Abstract
The retinal determination (RD) gene network encodes a group of transcription factors and cofactors necessary for eye development. Transcriptional and posttranslational regulation of RD family members is achieved through interactions within the network and with extracellular signaling pathways, including epidermal growth factor receptor/RAS/mitogen-activated protein kinase (MAPK), transforming growth factor β/DPP, Wingless, Hedgehog, and Notch. Here we present the results of structure-function analyses that reveal novel aspects of Eyes absent (EYA) function and regulation. We find that the conserved C-terminal EYA domain negatively regulates EYA transactivation potential, and that GROUCHO-SINE OCULIS (SO) interactions provide another mechanism for negative regulation of EYA-SO target genes. We have mapped the transactivation potential of EYA to an internal proline-, serine-, and threonine-rich region that includes the EYA domain 2 (ED2) and two MAPK phosphorylation consensus sites and demonstrate that activation of the RAS/MAPK pathway potentiates transcriptional output of EYA and the EYA-SO complex in certain contexts. Drosophila S2 cell two-hybrid assays were used to describe a novel homotypic interaction that is mediated by EYA's N terminus. Our data suggest that EYA requires homo- and heterotypic interactions and RAS/MAPK signaling responsiveness to ensure context-appropriate RD gene network activity.
Proper development of an organism requires a formidable amount of cell-cell communication, wherein successful information transfer is effected by signaling cascades that ultimately alter the gene expression profile of a cell. Additionally, cells must integrate signals from multiple pathways to coordinate morphogenesis and differentiation in a spatially and temporally appropriate manner. Examples of this combinatorial control paradigm have been elucidated by using the Drosophila eye as a model system and have demonstrated that unique combinations of general and tissue-specific transcription factors are required to specify and maintain distinct cell fates (15, 18, 43).
Components of the retinal determination (RD) gene network, which includes twin-of-eyeless (toy), eyeless (ey), eyes absent (eya), sine oculis (so), and dachshund (dac), are essential for eye fate specification in metazoans. The RD gene network collectively encodes a cohort of nuclear transcription factors and/or cofactors whose expression is regulated by a conserved hierarchy of transcriptional regulation, such that TOY activates ey expression, EY induces so and eya, and EYA turns on dac (9). In addition to assuming pivotal roles during visual system development, the RD genes function in a variety of other contexts, including gonadogenesis (2), myogenesis (19), limb formation (16, 44), neurogenesis (30), and the cell cycle (29). Consequently, null mutations are lethal and exhibit complex phenotypes that are reflective of the pleiotropic roles assumed by RD network proteins during development (6, 11, 27, 32). Additionally, the expression patterns of the RD genes are not wholly coincident (4), suggesting that reiterative deployment of the entire RD network module is not obligatory for the function of specific RD network proteins.
Much analysis of RD gene function and regulation has focused on the visual system, particularly in Drosophila, where gene activity can be manipulated without compromising viability or fertility of the animal. RD genes are best known for the “eyeless” phenotype associated with eye-specific hypomorphic mutations and for the ability to induce formation of ectopic eye tissue upon overexpression (5, 9, 13, 17, 37, 41). Use of these two assays has revealed a complex system of positive feedback loops superimposed on the defined linear hierarchy of transcriptional regulation, which most likely amplifies and stabilizes expression of the RD gene products.
eya is the founding member of a novel gene family characterized by a highly conserved C-terminal motif that contains both SO (31) and DAC (9) binding sites, termed the EYA domain (ED; amino acids [aa] 486 to 760) (Fig. 1A). Vertebrate homologs, such as murine EYA1-4, are strikingly similar in their ED, yet their N termini, with the exception of a small tyrosine-rich region, the ED2 (Fig. 1A), are largely divergent (45, 47). so and vertebrate Six family genes encode nuclear proteins with a homeobox-type DNA binding domain and the conserved SIX domain (23). The latter is an important mediator of EYA-SO interactions (37). DAC, a novel nuclear protein, and its vertebrate DACH counterparts contain two regions of high homology: DACH-box-N and DACH-box-C (32). Like SO, DAC has been shown to physically interact with the ED, via DACH-box-C (19).
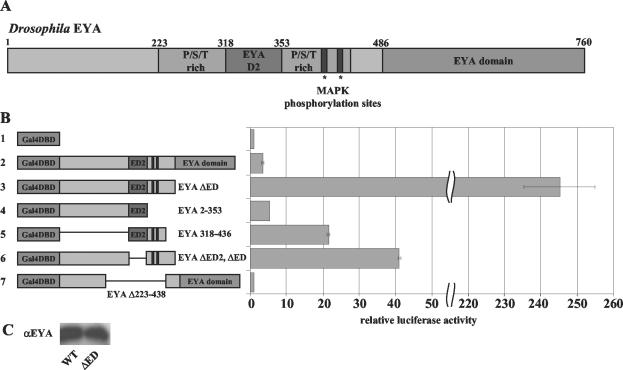
FIG. 1.
The N-terminus of EYA is a potent transactivator. (A) Drosophila EYA contains two conserved regions, the ED2 and ED. The P/S/T-rich region includes both the ED2 and the two MAPK phosphoacceptor sites. (B) Gal4DBD-EYA fusions were used to assay the ability of EYA to activate transcription from a UAS-luciferase reporter gene. Transactivation potentials were calculated by taking the luciferase/β-galactosidase activity ratio for each construct and were plotted relative to the activity of the Gal4DBD vector alone. As shown in panel B, for construct 2, full-length EYA can activate transcription 3.5-fold above Gal4DBD alone. The N terminus of EYA (construct 3), a construct that lacks the ED, is a potent transactivator, activating transcription over 250-fold (note scale change on axis). Gal4DBD-EYA 1-353 (construct 4), a truncation that contains ED2 but removes part of the P/S/T-rich region, reduces transactivation potential to only fivefold. ED2 plus part of the P/S/T-rich region is able to activate transcription at low levels (construct 5), indicating that the entire N terminus is necessary for full transactivation potential. Deletion of the conserved ED2 within the N terminus of EYA (construct 6) sharply reduces transactivation to 41-fold above background, one-fifth the activity of the intact N terminus. Strikingly, deletion of the entire P/S/T-rich region (construct 7) results in complete loss of transactivation potential. (C) Deletion of the EYA domain does not affect protein expression levels. WT, wild type.
Analysis of loss-of-function eya mutants in Drosophila and in human patients suffering from branchio-oto-renal (BOR) syndrome underscores the importance of EYA-SO and/or EYA-DAC interactions in vivo. BOR syndrome, a disease characterized by craniofacial, ear, and kidney defects, arises from mutations in the human Eya1 gene (1, 8). In Drosophila, point mutations in the conserved ED appear to cause loss-of-function phenotypes by impairing EYA's ability to interact with SO and DAC (8). Similarly, a recent study has shown that human BOR alleles that map to the ED also have impaired interactions with SIX and DACH family members, emphasizing the evolutionarily conserved importance of interactions between these three RD family members in vivo (35).
The combined physical interaction, colocalization, and genetic data have led to a model whereby EYA and SO together constitute a functional transcription factor, with EYA providing the activation domain and SO contributing the DNA binding moiety. Consistent with this, in mammalian cell culture, SIX2, SIX4, and SIX5 are able to synergize with EYA to drive expression from a reporter construct (33). The functional consequences of EYA-DAC interactions remain less well understood. Although initial characterization of Drosophila dac and its vertebrate counterparts, Dach1 and Dach2, failed to identify a DNA binding domain, it has been postulated that DACH-box-N may encode a novel DNA binding motif similar to the winged helix/forkhead subgroup (24). Thus, like SO, DAC may play a role in recruiting EYA to the promoters of target genes (21).
