KE mutant is defective in binding cdks, the GH mutant is defective in pRB binding, and the 254/255 mutant is defective in binding the p160 coactivators (66). Each of these cyclin D1 mutants inhibited liganded PPARγ reporter activity to the same level as cyclin D1 wt (Fig. 1B). The T286A mutant, which evades GSK3-β phosphorylation in vitro and remains nuclear throughout the cell cycle in cultured cells (4), repressed PPARγ activity to a level similar to that with cyclin D1 wt. Sequential C-terminal deletion of cyclin D1 to N178 or amino-terminal deletion of the cdk- and pRB-binding regions maintained repression. N-terminal deletion from 143 to 179 (C4) abolished repression, indicating a requirement for residues between 143 and 179 for full repression (Fig. 1C). Together these studies suggest that cyclin D1 inhibits liganded PPARγ activity through a cdk-independent mechanism that requires the region of cyclin D1 between residues 143 and 179. To assess the effect of cyclin D1 on expression levels of PPARγ from the viral expression plasmid, Western blotting was conducted with 293 cells either transfected with pCMV-PPARγ alone or in the presence of cotransfected cyclin D1 expression vector. The relative abundance of PPARγ from the expression plasmid, assessed by the Flag epitope of PPARγ, was not affected by cyclin D1 (Fig. 1E).
FIG. 1.
The region of cyclin D1 required for repression of PPARγ activity. (A) Cyclin D1−/− 3T3 cells were transfected with reporter plasmid (AOX)3TK LUC carrying response elements specific for PPARγ (1 μg). prl-TK LUC was cotransfected as an internal control. Cells were cotransfected with the expression vector encoding human PPARγ in either the presence or absence of cyclin D1 (pCMV-cyclin D1) or (B and C) expression vectors for cyclin D1 mutants. The effect of the coexpressed cyclin D1 construction on ligand-induced luciferase reporter activity is shown as mean ± standard error of the mean. Activity of the parental prl-TK Luc reporter, which lacks the (AOX)3 element, was not affected by cyclin D1. (D) Western blotting for the Flag epitope of the cyclin D1 constructs. (E). Western blot of cells transfected with the pCMV-cyclin D1 and PPARγ expression vector.
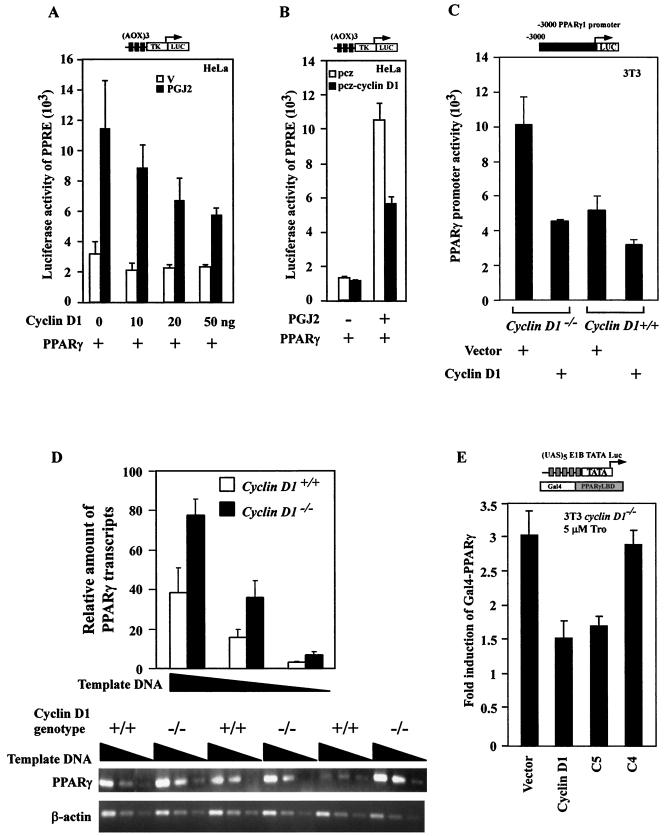
We examined further the role of cyclin D1 in regulating PPARγ-dependent gene activity using HeLa cells as previously described (54), transfecting cells with the wt PPARγ and a reporter gene (AOX)3LUC (Fig. 2A). Addition of 15d-PGJ2, which is also capable of activating PPARγ, induced reporter activity 3.5-fold. Coexpression of cyclin D1 inhibited 15d-PGJ2-induced PPARγ-dependent reporter activity by 60% (Fig. 2A). Cyclin D1 repressed ligand-induced (AOX)3LUC reporter activity at several different concentrations of vector (Fig. 2A) and ligand (1, 2, 5, 5, and 10 μm) (data not shown). Transfection of cells with a tetracycline-regulated expression vector for cyclin D1 repressed 15d-PGJ2-induced (AOX)3LUC reporter activity by 60% (Fig. 2B). 15d-PGJ2-induced (AOX)3LUC reporter activity was unaffected by either cyclin D2 or cyclin D3. The activities of the viral promoters (CMV-LUC and RSV-LUC) and the cyclin E, c-Jun, and JunB luciferase reporter vectors were not regulated by cyclin D1 (39; also data not shown). We next assessed the PPARγ1 promoter, since it is a PPARγ-responsive reporter gene (15). PPARγ1 promoter activity was twofold more active in cyclin D1−/− 3T3 compared with wt 3T3 cells, normalized for transfection efficiency. The PPARγ1 promoter was repressed two- to threefold by cyclin D1 overexpression in cyclin D1−/− 3T3 cells (Fig. 2C). The relative abundance of PPARγ1 mRNA was assessed by semiquantitative RT-PCR using liver cell extracts from either cyclin D1 wt or cyclin D1−/− mice. The relative abundance of PPARγ1 mRNA was increased twofold in the cyclin D1−/− mice (Fig. 2D). To determine whether cyclin D1 was capable of inhibiting the transactivation domain of PPARγ, a heterologous reporter system was used. To assess DNA-binding independent PPARγ activity, the PPARγ LBD linked to the Gal4 DNA binding domain was used with a reporter construct consisting of a multimeric DNA binding site for the Gal4 DNA binding domain linked to the luciferase reporter gene. The heterologous reporter system was introduced into cyclin D1-deficient (cyclin D1−/−) 3T3 cells, and luciferase activity was normalized to that of a cotransfected Renilla luciferase reporter gene. Cotransfection of the expression vector for cyclin D1 into cyclin D1−/− 3T3 cells inhibited PPARγ-Gal4 activity 50% (Fig. 2E). As with repression of liganded (AOX)3LUC reporter activity, the region from 143 to 179 was required for repression of PPARγ-Gal4 activity.