In addition to the complex interactions observed within the RD gene network, numerous signaling pathways, including epidermal growth factor receptor (EGFR)/RAS/mitogen-activated protein kinase (MAPK) (20), transforming growth factor-β (TGFβ)/Dpp (10, 38), Wingless (3), Hedgehog (12), and Notch (2, 28), have been shown to interact genetically with members of the RD network. However, with the exception of the EGFR pathway (20), little is known about how extracellular signaling pathways regulate RD gene expression and/or activity.
In order to better understand RD gene network regulation, we have performed an extensive structure-function analysis of the EYA protein. Using a series of Drosophila S2 cell-based transcriptional activation assays, we have defined a proline-, serine-, and threonine-rich (P/S/T-rich) region of EYA, encompassing both the MAPK phosphorylation consensus sites and the tyrosine-rich ED2, that is necessary for EYA transactivation potential in cell culture and for ectopic eye induction ability in vivo. We demonstrate that RAS/MAPK signaling can positively regulate EYA transactivation and that GROUCHO is a potent repressor of EYA-SO-mediated transcription through its interactions with SO. Finally we show that EYA is able to self-associate, adding yet another layer of functional complexity to the elaborate hierarchy of interactions that exists among the RD gene products.
MATERIALS AND METHODS
Construction of transactivation assay expression plasmids.
pUAST-luciferase was constructed by amplifying the luciferase cDNA from pGL3luc (Promega) with primers LUC1 (5′-TTGGAATTCCAACATGGAAGACGCCAAAAAC-3′) and LUC2 (5′-TTGGGTACCTTACACGGCGATCTTTCCGC-3′), digestion with EcoRI and KpnI, and insertion into the EcoRI-KpnI sites of pUAST. All eya constructs were made with the eya1 cDNA. pRmHa3-Gal4DBD-eyawt, S-A, and S-D/E full-length fusion constructs have a similar design. The MAPK consensus sequence is defined as P-X-S/T-P, and Drosophila eya contains two adjacent phosphorylation sites at aa S402 and S407. The construct referred to as pRmHa3Gal4DBD-eyaS-A contains two S-A point mutations in place of the phosphoacceptor residues, and conversely, the pRmHa3-Gal4DBD-eyaS-D/E construct contains S-D and S-E point mutations at these sites. A three-piece ligation was performed to insert a 660-bp, N-terminal eya PCR-amplified fragment and a 1.6-kb, C-terminal eya restriction fragment containing either wild-type or mutated MAPK sites into pRmHa3-Gal4DBD, cut with KpnI and SalI. The N-terminal PCR product was generated with primers EYA1 (5′-TGGGTACCTTGTATAATGTGCCGTGCTATC-3′) and EYA2 (5′-CGAAGAGTTGACCGCCACTG-3′) and was digested with KpnI and BamHI. BamHI-SalI restriction fragments from pRmHa3-eya, pRmHa3-eyaS-A, and pRmHa3-eyaD/E (constructs described previously in reference 20) were then combined with the digested eya PCR product in a ligation with pRmHa3-Gal4DBD, cut with KpnI and SalI.
Similarly, three truncated pRmHa3-Gal4DBD-eya constructs that lack the conserved EYA domain (aa 486 to 760) were made, which contain each of the MAPK site variants described above. Primers EYA1 and EYA3 (5′-TTGGTCGACTTACACACTGCTGCCTCCGCTC-3′) were used to amplify a 1.3-kb product from a pRmHa3-eya, pRmHa3-eyaS-A, or pRmHa3-eyaD/E template. PCR products were digested with KpnI and SalI and ligated into the pRmHa3-Gal4DBD vector. A construct encoding the first 353 aa of EYA, including the N-terminal portion of the P/S/T-rich region, was also generated. This construct, pRmHa3-Gal4DBD-eya 1-353, was generated with primers EYA1 and EYA1545A (5′-TTGGTCGACGTAGTTGGCCGGACTGTA-3′). The 920-bp PCR product and pRmHa3-Gal4DBD were digested with KpnI and SalI and ligated directionally. An internal deletion construct that lacks the entire P/S/T-rich region, pRmHa3-Gal4DBD-eya Δ223-438, was made by inserting an annealed, double-stranded linker with 5′ BamHI and 3′ KpnI sticky ends into pRmHa3-myc-eya, cut with the aforementioned enzymes. Primers EYAD1 (5′-GATCCATTTTGTACGGTACC-3′) and EYAD2 (5′-CGTACAAAATG-3′) were annealed and used in the directional ligation described above to generate pRmHa3-myc-eya Δ223-438. pRmHa3-Gal4DBD-eya Δ223-438 resulted from a two-piece ligation between pRmHa3-Gal4DBD-eya, cut with BamHI and SalI, and a complementary restriction fragment from pRmHa3-myc-eya Δ223-438. A construct encoding a truncated EYA in which ED2 (aa 318 to 353) is also deleted, pRmHa3-Gal4DBD-eya ΔED2 ΔED, was made by the Stratagene Quikchange site-directed mutagenesis protocol. Primers dEya 1351/1455 (S) (5′-CAGCTGTACAGCAGTCCGTCACCGTATGCGGTCAGC3′) and dEya1351/1455(A) (5′-GCTGACCGCATACGGTGACGGACTGCTGTACAGCTG-3′) were used to generate pBSSK-eya ΔED2. A 600-bp BamHI-SacII eya ΔED2 fragment was then shuttled into the truncated pRmHa3-Gal4DBD-eya construct in a directional ligation. pRmHa3-Gal4DBD-eya 318-436 was made with primers KpnI-EYA D2 (S) (5′-TTGGGTACCTACGCCGGCTACAACAACTTC-3′) and EYA3 to amplify a 350-bp product. The PCR product and pRmHa3-Gal4DBD were digested with KpnI and SalI and used in two-piece ligations.
Development of S2-2H assay.
The S2 cell two-hybrid (S2-2H) assay was developed as follows. A vector containing the DNA binding domain of yeast Gal4 (aa 1 to 147), pRMHa3-Gal4DBD, was constructed as follows. Gal4DBD was amplified from pCasperUbGal4 with the primers 5′-TGGAATTCCAACATGAAGCTACTGTCTTCTATCG-3′ and 5′-TGGGTACCCGATACAGTCAACTGTCTTTG-3′, which contain 5′ and 3′ EcoRI and KpnI sites, respectively. The digested PCR product was ligated into pRmHa3, a pUC9-derived vector containing a metallothionein-responsive promoter upstream of the multiple cloning site (MCS).
pRmHa3-Gal4AD was made by amplification of the Gal4 activation domain (aa 768 to 874) from pCasperUbGal4 via PCR with primers Gal4AD S2302 (5′-TGGAATTCCAACATGGCCAATTTTAATCAAAGTG-3′) and Gal4AD A2673 (5′-TTGGTACCGTATCTTCATCATCGAATAGA-3′), cut with EcoRI and KpnI, and inserted into pRmHa3. To ensure nuclear expression of these constructs, a nuclear localization signal (NLS) was added with these two oligonucleotides, Gal4AD NLS S-50 (5′-TGTGACCCCCCCCAAGAAGAAGCGCAAGGTGGAGGACGATGGTAC-3′) and Gal4 AD NLS A-54 (5′-CATCGTCCTCCACCTTGCGCTTCTTCTTGGGGGGGGTCACAGTAC-3′). Oligo-nucleotides were annealed and then ligated into the KpnI site of pRmHa3-Gal4AD to make pRMHa3-Gal4AD-NLS, which we refer to as pRMHa3-Gal4AD. pRmHa3-Gal4AD-eya resulted from a three-piece ligation using KpnI-BamHI eya and BamHI-SalI eya restriction fragments and with pRmHa3-Gal4AD cut with KpnI and SalI. Primers SoS4 (5′-TTGGTACCTTACAGCATCCCGCCACAG-3′) and So1234 (5′-TTGGTCGACTCATAAGTGCTGGTACTC-3′) were used to amplify full-length so cDNA from pBSSK so (gift of G. Mardon), with unique 5′ KpnI- and 3′ SalI-cut sites. The digested PCR product was inserted into pRmHa3-Gal4DBD in a directional, two-piece ligation to make pRmHa3-Gal4DBD-so. To make pRmHa3-Gal4AD-so, so was cut out of pRmHa3-Gal4DBD-so with KpnI and SalI inserted into pRmHa3-Gal4AD cut with the same enzymes. To make dac constructs, DacS622 (5′-TTGGTACCGATTCTGTGACAAGTGAAC-3′) and DacA1370 (5′-AAG TGCTTCAGGAAGAGCTCG-3′) primers were used to amplify the 5′ region of dac from pBSSK-dac (gift of G. Mardon), adding a 5′ KpnI site. The PCR product was cut with KpnI and StuI, which is found internally in the amplified dac fragment. A three-piece ligation was performed with KpnI-StuI 5′dac and a StuI-SalI restriction fragment from pBSSK-dac, into the KpnI-SalI sites in pRmHa3-Gal4DBD. For the AD construct, full length dac was cut out of pRmHa3-Gal4DBD-dac with KpnI and SalI and ligated into those sites in pRmHa3-Gal4AD to create pRmHa3-Gal4AD-dac. For pRmHa3-Gal4DBD-eya ΔD2, the N-terminal eya ΔD2 construct described above was shuttled into full-length pRmHa3-Gal4DBD-eya by using BamHI and SacII. For the pRmHa3-Gal4DBD-eya domain, the eya domain was PCR amplified with primers EYADS (5′-TTGGGTACCGAACGGGTGTTCGTCTGG-3′) and EYA DA (5′-TTGGGATCCTCATAAGAAGCCCATGTC-3′), which contain KpnI and BamHI sites used to clone into the pRmHa3-Gal4DBD vector.
Construction of transcription assay expression plasmids.
To construct ARE-luciferase, luciferase cDNA was isolated as an XhoI-XbaI fragment (∼1.6 kb) from Promega pGL3-Luciferase and inserted into XhoI-XbaI sites of pBluescript SK+. To add the hsp70 TATA box, oligonucleotides EBS link 1 (5′-CCATATGATCTGCAGAGGGTATATAATGC-3′) and EBS link 2 (5′-TCGAGCATTATATACCCTCTGCAGATCATATGGGTAC-3′) were annealed and inserted into the KpnI-XhoI sites of pBSSK-luciferase. KpnI and XhoI sites were retained, and NdeI and PstI sites and a TATA box were inserted. ARE-luciferase was made by multimerizing the AREC3 (SIX4) binding site, as defined in reference 22, using oligonucleotides ARES (5′-TCGAGGGTGTCAGGTTGCG-3′) and AREA (5′-TCGACGCAACCTGACACCC-3′). Oligonucleotides were annealed and ligated and then cut with XhoI and SalI. A resultant 7-mer was cloned into the MCS of pBSSK and then shuttled into pBSSK-TATA-luciferase. To make pRmHa3-flag-eya, full-length eya1 cDNA was PCR amplified with primers EYAI 5397 (5′-TTGTATAATGTGCCGTCGTATC-3′) and EYA STOP (5′-TTTCATAAGAAGCCCATGTCGAGG-3′) and then digested with SmaI, which cuts the eya cDNA internally. The 0.7-kb blunt/SmaI fragment was inserted into SmaI-cut pBSSK + Flag vector (gift from R. Fehon) to produce an in-frame fusion of the Flag epitope with the second amino acid of EYA. A three-piece ligation was done to join the 5′ end of flag-eya (obtained as a SacI-SmaI fragment from pBSSK-flag-eya) with the 3′ end of eya (obtained from pRMHa3-eya) as a SmaI-SalI fragment. These were inserted into SacI-SalI-cut pRMHa3. Similarly, pBSSK-myc-eya was generated by PCR amplification with primers EYAI and EYA STOP and blunt ligated into the StuI site of pBSSKmyc. A three-piece ligation between the EcoRI-BamHI fragment from pBSSK-myc-eya and BamHI and SalI from pRmHa3-eya into the EcoRI and SalI sites of pRMHa3 resulted in pRMHa3-myc-eya. In order to construct pRMHa3-dac, first a ClaI fragment was cut out of pBSSK-dac (gift from G. Mardon) and cloned into pBSSK to remove most of the large 5′ untranslated region (UTR) (Cla-dac). A 3′ fragment was cloned into pBSSK by using BamHI and HindIII sites (HI/HIII-dac). A full-length construct was made by four-piece ligation with EcoRI-StuI from Cla-dac plus a StuI-BamHI fragment from pBSSK-dac plus the BamHI-XhoI fragment from HI/HIII-dac into the EcoRI-XhoI sites of pUAST. The entire full-length dac cDNA, including approximately 45 bp of the 5′ UTR and 245 bp of the 3′ UTR, was then excised with EcoRI and XhoI and ligated into the EcoRI-SalI sites of pRMHa3. pRMHa3-myc-so was generated by PCR amplification of full-length so cDNA by using primers So S805 (5′-TTACAGCATCCCGCCACAGAT-3′) and So A2268 (5′-AACTAGAATCATAAGTGCTGG-3′) and blunt end ligated into the StuI site of pBSSK-myc, resulting in an in-frame fusion of the MYC epitope to the second amino acid of SO. This was moved into pUAST by using EcoRI and XbaI. To move the full-length tagged so into pRMHa3, myc-so was cut out of pUAST-myc-so with XbaI, blunted with Klenow fragment, and then digested with EcoRI and subcloned into EcoRI-SmaI sites of pRMHa3. A full-length groucho expression construct, pMT-GROUCHO, was generated by PCR amplification of the N terminus of GROUCHO, using the groucho cDNA clone LD33829 as a template and primers Groucho-start (5′-ATGAATTCAACAACATGTATCCCTCACCGG-3′) and Groucho-A879 (5′-TGTGCGATACTTCTCACGATCGG-3′), digestion with EcoRI and XbaI, and ligation with an XbaI-XhoI fragment from LD33829 into EcoRI-SalI-cut pRMHa-3.
All regions generated by PCR were verified by sequencing, and all constructs were tested for expression and localization in S2 cells by immunohistochemistry. Further subcloning details are available upon request.
Co-IP and Western blots.
Transfected cells were harvested and then lysed by rocking at 4°C for 20 min in 1 ml of lysis buffer (100 mM NaCl, 50 mM Tris [pH 7.5], 2 mM EDTA, 2 mM EGTA, 1% NP-40, one Roche Complete, Mini protease inhibitor cocktail tablet per 10 ml). Clarified lysates were subjected to immunoprecipitation (IP) with anti-Flag-conjugated agarose beads (Sigma) for 1.5 h at 4°C. Beads were washed three times with lysis buffer. The immunoprecipitates were boiled in 40 μl of 2× sodium dodecyl sulfate (SDS) buffer, and Western blotting was carried out as previously described (34) with mouse anti-MYC (1:300), mouse anti-GRO (1:50), and Rb anti-Flag (1:5,000) antibodies.
For Western blots to analyze protein levels, half of the transfected cells were lysed as described below for β-galactosidase assays, and the other half were pelleted, vortexed briefly, and resuspended in 40 μl of 2× SDS buffer. Transfection efficiency was measured by β-galactosidase assays, and appropriate amounts of crude lysate were run on the gel. Efficiency was confirmed by Western blotting for β-galactosidase (Rb anti-β-galactosidase, 1:20,000). Protein levels were examined with GP anti-EYA (1:10,000), mouse anti-MYC (1:300), and Rb anti-Flag (1:5,000) antibodies.