FIG.2.
Cyclin D1 inhibits liganded PPARγ transactivation function. (A) The (AOX)3 luciferase reporter (1 μg) was transfected into HeLa cells with the expression vector encoding the human PPARγ in either the presence or absence of cyclin D1 (pCMV-cyclin D1). Comparison was made with the effect of the expression of equal amounts of empty expression vector cassette (pRC/CMV). 15d-PGJ2 (10 μM) was added as indicated. The results are shown as mean ± standard error of the mean throughout. (B) (AOX)3LUC reporter activity in HeLa cells transfected with tetracycline-inducible vector pcz-cyclin D1. (C). PPARγ1 promoter activity in cyclin D1−/− or cyclin D1+/+ 3T3 cells cotransfected with pCMV-cyclin D1 or control vector as indicated. (E) Semiquantitative RT-PCR for PPARγ1 from mRNA of livers of cyclin D1−/− or cyclin D1+/+ mice. The PPARγ LBD construct linked to the Gal4 DNA binding domain was assessed for activity using the heterologous reporter (UAS)5E1BTATA LUC in the presence or absence of the expression vectors for cyclin D1 in cyclin D1−/− 3T3 cells. The relative transactivation level was shown as luciferase activity represented by light units measured in cells cotransfected with a specific receptor expression plasmid. Reporter gene activity was normalized to prl-TK LUC activity.
PPARγ expression is reduced in human benign breast disease and cancers correlating with increased cyclin D1 abundance.
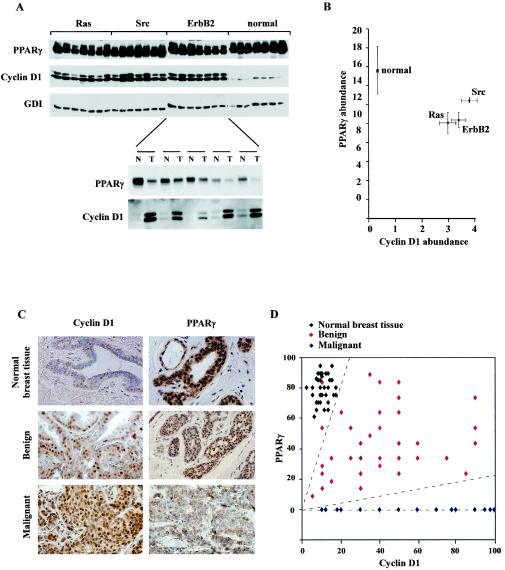
We determined PPARγ abundance in murine mammary tumors, reasoning that expression of cyclin D1 and PPARγ may be reciprocal. In mammary tumors induced by mammary tissue-targeted oncogenes (ErbB2, Ras, and Src), the relative abundance of cyclin D1 was increased and that of PPARγ was decreased compared with results for normal mammary epithelium (Fig. 3A and B). Cyclin D1 abundance was also increased in the mammary tumor compared with that in the adjacent mammary epithelium in the same animal, with reciprocal changes in PPARγ expression (Fig. 3A and B).
FIG. 3.
PPARγ and cyclin D1 abundance in mammary tumors induced by distinct oncogenes and human benign and malignant breast disease. (A) Expression of PPARγ and cyclin D1 in murine mammary tumors. Western blotting of normal mammary epithelium or mammary tumors of MMTV-directed oncogene mice (MMTV-Ras [Ras], MMTV-Src [Src], or MTV-ErbB2 [ErbB2]) is shown. Abundance of PPARγ, cyclin D1, or the loading control GDI is shown. MMTV-ErbB2 mammary tumors (T) and adjacent normal mammary epithelium (N) are represented below. (B). The mean levels of cyclin D1 and PPARγ protein are shown for n > 7 tumors induced by each oncogene, compared with normal mammary epithelium. (C). Representative immunostaining of normal mammary epithelium, benign breast disease, and ERα+ infiltrating breast cancers (n = 40) (magnification, ×388). (D). The percent immunopositive staining for PPARγ and cyclin D1 is shown for premalignant lesions (n = 36) with comparison to normal mammary epithelium.
Immunohistochemical assessment was made of cyclin D1 and PPARγ in normal breast epithelium, benign breast lesions, and infiltrating ERα+ breast cancers (Fig. 3C). PPARγ was detectable in normal mammary epithelium as previously reported (32). Preimmune sera showed no nuclear staining of breast epithelial cells (not shown). The majority of human breast cancers were cyclin D1 immunopositive and PPARγ negative. The samples of human benign disease included a spectrum of histopathological subtypes (nonproliferative, proliferative [epithelial cell hyperplasia with or without atypia, sclerosing adenosis, papilloma, and fibroadenoma]). The benign breast lesions displayed increased cyclin D1 immunopositivity and reduced PPARγ staining compared with normal mammary epithelium (Fig. 3D). Thus, in human ERα-positive breast tumors, reduced PPARγ abundance is found in conjunction with increased cyclin D1 levels.
Inhibition of PPARγ ligand-induced differentiation by cyclin D1.