Generation of transgenic lines.
pRmHa3-flag-eya and pRmHa3-myc-eya ΔD2 were cut with SmaI and SalI, and a directional ligation was performed to create pRmHa3-flag-eya ΔD2. Likewise, a SmaI-SalI double digest was performed on pRmHa3-myc-eya Δ223-438 to generate pRmHa3-flag-eya Δ223-438. EcoRI-SalI double digests were used to excise the flag-eya, flag-eya ΔD2, and flag-eya Δ223-438 cDNAs from the pRmHa3 vector for insertion into pUAST, cut with EcoRI and XhoI. pUAST-flag-eya, pUAST-flag-eya ΔD2, and pUAST-flag-eya Δ223-438 were subsequently used to generate transgenic lines as previously described (39).
S2 cell transactivation and transcription assays.
Drosophila S2 cells were transiently transfected with calcium phosphate as described previously (36). Each construct (2.5 μg) indicated was transfected for 6 h along with 2.5 μg of pUAST-luciferase or 10 μg of ARE-luciferase as the reporter gene and with 1 μg of pActin 5.1-V5His-lacZ (Invitrogen) to normalize for transfection efficiency. Cells were allowed to recover for 17 h, whereupon expression was induced by addition of 0.1 M CuSO4. After 24 h, cells were harvested by spinning at 200 × g for 1 min and lysed by rocking at 4°C for 20 min in 250 μl of lysis buffer (Tropix/Applied Biosystems). Samples were subsequently microcentrifuged at 20,800 × g for 1 min at 4°C, and supernatants were transferred to fresh tubes. Luciferase and β-galactosidase activities were quantified with a Tropix/Applied Biosystems luciferase assay kit or Galacto-Star assay kit. Assays were performed in triplicate on whole-cell extracts according to the manufacturer's instructions (TROPIX/Applied Biosystems). A minimum of four independent transfections were performed for each condition. The average luciferase or β-galactosidase signal for Gal4DBD/pUAST-luciferase or ARE-luciferase alone was set to 1, and the experimental averages were normalized relative to this value. Data were analyzed and graphed with Microsoft Excel. Error bars denote one standard deviation above and below the mean for each construct. In Fig. 3, 5, and 6, SO is always tagged with the Flag epitope. In Fig. 3, EYA constructs are tagged with the Flag epitope, and in Fig. 5 and 6, EYA constructs are tagged with the MYC epitope.
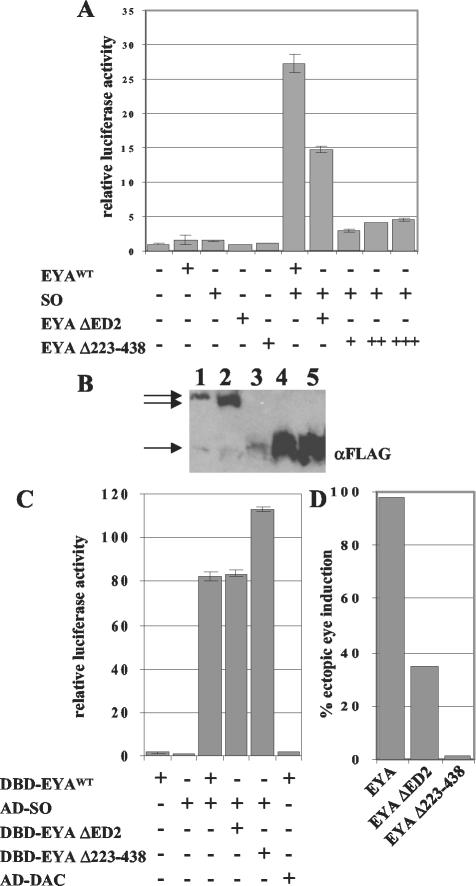
FIG. 3.
ARE-luciferase is responsive to the EYA-SO transcription factor. (A) EYA and SO alone do not affect transcription of the ARE-luciferase reporter gene, but together can activate transcription 27-fold. Full-length EYA ΔED2 is unable to fully activate transcription of this gene, and neither is a construct missing the entire P/S/T-rich region, EYA Δ223-438. This construct was not expressed at the same level as wild-type EYA (EYAWT), so we transfected two (++) and three (+++) times the amount of plasmid to raise protein levels to and above the levels of EYA. These levels still do not activate ARE-luciferase. (B) Quantitative Western blotting shows that EYA (lane 1) and EYA ΔED2 (lane 2) are expressed at similar levels, while the same amount of EYA Δ223-438 plasmid (lane 3) is not. However, transfection of two and three times more EYA Δ223-438 plasmid results in robust expression, as shown in lanes 4 and 5. (C) EYA and SO show a strong interaction in an S2-2H assay. The deletions in EYA do not affect interactions with SO, and in fact, EYA Δ223-438 appears to have a stronger interaction with SO. DAC does not interact with EYA in S2-2H assays. (D) Overexpression of EYA using the 57A1dpp-Gal4 driver causes ectopic eye induction in over 98% of animals (n = 413). Overexpression of EYA ΔED2 results in ectopic eyes in only 35% of animals (n = 506). Deletion of the entire P/S/T-rich region, EYA Δ223-438, results in a protein that can only rarely induce ectopic eyes, seen in only 1.5% of animals examined (n = 204).
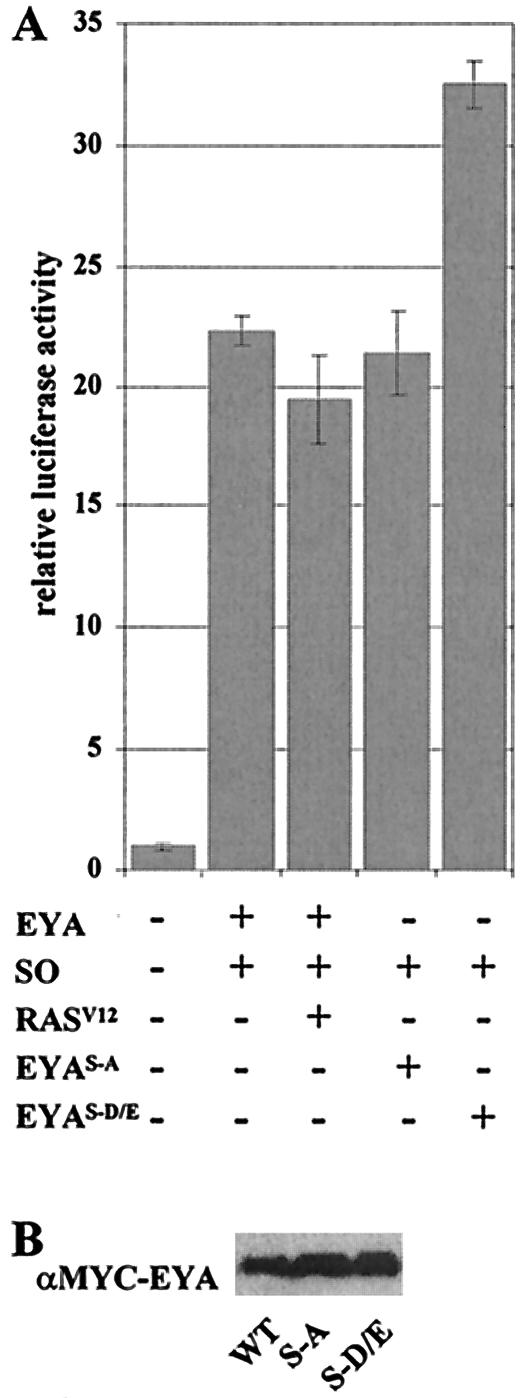
FIG. 5.