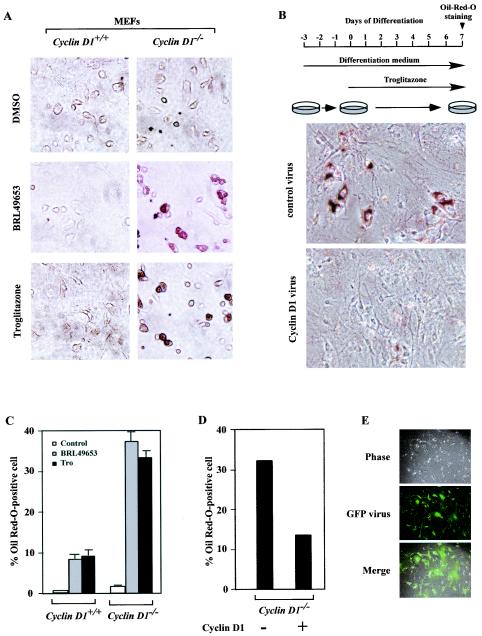
Although cyclin D1 inhibited PPARγ-responsive reporter gene activity, it was important to determine whether cyclin D1 inhibited the in vivo function of liganded PPARγ. Liganded PPARγ is both necessary and sufficient for the induction of Oil Red-O positive staining as a marker of adipocyte phenotype in immortalized 3T3 cells (40). We compared PPARγ ligand-regulated adipogenesis in cyclin D1+/+ versus cyclin D1−/− MEFs prior to immortalization. 3T3-L1 adipogenesis medium was not capable of inducing MEF differentiation into adipocytes. Addition of the PPARγ agonists rosiglitazone (0.2 μM) or troglitazone (5 μM) induced a modest (8% positive) but significant lipid droplet accumulation in cyclin D1+/+ wt MEFs. In contrast, more than 35% of the cyclin D1-deficient MEFs showed Oil Red-O positive staining (Fig. 4A and C).
FIG. 4.
Cyclin D1 deficiency enhances PPARγ function. (A) MEFs derived from either cyclin D1+/+ or cyclin D1−/− mice were treated with vehicle, BRL-49653 (1 μM), and troglitazone (5 μM), and lipid accumulation was assessed. cyclin D1−/− MEFs exhibit lipid accumulation by OilRed-O staining in the presence of BRL-49653 or troglitazone, shown as mean ± standard error of the mean for three experiments in panel C. (B) cyclin D1−/− MEFs were infected with either a parental retrovirus or retroviruses encoding cyclin D1. Two days postconfluence, cells were treated with differentiation media and troglitazone (5 μM). Seven days later, cells were stained with Oil Red-O to visualize the degree of lipid accumulation. The results were quantitated for two separate experiments (D). Representative infection of cyclin D1−/− MEFs with viral GFP expression construct is shown. Tro, troglitazone; DMSO, dimethyl sulfoxide.
These findings suggest that the deficiency of cyclin D1 or factors regulated by cyclin D1 contributes to the adipocyte phenotype induced by the specific PPARγ ligands. To determine whether cyclin D1 deficiency of the cyclin D1−/− MEFs was the key regulator of the PPARγ ligand-induced adipocyte phenotype, cyclin D1−/− MEFs were infected with a retroviruses encoding cyclin D1 prior to the differentiation protocol. Expression of cyclin D1 inhibited rosiglitazone (0.2 μM)-induced or troglitazone (5 μM)-induced lipid accumulation in the cyclin D1−/− MEFs (Fig. 4B and D). The transduction efficiency of the MEFs was >90%, assessed by transduction with the viral vector encoding GFP (Fig. 4E). These studies provide genetic evidence that cyclin D1 is necessary and sufficient to regulate the adipogenic differentiation function of PPARγ.
Reduction of cyclin D1 by ponasterone-regulated cyclin D1 antisense transgenic mice induces PPARγ abundance.
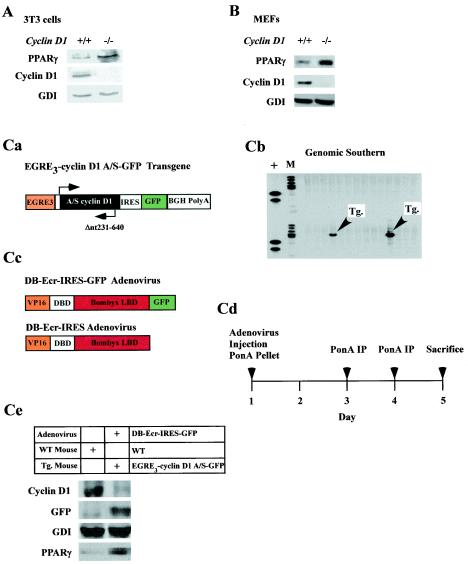
The correlative reciprocal expression profile of cyclin D1 and PPARγ in mammary epithelium was assessed further to determine whether cyclin D1 inhibited PPARγ expression. PPARγ levels were increased in untreated cyclin D1−/− 3T3 cells (Fig. 5A) and cyclin D1−/− MEFs (Fig. 5B), suggesting that cyclin D1 may inhibit PPARγ expression, consistent with our finding that cyclin D1 repressed the PPARγ1 promoter. To examine further whether cyclin D1 inhibited PPARγ abundance in vivo, we generated transgenic mice in which the murine cyclin D1 antisense cDNA linked to a GFP transgene was regulated under control of the ecdysone enhancer (Fig. 5Ca). We had previously used ponasterone-inducible cyclin D1 antisense to reduce cyclin D1 abundance in cultured cells (63). Transgene transmission was confirmed by genomic Southern blotting (Fig. 5Cb). To induce expression of the transgene, an ecdysone receptor (Bbyx), adenoviruses were used that expresses GFP from a second cistron (DB-Ecr-IRES-GFP, Fig. 5Cc). The entire experiment was conducted on two separate occasions, and representative results are shown. Comparison was made with equal titer of an adenovirus for the ecdysone receptor that does not express GFP.
FIG. 5.
Transgenic inducible cyclin D1 antisense regulates PPARγ abundance in vivo. Western blot of cyclin D1−/− 3T3 (A) and cyclin D1−/− MEFs (B) with antibodies as indicated. (Ca) An ecdysone enhancer-driven cyclin D1 antisense transgene (schematic shown), was used to generate transgenic mice (Tg) confirmed by genomic Southern blot (Cb). (Cc) Adenoviral vectors encoding receptors for the Bombyx LBD either with or without a second cistron for GFP were used to infect the cyclin D1 antisense transgenic mice (Cd). (Ce) Transgenic mice were treated with ponasterone to induce transgene expression, and Western blotting of hepatic extracts was performed at 5 days for the loading control (GDI), cyclin D1 abundance and GFP from the second cistron of the cyclin D1 antisense, and PPARγ.