The EYA-SO transcription factor is regulated by phosphorylation. (A) As shown in Fig. 3, the EYA-SO transcription factor can activate expression of ARE-luciferase. This expression is not affected by addition of RASV12 nor upon mutation of the MAPK phosphoacceptor sites to alanine (EYAS-A). A striking increase in activation is seen when the EYAS-D/E phosphomimetic mutant is used, showing that phosphorylation acts to increase EYA transactivation potential. That RASV12 itself does not produce the same increase on wild-type EYA (WT) in this assay suggests that RAS signaling may have multiple effects on the RD gene network and in particular may negatively regulate SO. (B) EYA phosphoacceptor mutations do not affect protein expression levels.
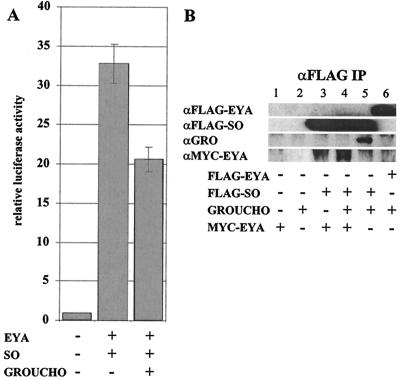
FIG. 6.
The EYA-SO transcription factor is negatively regulated by interactions with GROUCHO. (A) When coexpressed with EYA and SO, GRO is a repressor of the EYA-SO transcription factor. (B) Lanes 1 and 2 show that MYC-EYA and GRO are not pulled down by anti-Flag beads. In lane 3, IP of SO can co-IP EYA. Lane 4 shows that IP of SO can co-IP EYA but not GRO; however, in lane 5, we see that without EYA, GRO can associate with SO. Thus, co-IP of GRO with SO is disrupted by cotransfection of EYA. This result does not seem to be due to direct competition between EYA and SO for GRO, because IP of EYA cannot co-IP GRO. All proteins were expressed at similar levels in crude cell lysates (data not shown).
RESULTS
EYA functions as a transactivator.
To address the question of whether EYA can function as a transcriptional coactivator, we took advantage of the well-characterized yeast transcription factor Gal4 and its target sequence, UAS (14), to design an assay for transactivation in Drosophila S2 cells. The DNA binding domain of Gal4 (Gal4DBD) was fused in frame to the eya coding region (Gal4DBD-EYA) (Fig. 1B) and subcloned into a vector containing a metallothionein promoter, which allows inducible expression in S2 cells. Gal4DBD-EYA fusion proteins were tested for their ability to activate expression of a UAS-luciferase reporter gene; cotransfection of a constitutively expressed actin-LacZ plasmid enabled us to normalize activity levels based on transfection efficiency. Immunohistochemistry with anti-Gal4DBD antibodies confirmed the expression and nuclear localization of all Gal4DBD-EYA fusion proteins in transfected S2 cells (data not shown).
First, we tested the full-length eya coding region fused in-frame to the Gal4DBD (Fig. 1B). As shown in Fig. 1B (constructs 1 and 2), Gal4DBD-EYA exhibits 3.5-fold-greater activity than the Gal4DBD alone. A series of deletion and truncation constructs were designed to define a minimal and sufficient domain for this activity. Strikingly, a fusion protein expressing the N-terminal 485 aa of EYA but lacking the conserved ED, Gal4DBD-EYA ΔED, displays an approximately 70-fold increase in transactivation potential relative to the full-length Gal4DBD-EYA construct (Fig. 1B, construct 3 versus construct 2 [note scale]). The converse fusion protein expressing only the C-terminal ED, Gal4DBD-EYA Domain (aa 486 to 760), does not transactivate (data not shown). This difference is likely not due to changes in protein stability, as EYA and EYA ΔED are expressed at similar levels (Fig. 1C). The discrepancy in activity levels of the full-length versus Gal4DBD-EYA ΔED chimeras suggests that the ED may function as an autoregulatory inhibitor in this context.
A P/S/T-rich region is critical for EYA transactivation.
EYA ΔED was dissected further to determine the regions critical for transactivation. EYA ΔED contains the conserved ED2 (aa 318 to 353), a tyrosine-rich region that has not been functionally characterized (Fig. 1A). ED2 lies within a larger P/S/T-rich region (aa 223 to 438) that includes two consensus MAPK phosphorylation sites previously shown to be important for EYA regulation in vivo (20).
We found that Gal4DBD-EYA 2-353, an N-terminal construct that contains the ED2 but truncates the P/S/T-rich region, exhibits very low transactivation activity, suggesting a critical requirement for the latter domain (Fig. 1B, construct 4). Consistent with these results, the fusion protein that contains the last two-thirds of the P/S/T-rich region, Gal4DBD-EYA 318-436, exhibits a 22-fold increase in transactivation potential relative to the Gal4DBD-alone control (Fig. 1B, construct 5 versus construct 1). Therefore the P/S/T-rich region of EYA is critical but not entirely sufficient for transcriptional coactivation, and upstream regions of the protein (aa 2 to 223) are required to achieve maximal transactivation levels.
Interestingly, deletion of the conserved ED2 (Gal4-DBD-EYA ΔED2, ΔED; aa 2 to 317 and 353 to 485; Fig. 1B, construct 6) results in a 6-fold reduction in transactivation potential relative to that of EYA ΔED (Fig. 1B, construct 3), yet shows a 41-fold increase relative to the Gal4DBD control (Fig. 1B, construct 1). Although ED2 is not essential for EYA transactivation per se, our results suggest that it is needed to achieve maximal levels and ascribes a function to this previously uncharacterized domain.
Although the full-length EYA is a weaker transactivator in this assay than any of the “active” deletion constructs that lack the C-terminal EYA domain, we wanted to determine whether the P/S/T-rich region is necessary for the transactivation potential of full-length EYA. To address this question, the entire P/S/T-rich region, including ED2, was deleted to generate Gal4DBD-EYA Δ223-438. We found that this internal deletion abrogates transcriptional activation in full-length EYA (Fig. 1B, construct 7 versus construct 3). An identical result, namely a complete lack of transactivation, was obtained when the C-terminal EYA domain was also deleted from construct 7 (data not shown).
RAS/MAPK signaling increases EYA transactivation potential.
Because the P/S/T region important for EYA transactivation potential also contains the MAPK phosphorylation sites (Ser402 and Ser407) shown to affect EYA activity in vivo, we asked whether RAS signaling modulates EYA's transactivation potential. To examine this possibility, we cotransfected activated ras (rasV12) with the Gal4DBD-EYA fusion constructs. We did not observe an increase in the transactivation potential of Gal4DBD-EYA (data not shown), but observed a variable but significant increase in Gal4DBD-EYA ΔED transactivation potential in the presence of RASV12 (Fig. 2). In repeated trials, this increase in transactivation ranged from 10 to 66%, the latter of which is depicted in Fig. 2.
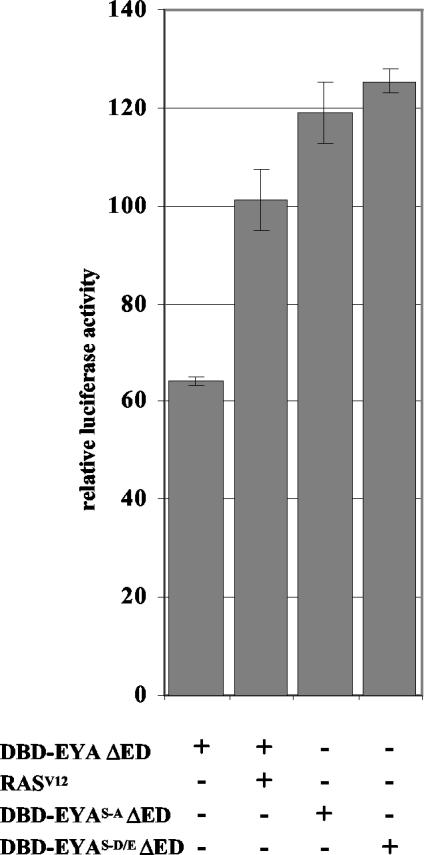
FIG. 2.