The adenoviral receptors encoding the ecdysone receptor were introduced into transgenic cyclin D1 antisense mice or control strain-matched mice, and activity of the receptors was induced using ponasterone pellets (Fig. 5Cd). Serum samples demonstrated the induction of ecdysone enhancer-driven SEAP or luciferase activity in parallel experiments (data not shown). Analysis was performed on the livers of transgenic and antisense transgenic mice. PPARγ is expressed in hepatocytes, and the expression of PPARγ is induced by TZDs in vivo (13). It has been hypothesized that the insulin-sensitizing effects of TZDs may result in part from effects on PPARγ expression and function in muscle and liver (12, 13). The induction of the GFP from the second cistron of the cyclin D1 antisense transgene was observed by Western blotting (Fig. 5Ce). Analysis of hepatic tissue evidenced the presence of GFP by immunofluorescence and by Western blotting with mice transduced with the DB-Ecr-IRES-GFP adenovirus, used as a positive control for GFP in parallel experiments (data not shown). The abundance of cyclin D1 protein was reduced by 90% for the cyclin D1 antisense transgenic mice compared with results for control mice (n = 2). The reduction in cyclin D1 protein levels correlated with the induction in PPARγ abundance in vivo (Fig. 5Ce), providing further evidence that cyclin D1 inhibits PPARγ expression in vivo.
Cyclin D1 inhibits PPARγ ligand-induced gene expression without affecting C/EBPα or C/EBPβ.
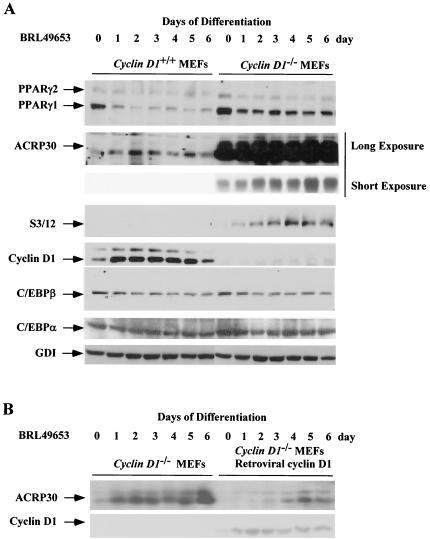
The expression of genes known to be either upstream (C/EBPβ) (60) or downstream (adipocyte complement related protein of 30 kDa [ACRP30], C3/12) of PPARγ in the adipocyte differentiation program was next assessed. C/EBPα-mediated adipocyte differentiation requires PPARγ; however, PPARγ-mediated differentiation is independent of CEBPα, placing PPARγ as a downstream effector of CEBPα (40). Analysis was performed during differentiation of cyclin D1+/+ and cyclin D1−/− MEFs. Normalization of protein loading was performed using GDI (25). Cells were harvested every day for 7 days and lysed, and the cell lysates were subjected to Western blotting for ACRP30 and C3/12. Prior to the induction of differentiation by the PPARγ ligands BRL49653 (Fig. 6) and troglitazone (data not shown), cyclin D1 was detectable in the cyclin D1+/+ but not the cyclin D1−/− MEFs. PPARγ levels were increased two- to threefold in the cyclin D1−/− MEFs compared with results in the cyclin D1 +/+ MEFs (Fig. 6), consistent with the finding that cyclin D1 inhibits both PPARγ expression and transactivation. In the undifferentiated state, cyclin D1+/+ and cyclin D1−/− MEFs expressed similar levels of C/EBPα and C/EBPβ.
FIG. 6.
Enhanced PPARγ-responsive gene expression in cyclin D1−/− cells. (A) MEFs derived from either cyclin D1+/+ or cyclin D1−/− mice were treated with BRL-49653 (1 μM). Western blotting was conducted for PPARγ and the PPARγ-responsive genes, ACRP30 (11) and S3/12. Levels of cyclin D1, C/EBPβ, C/EBPα, and the loading control (GDI) are shown. (B) Western blot analysis of cyclin D1−/− MEFs or cyclin D1−/− MEFs infected with a cyclin D1 retrovirus. MEFs were treated for the time points indicated.
ACRP30 (adiponectin/apM1/AdipoQ/GBP28) is an adipocyte-secreted protein induced during fat cell differentiation (5) induced by the PPARγ ligand TZD (13). At day 6, the levels of the adipocyte differentiation markers downstream of PPARγ (ACRP30 and S3/12) were substantially higher in the cyclin D1−/− MEFs than in the cyclin D1+/+ MEFs. The levels of ACRP30 was >100-fold higher in the cyclin D1−/− MEFs than in the wt MEFs. In both wt and cyclin D1−/− MEFs, the levels of ACRP30 were increased 10-fold by BRL49653 or troglitazone. Upon induction of differentiation, both cyclin D1+/+ and cyclin D1−/− MEFs showed a similar pattern of C/EBPα and C/EBPβ expression. These studies suggest that inhibition of adipocyte differentiation by cyclin D1 occurs distal to C/EBPs and involves inhibition of PPARγ function. Transduction of cyclin D1−/− MEFs with a cyclin D1 retrovirus inhibited the induction of differentiation by BRL49653 and reduced the time-dependent induction of ACRP30 (Fig. 6B).
Adipogenic steatosis in cyclin D1−/− mouse liver.
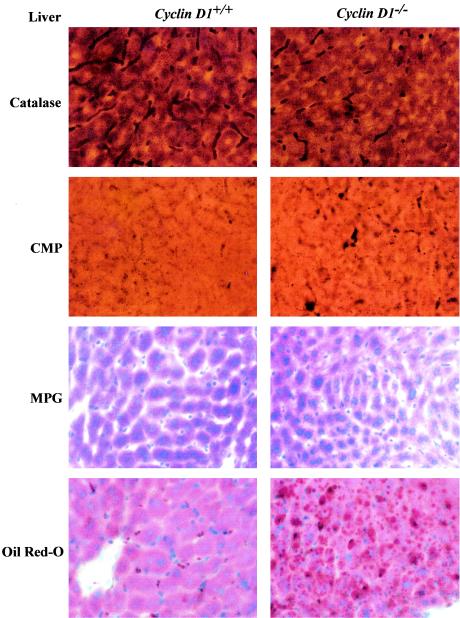
In previous studies, adenoviral delivery of PPARγ1 overexpression in the murine liver induced hepatic steatosis (65). If repression of PPARγ1 activity by cyclin D1 was functionally significant in vivo, it would be predicted that the cyclin D1−/− mice would demonstrate features of hepatic steatosis. Detailed morphometric and histological analysis was therefore conducted of the livers from cyclin D1−/− and wt littermate controls. Acid phosphatase cytochemistry revealed that the Kupffer cells in the cyclin D1−/− mice were enlarged, appeared increased in numbers (Fig. 7), and developed an extensive lysosomal system filled with lipid deposits (Fig. 7). In addition, acid phosphatase cytochemistry revealed that the lysosomes of hepatocytes appeared larger and appeared to be distributed more widely in the cytoplasm in contrast to the wt hepatocytes lysosomes, which were more concentrated near the bile canaliculus. Neutral lipid staining using Oil Red-O showed a substantial increase in neutral lipid droplets in the cyclin D1−/−. Ito cells from cyclin D1−/− mice were increased in numbers and enlarged, with more numerous and larger lipid spheres, compared with Ito cells of wt littermate controls. Changes were also found in hepatocytes and included increased numbers of microperoxisomes, and cytoplasmic lipid spheres were noted (Fig. 7). Thus, the livers of cyclin D1−/− mice display the features of hepatic steatosis consistent with increased PPARγ activity.
FIG. 7.
Morphological analysis of hepatic cells from cyclin D1 wt and cyclin D1−/− mice. Hepatic tissues were stained for catalase, CMP, methyl-green pyronin (MPG), and Oil Red-O for staining of neutral lipids as described in Materials and Methods.
Three-dimensional modeling of the cyclin D1 143-179 region.
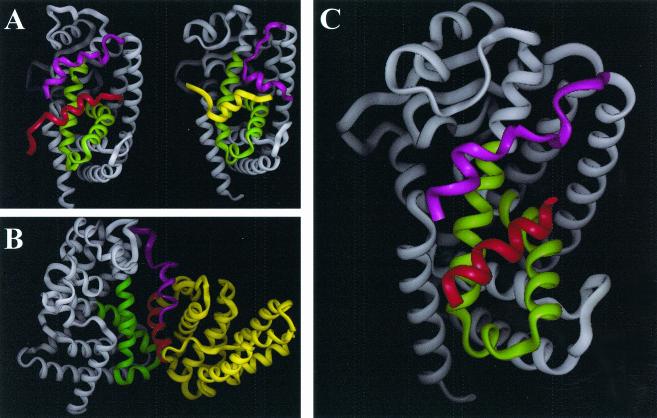
In view of the importance of the region of cyclin D1 from 143 to 179 in repression of PPARγ transactivation, we determined the predicted structure of this domain using the homology modeling program PROMOD (37) (see Materials and Methods). The coordinates for the three-dimensional model of cyclin D1 were generated based on the sequence alignment between cyclin D1 and cyclin A and the published crystal structures of cyclin A (6). We generated models from the published crystal structure of PPARα corepressor (62) (Fig. 8A, left) and PPARγ-coactivator (61) (Fig. 8A, right). The coactivator is shown in yellow. PPARγ is shown in grey with the region of PPARγ contacting the coactivator shown in green. In the homology model of cyclin D1, the region from 142 to 179 forms a helix-loop-helix, shown in Fig. 8B in yellow and red. The cluster of hydrophobic residues (aa 137 to 148, LLXXXLLLVXXL) was then modeled binding to the same region where corepressors and coactivators bind to PPARγ (61). Using these two PPAR structures and the homology model of cyclin D1, a three-dimensional model of the complex between PPARγ and cyclin D1 was generated (Fig. 8C) (INSIGHT; Accelrys Inc., San Diego, Calif.). The hydrophobic cluster region of the cyclin D1 HLH is shown in red in Fig. 8C.
FIG. 8.
Prediction model of cyclin D1 helix-loop-helix region binding to PPARγ. (A) Ribbon model of left panel: nuclear corepressor (red) binding to PPARα (grey, with helix 12 in magenta and coactivator binding region of PPARα in green); right panel, nuclear receptor coactivator (yellow) binding to PPARγ (grey, with helix 12 in magenta and coactivator binding region of PPARγ in green). (B). Ribbon model of PPARγ and cyclin D1 helix-loop-helix region. Predicted cyclin D1 helix-loop-helix structure is in yellow with component of hydrophobic cluster in red. (C). PPARγ (grey, with helix 12 in magenta and coactivator binding region of PPARγ in green) and cyclin D1 hydrophobic cluster of helix-loop-helix motif colored red. (The position alignment, determined by energy minimization using the AMBER force field, displays the cyclin D1 helix-loop-helix motif in an alignment similar to that of a nuclear receptor corepressor).
DISCUSSION
The present studies demonstrate for the first time in vivo antagonism between a collaborative oncogene, cyclin D1, and a nuclear receptor, PPARγ. Cyclin D1 inhibited PPARγ-dependent reporter activity and repressed the trans activity of PPARγ when linked to a heterologous DNA binding domain. Consistent with previous findings that PPARγ expression is induced by PPARγ (12), cyclin D1 also inhibited PPARγ expression and promoter activity. The reduction in cyclin D1 abundance in cyclin D1−/− 3T3 cells, cyclin D1−/− MEFs, and cyclin D1 antisense transgenic mice correlated with the induction of PPARγ expression (Fig. 5). The induction of adipocyte differentiation by PPARγ-specific ligands was substantially enhanced in cyclin D1-deficient cells. The reintroduction of cyclin D1 into cyclin D1-deficient cells abolished the adipogenic phenotype, consistent with a key role for cyclin D1 as an inhibitor of PPARγ-specific functional activity. Since cyclin D1 abundance is regulated by diverse oncogenic and mitogenic stimuli, the inhibition of PPARγ transactivation by cyclin D1 may have important implications for signal transduction and tumorigenesis.