RAS/MAPK signaling activates EYA transactivation. Transactivation via Gal4DBD-EYA ΔED is increased significantly but variably upon the addition of RASV12. Mutations of the MAPK phosphoacceptors to alanine (EYAS-A) to prevent phosphorylation, or to aspartic or glutamic acid (EYAS-D/E) to mimic phosphorylation, both increase Gal4DBD-EYA ΔED transactivation activity, suggesting that these sites are important for regulation of EYA transactivation potential. Taken with the increase seen upon addition of RASV12, we conclude that RAS signaling can increase EYA transactivation potential.
Because we had previously shown that site-specific mutations in the MAPK consensus phosphoacceptor residues affect EYA function in vivo (20), we wanted to test whether these mutations might also influence transactivation in the Gal4DBD assay system. Mutation of the two MAPK consensus sites S402 and S407 to alanine (EYAS-A) significantly reduces EYA activity in ectopic eye induction assays. Conversely, mutation of the phosphoacceptor residues to aspartic or glutamic acid (EYAS-D/E) produces a hyperactive protein, presumably by mimicking its phosphorylated state (20). Surprisingly, we found that both EYAS-A and EYAS-D/E point mutations increased transactivation activity of Gal4DBD-EYA ΔED to slightly higher levels than were seen upon addition of RASV12 (Fig. 2).
Because EYA transactivation potential is increased upon cotransfection of rasV12 or mutation of the MAPK phosphoacceptor residues, it seems likely that these residues are important for proper regulation of EYA transactivation. Our finding that in this system both EYAS-A and EYAS-D mutations result in increased transactivation relative to wild-type EYA may reflect the complexity of the consequences of RAS/MAPK signaling. However, it is important to note that the chimeric Gal4DBD-EYA fusion proteins and/or the truncations we engineered may have an altered conformation relative to native EYA, such that MAPK phosphorylation events have different consequences in this context.
ARE-luciferase is responsive to the EYA-SO transcription factor.
The synthetic assay system described above enabled us to map the regions crucial for EYA transactivation activity to the ED2 and the surrounding P/S/T-rich region. RAS/MAPK-dependent effects on EYA transactivation levels indicate that RAS signaling can activate EYA transactivation but that such effects may not afford themselves to straightforward interpretation. In order to examine this question in a more native system, we designed a transcription assay system to measure the transactivation potential of EYA-SO complexes.
Currently, the genomic DNA target sequences bound by Drosophila SO are not known, but several studies in mammalian cells and tissues have identified SIX family response elements. In particular, the Na, K-ATPase α1 subunit gene (ATP1a1) regulatory element (ARE) has been shown to respond to SIX family members SIX2, SIX4 and SIX5 in vivo and in vitro (22). Because SO and SIX2 belong to the same SIX family subgroup (SIX1/2) (23) and their homeodomains are 93% identical (40), we reasoned that they are likely to bind similar sequences. We therefore multimerized the core ARE binding site (see Materials and Methods) and placed this enhancer in front of a minimal promoter followed by luciferase cDNA, which we will refer to as ARE-luciferase.
We found that ARE-luciferase is responsive to cotransfection of eya and so, and together they activate ARE-luciferase 27-fold over the reporter alone (Fig. 3A). ARE-luciferase is not appreciably activated alone nor upon transfection of eya or so individually (Fig. 3A). Therefore, ARE-luciferase activation provides a measure of EYA's efficacy as a transcriptional coactivator when bound to SO. We also asked whether the addition of DAC affects ARE-luciferase transcription and found that DAC did not affect the transactivation potential of the EYA-SO transcription factor (data not shown).
The P/S/T-rich region of EYA is necessary for transactivation but not for EYA-SO interactions.
We used the ARE-luciferase reporter and full-length EYA and SO to ask whether the ED2 and the P/S/T-rich region determined to be critical in the synthetic Gal4DBD assay system are essential in a more physiologically relevant context. In this assay, we used otherwise full-length EYA constructs that lack either the ED2 or the entire P/S/T-rich region, EYA ΔED2 and EYA Δ223-438. As shown in Fig. 3A, deletion of the ED2 reduces transactivation to only 14-fold relative to that in controls, while deletion of the entire P/S/T-rich region, EYA Δ223-438, virtually eliminates transcriptional coactivation.
One possible explanation for this lack of activity might be a decrease in protein expression levels in the EYA deletion constructs. In order to test this, we performed quantitative Westerns blots to assay tagged EYA protein levels. Transient transfections were normalized for efficiency by using β-galactosidase assays, and appropriate amounts of lysate were loaded. We found that while the ΔED2 deletion did not alter protein expression levels (Fig. 3B, lane 2), EYA Δ223-438 was less abundantly expressed than full-length EYA (Fig. 3B, lane 3). In order to correct for this difference, we transfected sufficient EYA Δ223-438 plasmid to produce protein levels above that of full-length EYA (Fig. 3B, lanes 4 and 5). Using these larger amounts of EYA Δ223-438 plasmid, we still observed only low levels of transactivation (Fig. 3A), indicating that deletion of this region of EYA compromises activity.
To confirm that the loss of transactivation potential seen in these deletions was not due to a loss of EYA-SO binding, we developed a system to test for direct protein-protein interactions in S2 cells, which we term S2-2H assays (see Materials and Methods for details). Use of Drosophila cultured cells, rather than the more common yeast or mammalian cell-based systems (14, 15), allows interactions between Drosophila proteins to be assayed in a more physiologically native environment, thereby increasing the probability that necessary cofactors and/or protein modifications are present.
Using our S2-2H assay, Gal4DBD-EYA, EYA ΔED2, and EYA Δ223-438 fusions were each tested for interaction with a Gal4AD-SO fusion protein. As shown in Fig. 3C, Gal4AD-SO is able to interact with all three EYA proteins and demonstrates even stronger interactions with Gal4DBD-EYA Δ223-438. This, combined with our Western analysis, indicates that the reduced transactivation observed upon deletion of the ED2 and the P/S/T-rich region (Fig. 3A) reflects a change in activity levels of the EYA-SO transcription factor rather than simply the loss of EYA protein or loss of the ability to form an EYA-SO complex.
We confirmed that the P/S/T-rich region and ED2 are necessary for EYA activity in vivo by assaying the ability of these deletions to induce ectopic eyes when overexpressed with the UAS/Gal4 system (7). We found that when driven by the 57A1dpp-Gal4 driver, EYA ΔED2 and EYA Δ223-438 exhibited drastically reduced activity relative to wild-type EYA (Fig. 3D), although all constructs were expressed at comparable levels as assayed by Western blots of Ub-Gal4-driven expression of UAS-EYA constructs in embryos (data not shown). Thus, ED2 and the P/S/T-rich region are critical for EYA function in vivo.
DAC does not interact with EYA or SO in S2-2H assays.