Repression of PPARγ transactivation by cyclin D1 was independent of the cdk and pRb binding functions and required a C-terminal region of cyclin D1 that is predicted to form a helix-loop-helix structure. The mechanism by which cyclin D1 regulated PPARγ activity is thus distinct from that regulating the ERα through recruiting a p160 coactivator SRC-1 in vitro (66), as an SRC-1 binding point mutation of cyclin D1 maintained wt repression of liganded PPARγ activity. Herein, genetic deletion of cyclin D1 enhanced PPARγ ligand-mediated differentiation of MEFs into adipocytes. Although interactions between cyclin D1 and nuclear receptors have been previously described, the present studies provide strong genetic evidence for a functional interaction between cyclin D1 and a nuclear receptor. Ectopic expression of PPARγ induced differentiation of NIH-3T3 fibroblast cells into fat-laden adipocyte cells (51). In the present studies, the cyclin D1−/− MEF adipogenic phenotype induced by PPARγ ligands was reversed by cyclin D1 overexpression. ACRP30 was increased 50-fold in the cyclin D1−/− MEFs, consistent with the enhanced induction of the adipogenic phenotype. The relatively modest increase in PPARγ abundance, together with the dramatic enhancement of PPARγ activity in the cyclin D1−/− MEFs, is consistent with the reporter gene studies in which cyclin D1 inhibited PPARγ trans activity.
These studies raise the possibility that reduced PPARγ expression, together with increased cyclin D1, may be a genetic feature of the transition from normal breast epithelium, to benign breast disease and adenocarcinoma. PPARγ immunopositivity was decreased in benign breast disease compared with normal mammary epithelium and was reduced further in adenocarcinomas. Cyclin D1 immunopositivity increased from normal epithelium to benign disease and adenocarcinomas. The reduction in PPARγ expression in the cyclin D1-infected MEFs, together with the finding of increased levels of PPARγ mRNA and protein in cyclin D1−/− livers by microarray (not shown) and Western blotting, suggest that cyclin D1 inhibits PPARγ expression. The overexpression of cyclin D1 with ERα reflects poor prognosis in human breast cancer. Given the repression of PPARγ function and expression by cyclin D1 and the cytoinhibitory role of PPARγ in breast epithelium, these studies raise the question of whether reduced PPARγ may contribute to poor prognosis in a subset of patients. The reduction in PPARγ staining in proliferative breast disease suggests that further studies of PPARγ as a prognostic indicator and candidate target for prevention or therapy of human breast cancer warrants consideration.
The present studies are important in demonstrating a functional antagonism between a collaborative oncogene, cyclin D1, and a candidate tumor suppressor, PPARγ. Several lines of evidence suggest that PPARγ may function as a tumor suppressor (42). Consistent with a role for PPARγ as an inhibitor of tumorigenesis, heterozygous mutations of PPARγ were detected in 4 of 55 patients with colon cancer (45). In follicular thyroid cancer, a fusion oncoprotein has been described, formed by a chromosomal translocation between PPARγ1 and PAX8 with a deletion in its C-terminal activation domain. The PPARγ fusion protein functioned as a powerful dominant-negative of wt PPARγ and was not observed in benign follicular adenomas (24). The addition of PPARγ ligands (TZD or 15d-PGJ2) inhibited breast and colonic cellular proliferation (7, 14, 32, 52). In contrast, cyclin D1 abundance is induced by diverse oncogenic and mitogenic signals in breast and colonic epithelial cells and functions as a collaborative oncogene (18, 38). Cyclin D1 antisense inhibits the growth of murine mammary tumors derived from MMTV-ErbB2 mice (25), and cyclin D1−/− mice are resistant to the induction of tumor formation by ErbB2 (64). Since PPARγ inhibits the expression of several genes promoting tumor invasion (those encoding iNOS, gelatinase B, matrix metalloproteinases [10, 21, 30], and UPA) (data not shown), cyclin D1 antagonism of PPARγ function may enhance expression of tumor invasion genes.
The functional antagonism between PPARγ and cyclin D1 may also have implications for signal transduction cross talk. Cyclin D1 is induced by diverse mitogenic signaling pathways, including those of Src, Rac mutants, Dbl proteins, and β-catenin, and the NF-κB signaling pathway (1, 2, 17, 26, 49, 55, 56, 58, 59). PPARγ activity is also induced by a large number of synthetic and natural ligands, including prostaglandins and fatty acids (42, 50). The inhibition of PPARγ function by cyclin D1 may contribute to altered metabolism and altered inflammatory responses and remains to be further explored.
Acknowledgments
We thank R. Evans, V. Fantl, C. Glass, P. Sicinski, and M. Lazar for reagents and helpful discussion. We thank M. Caparas for assistance with the manuscript.
This work was supported in part by awards from the Susan Komen Breast Cancer Foundation, Breast Cancer Alliance Inc., R01CA70896, R01CA75503, R01CA86072, R01CA86071 (R.G.P.), R03AG20337 (C.A.), NIH CA06576 (P.N.), and R01DK55758 (P.E.S.). R.G.P was a recipient of the Irma T. Hirschl and Weil Caulier award and was the Diane Belfer Faculty Scholar in Cancer Research. M.D. was a recipient of the New York State mentored EMPIRE award. Work conducted at the Lombardi Cancer Center was supported by the NIH Cancer Center Core grant.
REFERENCES
- 1.Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 3.Albanese, C., A. Reutens, M. D'Amico, B. Boumediene, M. Fu, T. Link, R. Nicholson, R. A. Depinho, and R. G. Pestell. 2000. Sustained mammary gland directed ponasterone A-inducible expression in transgenic mice. FASEB J. 14:877-884. [DOI] [PubMed] [Google Scholar]
- 3a.Albanese, C., K. Wu, M. D’Amico, C. Jarrett, D. Joyce, J. Hughes, J. Hulit, T. Sakamaki, M. Fu, A. Ben-Ze’ev, J. F. Bromberg, C. Lamberti, U. Verma, R. B. Gaynor, S. W. Byers, and R. G. Pestell. 2003. IKKα regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14:585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alt, J. R., J. L. Cleveland, M. Hannink, and J. A. Diehl. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14:3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, A. H., T. Combatsiaris, X. L. Du, M. Brownlee, and P. E. Scherer. 2001. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 7:947-953. [DOI] [PubMed] [Google Scholar]
- 6.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. H. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockman, J. A., R. A. Gupta, and R. N. Dubois. 1998. Activation of PPARgamma leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology 115:1049-1055. [DOI] [PubMed] [Google Scholar]
- 8.Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, J. Wang, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, H. Gohlke, R. J. Radmer, Y. Duan, J. Pitera, I. Massova, G. L. Seibel, U. C. Singh, W. P. K., and K. P. A. 2002. AMBER 7. University of California, San Francisco, Calif.
- 9.Chalandon, Y., X. Jiang, G. Hazlewood, S. Loutet, E. Conneally, A. Eaves, and C. Eaves. 2002. Modulation of p210BCR-ABL activity in transduced primary human hematopoietic cells controls lineage programming. Blood 99:3197-3204. [DOI] [PubMed] [Google Scholar]
- 10.Chawla, A., Y. Barak, L. Nagy, D. Liao, P. Tontonoz, and R. M. Evans. 2001. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 7:48-52. [DOI] [PubMed] [Google Scholar]
- 11.Combs, T. P., J. A. Wagner, J. Berger, T. Doebber, W. J. Wang, B. B. Zhang, M. Tanen, A. H. Berg, S. O'Rahilly, D. B. Savage, K. Chatterjee, S. Weiss, P. J. Larson, K. M. Gottesdiener, B. J. Gertz, M. J. Charron, P. E. Scherer, and D. E. Moller. 2002. Induction of adipocyte complement-related protein of 30 kilodaltons by PPARgamma agonists: a potential mechanism of insulin sensitization. Endocrinology 143:998-1007. [DOI] [PubMed] [Google Scholar]
- 12.Davies, G. F., R. L. Khandelwahl, and W. J. Roesler. 1999. Troglitazone induces expression of PPARγ in liver. Mol. Cell. Biol. Res. Commun. 2:202-208. [DOI] [PubMed] [Google Scholar]
- 13.Davies, G. F., P. J. McFie, R. L. Khandelwahl, and W. J. Roesler. 2002. Unique ability of troglitazone to upregulate peroxisome proliferator-activated receptor-g expression in hepatocytes. J. Pharmacol. Exp. Ther. 300:72-77. [DOI] [PubMed] [Google Scholar]
- 14.Elstner, E., C. Muller, K. Koshizuka, E. A. Williamson, D. Park, H. Asou, P. Shintaku, J. W. Said, D. Heber, and H. P. Koeffler. 1998. Ligands for peroxisome proliferator-activated receptorγ and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc. Natl. Acad. Sci. USA 95:8806-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajas, L., K. Schoonjans, L. Gelman, J. B. Kim, J. Najib, G. Martin, J. C. Fruchart, M. Briggs, B. M. Spiegelman, and J. Auwerx. 1999. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol. Cell. Biol. 19:5495-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganter, B., S.-L. Fu, and J. S. Lipsick. 1998. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 17:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin. 1999. NF-κB controls cell growth and differentiation through the transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D., and R. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 19.Hoppe, U. C., E. Marban, and D. C. Johns. 1999. Adenovirus-mediated inducible expression in vivo by a hybrid ecdysone receptor. Mol. Ther. 1:159-164. [DOI] [PubMed] [Google Scholar]
- 20.Inoue, K., and C. J. Sherr. 1998. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol. Cell. Biol. 18:1590-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, C., A. T. Ting, and B. Seed. 1998. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature 391:82-86. [DOI] [PubMed] [Google Scholar]
- 22.Karnovsky, M. J. 1965. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J. Cell Biol. 27:137A-138A.
- 23.Kenny, F. S., R. Hui, E. A. Musgrove, J. M. Gee, R. W. Blamey, R. I. Nicholson, R. L. Sutherland, and J. F. Robertson. 1999. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin. Cancer Res. 5:2069-2076. [PubMed] [Google Scholar]
- 24.Kroll, T. G., P. Sarraf, L. Pecciarini, C. J. Chen, E. Mueller, B. M. Spiegelman, and J. A. Fletcher. 2000. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma. Science 289:1357-1360. [DOI] [PubMed] [Google Scholar]
- 25.Lee, R. J., C. Albanese, M. Fu, M. D'Amico, B. Lin, G. Watanabe, G. K. I. Haines, P. M. Siegel, M. C. Hung, Y. Yarden, J. M. Horowitz, W. J. Muller, and R. G. Pestell. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, R. J., C. Albanese, R. J. Stenger, G. Watanabe, G. Inghirami, G. K. I. Haines, M. Webster, W. J. Muller, J. S. Brugge, R. J. Davis, and R. G. Pestell. 1999. pp60v-src induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways: a role for cAMP response element-binding protein and activating transcription factor-2 in pp60v-src signaling in breast cancer cells. J. Biol. Chem. 274:7341-7350. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre, A. M., I. Chen, P. Desreumaux, J. Najib, J. C. Fruchart, K. Geboes, M. Briggs, R. Heyman, and J. Auwerx. 1998. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nat. Med. 4:1053-1057. [DOI] [PubMed] [Google Scholar]
- 28.Lillie, R. D. 1965. Histopathologic technic and practical histochemistry, 3rd ed. McGraw-Hill Book Co., Inc., New York, N.Y.