Because DAC did not affect EYA-SO-mediated transcription of ARE-luciferase, we asked whether DAC was able to interact with EYA or SO according to our S2-2H system. Gal4DBD and Gal4AD fusions were made with full-length EYA and DAC, and tests were performed in both directions. Surprisingly, no interaction was observed in either direction (data shown in one direction, Fig. 3C). We also asked whether SO might be required to nucleate an EYA-DAC complex, but did not observe an interaction (data not shown). To determine whether RAS/MAPK signaling might be required for formation of an EYA-DAC complex, we cotransfected RasV12 or used the EyaS-A and EyaS-D constructs described above, but still did not observe an EYA-DAC interaction (data not shown). SO and DAC also fail to interact in the S2-2H assay (data not shown).
EYA-EYA interactions are mediated via the N terminus.
Because the RD gene network is known to function in reiterative feedback loops, we wondered if EYA might interact with itself to potentiate or restrict function. Using our S2-2H assay, we found that full-length EYA shows a significant interaction with itself (using Gal4DBD-EYA and Gal4AD-EYA; Fig. 4), at sevenfold above background. We then used our series of deletion and truncation constructs fused to the Gal4DBD domain coexpressed with the Gal4AD-EYA fusion to ask which domain mediates this EYA-EYA interaction. In order to distinguish between the activity of the fusion alone and the S2-2H interaction, we transfected each alone and with AD-EYA (Fig. 4).
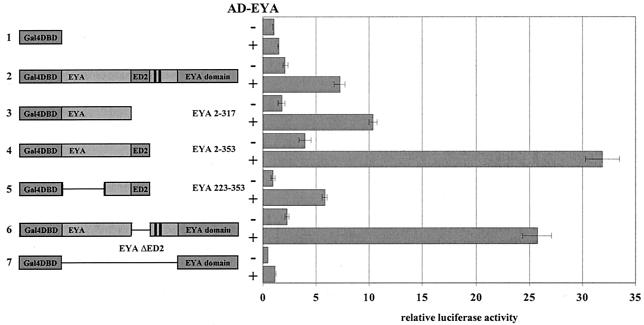
FIG. 4.
EYA-EYA interactions are mediated by the EYA N terminus. Using the S2-2H system, Gal4DBD-EYA fusions shown on the left were coexpressed with (+) and without (−) full-length Gal4AD-EYA. Full-length EYA interacts with itself sevenfold above the background level. This interaction is mediated by aa 223 to 317, because all constructs that contain this minimal region can interact with Gal4AD-EYA. Strikingly, Gal4DBD-EYA 2-353 and Gal4DBD-EYA ΔED2 (constructs 4 and 6) show a more than threefold-stronger interaction than that of full-length EYA. The Gal4DBD-ED fusion (construct 7) does not interact with Gal4AD-EYA, consistent with our finding that the EYA N terminus mediates this interaction.
As shown in Fig. 4, EYA 2-317 (construct 3) shows slightly greater interaction with AD-EYA than full-length EYA, while a construct including ED2, EYA 2-353 (construct 4), shows a striking increase in interaction relative to full-length EYA. EYA 223-353 (construct 5) shows interaction levels comparable to those of full-length EYA; however, deletion of ED2 in the context of full-length EYA also results in a striking increase in EYA-EYA interaction (Fig. 4, construct 6). The ED, which mediates EYA-SO interactions, does not interact above background level with full-length EYA (Fig. 4, construct 7). This result, coupled with the striking increase observed between EYA 2-353 and EYA, leads us to conclude that EYA-EYA interactions are likely mediated by amino acids in the N terminus and that ED2 (aa 318 to 353, Fig. 1A) and the regions immediately following it, including the MAPK phosphorylation sites, while not necessary for this interaction, may strongly potentiate it. We could not test other N-terminal EYA constructs in this assay, such as EYA ΔED and EYA ΔED2ΔED, because when expressed alone they strongly activate the reporter (Fig. 1B). Because this interaction maps to regions we describe above as crucial for full EYA transactivation potential (Fig. 1B) and for EYA function in vivo (Fig. 3D), it seems likely that the EYA-EYA interaction is functionally significant and could contribute to regulated transcriptional activity.
Phosphorylation increases EYA transactivation potential in the context of the EYA-SO transcription factor.
Having established an assay in which full-length EYA acts as a transcriptional coactivator when complexed to SO, we returned to the question of whether EYA activity is regulated by RAS/MAPK signaling. Although activated RAS did not affect EYA-SO mediated transcriptional regulation, nor did mutation of the EYA phosphoacceptor sites to alanine (Fig. 5A), we found that the EYAS-D/E-SO complex consistently exhibited a 50% increase in transactivation levels relative to the EYA-SO complex (Fig. 5A). This result is consistent with previously reported transgenic analyses, wherein overexpression of EYAS-D/E led to stronger and more penetrant phenotypes than did EYA (20). Analysis of protein levels in flies expressing EYAS-A or EYAS-D/E transgenes indicated no differences in protein stability (20), and we repeated these results in cell culture with quantitative Western blots as shown in Fig. 5B, where wild-type and mutant EYA proteins are expressed at the same levels. The S2-2H system was used to rule out the possibility that the transactivation increase seen with the EYAS-D/E-SO complex results from an increase in EYA-SO interactions (data not shown). Therefore, the increase in transactivation exhibited by the EYAS-D/E-SO complex suggests that phosphorylation directly increases EYA's transactivation potential.
Because we have seen this result only upon mutation of the phosphoacceptor site, but not in response to RAS/MAPK activation, it remains possible that a different signaling pathway mediates this phosphorylation event. However, previously reported genetic and biochemical evidence (20) indicates that RAS/MAPK signaling is responsible for phosphorylation of this site in vivo. Thus we favor the interpretation that EYA transactivation is potentiated by MAPK phosphorylation, as evidenced by the increased activity of the EYAS-D/E-SO complex, and that the lack of response to RAS stimulation likely reflects a more complex role for RAS within the RD gene network.
GROUCHO is a repressor of the EYA-SO transcription factor.
Recent studies of zebra fish (26), mice (46), and medaka fish (31) have revealed a functional role for SIX3 interactions with GROUCHO (GRO), a transcriptional corepressor. One of these studies also demonstrates weak interactions between SO and a murine GROUCHO homolog, GRG5 (46). Because SO belongs to a different class of SIX homologs and the SIX3/SIX6 families are distinct from other families in that they do not interact with EYA or EYA homologs, we wanted to ask if this SO-GRG5 interaction indicated a functional role for GRO in regulation of the EYA-SO transcription factor. We found that coexpression of GRO strongly reduces but does not eliminate activation via the EYA-SO transcription factor. As shown in Fig. 6A, EYA-SO activates transcription more than 33-fold, while coexpression of GRO abrogates activation to only 20-fold.
This corepressor function of GRO may be mediated by interactions with SO through the previously characterized engrailed homology 1 (eh1) domains within the SIX domain (26) or may be mediated through an eh1 domain found within the ED (Z. Paroush, personal communication). In order to address this question, we performed coimmunoprecipitation (co-IP) to look at direct interactions between GRO and EYA or SO. Strikingly, we found that GRO can co-IP with SO alone, but not in the presence of EYA (Fig. 6B, lanes 5 and 4). This is not due to competition with EYA for GRO binding, because IP of EYA cannot co-IP GRO (Fig. 6B, lane 6). We therefore propose an additional negative regulatory mechanism for EYA-SO targets, whereby in the absence of high levels of EYA, GRO interactions with SO lead to repression and downregulation of target genes. Thus, SO may function both as a transcriptional activator and repressor dependent upon the context-specific expression levels of particular cofactors.