- 29.Lukas, J., J. Bartkova, and J. Bartek. 1996. Convergence of mitogenic signalling cascades from diverse classes of receptors at the cyclin D-cyclin dependent kinase-pRb-controlled G1 checkpoint. Mol. Cell. Biol. 16:6917-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx, N., F. Mach, A. Sauty, J. H. Leung, M. N. Sarafi, R. M. Ransohoff, P. Libby, J. Plutzky, and A. D. Luster. 2000. Peroxisome proliferator-activated receptor-gamma activators inhibit IFN-gamma-induced expression of the T cell-active CXC chemokines IP-10, Mig, and I-TAC in human endothelial cells. J. Immunol. 164:6503-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahon, C., T. Suthiphongchai, J. DiRenzo, and M. E. Ewen. 1999. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc. Natl. Acad. Sci. USA 96:5382-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, E., P. Sarraf, P. Tontonoz, R. M. Evans, K. J. Martin, M. Zhang, C. Fletcher, S. Singer, and B. M. Spiegelman. 1998. Terminal differentiation of human breast cancer through PPARγ. Mol. Cell 1:465-470. [DOI] [PubMed] [Google Scholar]
- 33.Neuman, E., M. H. Ladha, N. Lin, T. M. Upton, S. J. Miller, J. DiRenzo, R. G. Pestell, P. W. Hinds, S. F. Dowdy, M. Brown, and M. E. Ewen. 1997. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol. Cell. Biol. 17:5338-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novikoff, A. B., P. M. Novikoff, C. Davis, and N. Quintana. 1972. Studies on Microperoxisomes. II. A cytochemical method for light and electron microscopy. J. Histochem. Cytochem. 2:1006-1022. [DOI] [PubMed] [Google Scholar]
- 35.Novikoff, P. M., and A. B. Novikoff. 1972. Peroxisomes in absorptive cells of mammalian small intestine. J. Cell Biol. 53:532-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novikoff, P. M., and A. Yam. 1978. Sites of lipoprotein particles in normal rat hepatocytes. J. Cell Biol. 76:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peitsch, M. C. 1996. PROMOD and SWISS-MODEL: internet-based tools for automated comparative protein modeling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 38.Pestell, R. G., C. Albanese, A. T. Reutens, J. E. Segall, R. J. Lee, and A. Arnold. 1999. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocrine Rev. 20:501-534. [DOI] [PubMed] [Google Scholar]
- 39.Reutens, A. T., M. Fu, G. Watanabe, C. Albanese, M. J. McPhaul, S. P. Balk, O. A. Janne, J. J. Palvimo, and R. G. Pestell. 2001. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 15:797-811. [DOI] [PubMed] [Google Scholar]
- 40.Rosen, E. D., C.-H. Hsu, X. Wang, S. Sakai, M. W. Freeman, F. J. Gonzalez, and B. M. Spiegelman. 2002. C/EBP induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 16:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosen, E. D., P. Sarraf, A. E. Troy, G. Bradwin, K. Moore, D. S. Milstone, B. M. Spiegelman, and R. M. Mortensen. 1999. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4:611-617. [DOI] [PubMed] [Google Scholar]
- 42.Rosen, E. D., and B. M. Spiegelman. 2001. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 276:37731-37734. [DOI] [PubMed] [Google Scholar]
- 43.Saez, E., P. Tontonoz, M. C. Nelson, J. G. Alvarez, U. T. Ming, S. M. Baird, V. A. Thomazy, and R. M. Evans. 1998. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat. Med. 4:1058-1061. [DOI] [PubMed] [Google Scholar]
- 44.Sarraf, P., E. Mueller, D. Jones, F. J. King, D. J. DeAngelo, J. B. Partridge, S. A. Holden, L. B. Chen, S. Singer, C. Fletcher, and B. M. Spiegelman. 1998. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat. Med. 4:1046-1052. [DOI] [PubMed] [Google Scholar]
- 45.Sarraf, P., E. Mueller, W. M. Smith, H. M. Wright, J. B. Kum, L. A. Aaltonen, A. de la Chapelle, B. M. Spiegelman, and C. Eng. 1999. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol. Cell 3:799-804. [DOI] [PubMed] [Google Scholar]
- 46.Scherer, P. E., P. E. Bickel, M. Kotler, and H. F. Lodish. 1998. Cloning of cell-specific secreted and surface proteins by subtractive antibody screening. Nat. Biotechnol. 16:581-586. [DOI] [PubMed] [Google Scholar]
- 47.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 48.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 49.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiegelman, B. M. 1998. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes 47:507-514. [DOI] [PubMed] [Google Scholar]
- 51.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 52.Wang, C., M. Fu, M. D'Amico, C. Albanese, J. N. Zhou, M. Brownlee, M. P. Lisanti, V. K. Chatterjee, M. A. Lazar, and R. G. Pestell. 2001. Inhibition of cellular proliferation through IkappaB kinase-independent and peroxisome proliferator-activated receptor gamma-dependent repression of cyclin D1. Mol. Cell. Biol. 21:3057-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 54.Wang, X. L., J. Oosterhof, and N. Duarte. 1999. Peroxisome proliferator-activated receptor gamma C161→T polymorphism and coronary artery disease. Cardiovasc. Res. 44:588-594. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe, G., A. Howe, R. J. Lee, C. Albanese, I.-W. Shu, A. N. Karnezis, L. Zon, J. Kyriakis, K. Rundell, and R. G. Pestell. 1996. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc. Natl. Acad. Sci. USA 93:12861-12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 58.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression and actin organisation by multiple, PAK-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westwick, J. K., R. J. Lee, Q. T. Lambert, M. Symons, R. G. Pestell, C. J. Der, and I. P. Whitehead. 1998. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J. Biol. Chem. 273:16739-16747. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 61.Xu, H. E., M. H. Lambert, V. G. Montana, K. D. Plunket, L. B. Moore, J. L. Collins, J. A. Oplinger, S. A. Kliewer, R. T. Gampe, Jr., D. D. Mckee, J. T. Moore, and T. M. Willson. 2001. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 98:13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, H. E., T. B. Stanley, V. G. Montana, M. H. Lambert, B. G. Shearer, J. E. Cobb, D. D. Mckee, C. M. Galardi, K. D. Plunket, R. T. Nolte, D. J. Parks, J. T. Moore, S. A. Kliewer, T. M. Willson, and J. B. Stimmel. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPAR alpha. Nature 415:813-817. [DOI] [PubMed] [Google Scholar]
- 63.Yu, B., M. E. Lane, R. G. Pestell, C. Albanese, and S. Wadler. 2000. Downregulation of cyclin D1 alters cdk4 and cd2 specific phosphorylation of retinoblastoma protein. Mol. Cell. Biol. Res. Commun. 3:352-359. [DOI] [PubMed] [Google Scholar]
- 64.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 65.Yu, S., K. Matsusue, P. Kashireddy, W.-Q. Cao, V. Yeldandi, A. V. Yeldandi, M. S. Rao, F. J. Gonzalez, and J. K. Reddy 2003. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J. Biol. Chem. 278:498-505. [DOI] [PubMed] [Google Scholar]
- 66.Zwijsen, R. M. L., R. S. Buckle, E. M. Hijmans, C. J. M. Loomans, and R. Bernards. 1998. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes Dev. 12:3488-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zwijsen, R. M. L., R. Klompmaker, E. B. H. G. M. Wientjens, P. M. P. Kristel, B. van der Burg, and R. J. A. M. Michalides. 1996. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol. Cell. Biol. 16:2554-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwijsen, R. M. L., E. Wientjens, R. Klompmaker, J. van der Sman, R. Bernards, and R. J. A. M. Michalides. 1997. CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405-415. [DOI] [PubMed] [Google Scholar]