DISCUSSION
The RD gene network encodes proteins that operate in multiple contexts to effect differentiation of various cell types. Input from extracellular signaling pathways, such as the RAS/MAPK cascade, may provide instructive, context-dependent cues that regulate the expression and/or activity of RD gene family members. We have shown that Drosophila EYA is a potent transactivator, either on a heterologous promoter or in conjunction with SO, and that this activity maps to an internal P/S/T-rich region encompassing the ED2 and MAPK consensus sites. This activity is negatively regulated by the ED and positively modulated by phosphorylation, likely through RAS/MAPK signaling. We also provide evidence for direct EYA-EYA interactions and demonstrate that the ED2 may be critical in this context. Together our results suggest a complex cooperation and interplay among the distinct structural motifs of EYA that reflects the importance of proper regulation of the RD gene network.
Our transcription assay results correlate well with those obtained in mammalian cell culture studies of murine EYA homologs, mEYA1 to -4, which showed that their N termini can function as transactivators on a heterologous promoter (44). Our functional dissection of Drosophila EYA enables us to propose a role for a second, and previously uncharacterized, conserved domain in EYA, ED2, in mediating EYA transactivation potential. The P/S/T-rich region surrounding ED2, which includes two MAPK phosphorylation consensus sites, is absolutely necessary for EYA transactivation, and both ED2 and the P/S/T-rich region are essential for EYA function in vivo.
Ectopic expression of EYA is associated with a wide range of deleterious phenotypes (20), suggesting that EYA activity must be precisely regulated to ensure appropriate growth and development. Our observation that the conserved EYA domain functions as an autoregulatory inhibitor of EYA transactivation potential suggests that regulation of and by this domain is critical for proper EYA function. Relief of this inhibition in vivo may require cofactor binding or protein modification. Alternatively, the inhibition mediated by the ED might be dependent on interactions with an unidentified negative regulator of EYA. In this context, it would be interesting to ask whether any of the BOR alleles that map to the ED (35) affect the transactivation potential of human EYA1.
Another negative regulatory component of the EYA-SO transcription factor arises from our finding that striking repression is achieved in the presence of GROUCHO. Furthermore, we provide evidence that SO-GRO interactions are disrupted by EYA, providing an intriguing model for SO target gene regulation. Because EYA and SO are not entirely coexpressed and GRO is widely expressed (25), a SO-GRO complex may provide tight regulation of EYA-SO targets, functioning as an “off” switch in the absence of EYA. This may explain the lack of ectopic eye induction seen upon overexpression of SO alone (37) compared to that with EYA alone (5) and may be a mechanism for the cooperativitity observed when EYA and SO are coexpressed (37), since EYA is necessary to overcome SO-GRO-mediated repression.
Our finding that DAC does not interact with EYA or SO in S2-2H assays is surprising, but it does not preclude their interaction in vivo. It is possible that the EYA-DAC interaction requires cofactors or modifications not made in Drosophila S2 cell culture or that some factor present in S2 cells inhibits the interaction. As well, the use of Gal4DBD and Gal4AD fusions may in some way disrupt an EYA-DAC interaction. It will be interesting to ask whether cotransfection of one known mammalian cofactor, CREB binding protein (21), can nucleate an EYA-DAC interaction in S2 cells. Alternatively, EYA may indirectly associate with DAC in the context of an as-yet-uncharacterized macromolecular complex that may vary according to the particular transcriptional target being regulated.
The observation that EYA interacts with itself reveals yet another potential mechanism for complex and reiterative interactions within the RD gene network. This interaction appears to be mediated by the N-terminal half of EYA, a region that we have found necessary for full transactivation potential. Furthermore, we find that the ED2 potentiates EYA-EYA interactions. Because the EYA-EYA interaction maps to regions of EYA necessary for transactivation, we propose that homotypic interactions may contribute to EYA function as a transcription factor in vivo.
Determination of cell fate is dependent on both the presence of a particular complement of transcription factors and the appropriate activation state of these factors. Here we provide evidence that while RAS/MAPK activation is not necessary for EYA transactivation potential, it can potentiate EYA-mediated transactivation. This leads to an intriguing mechanism for modulation of the EYA-SO transcription factor, whereby in the absence of RAS/MAPK signaling, it is competent to activate some transcription, but in the presence of signal, this function is potentiated such that target genes may be activated to higher levels. An alternative but not mutually exclusive role for RAS/MAPK activation of EYA may be to allow the higher activation potential of EYA to overcome negative regulation of specific target genes.
Activation of EYA by RAS/MAPK signaling provides a direct point of cross talk between a signal transduction module and the RD gene network. Our results suggest that RAS/MAPK signaling may regulate multiple aspects of RD network function. In the context of Gal4DBD-EYA fusions acting on UAS-luciferase, RAS signaling clearly increases transactivation activity. Surprisingly, we found that mutation of the MAPK phosphoacceptor sites in this fusion protein to either alanine, to prevent phosphorylation, or a negatively charged residue, to mimic phosphorylation, results in higher transactivation levels. We believe that these results may be due the nature of this assay, which uses chimeric and truncated Gal4DBD-EYA ΔED fusions, since we did not observe the same effect in the context of full-length EYA working with SO to promote transcription. Rather, in the context of an EYA-SO complex acting on ARE-luciferase, RASV12 does not affect transcription, yet there is a consistently strong increase in activation when the phosphomimetic EYAS-D/E is used. One possible explanation for the lack of RAS responsiveness is that RAS signaling could simultaneously upregulate EYA but downregulate SO. This would provide a mechanism for fine-tuned transcriptional regulation, whereby RAS signaling can activate the EYA-SO transcription factor through phosphorylation of EYA but negatively regulates SO to prevent sustained high levels of activation. Such dual and conflicting inputs by the RAS/MAPK pathway are consistent with previous genetic observations. Specifically, our previous work has demonstrated a positive role for the pathway with respect to RD network function using EYA as a point of cross talk, whereas work by others has implicated the RAS pathway as antagonizing RD gene function, although in this case, the molecular mechanisms underlying the inhibitory regulation are unknown (28). Thus, it remains to be seen how RAS/MAPK signaling regulates SO or other members of the RD gene network or whether this direct interaction is unique to EYA.
In addition to RAS/MAPK signaling, Notch, Hedgehog, Wingless, and TGFβ/DPP signaling all play important roles in eye development (42). The integration of multiple signaling pathway inputs with our existing knowledge of RD gene network transcriptional regulatory loops suggests a mechanism for unique specification of multiple cell fates in the eye. Our work provides evidence that EYA is a crucial modulator of its own activity, through autoinhibition and homotypic interactions. We also show evidence that the RAS/MAPK pathway directly enhances the transactivation potential of EYA and that the corepressor GROUCHO inhibits EYA-SO-mediated transcription. It will be important to discover whether other signaling pathways interact directly with RD gene network proteins and how such inputs effect the expression of distinct cadres of target genes, thereby establishing and/or reinforcing unique cell fates.
Acknowledgments
We thank S. Artavanis-Tsakonas for GRO monoclonal antibody, Z. Paroush for gro cDNA, G. Mardon for so and dac plasmids, and R. Fehon and T. Orr-Weaver for helpful comments on the manuscript. We thank all members of the Rebay laboratory for their valuable advice on this project and the manuscript.
S.J.S. is a Howard Hughes Medical Institute Predoctoral Fellow. I.R. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and is a Rita Allen Foundation Scholar. This work was supported in part by National Institutes of Health grant RO1 EY-12549.
REFERENCES
- 1.Abdelhak, S., V. Kalatzis, R. Heilig, S. Compain, D. Samson, C. Vincent, D. Weil, C. Cruaud, I. Sahly, M. Leibovici, M. Bitner-Glindzicz, M. Francis, D. Lacombe, J. Vigneron, R. Charachon, K. Boven, P. Bedbeder, N. Van Regemorter, J. Weissenbach, and C. Petit. 1997. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 15:157-164. [DOI] [PubMed] [Google Scholar]
- 2.Bai, J., and D. Montell. 2002. Eyes Absent, a key repressor of polar cell fate during Drosophila oogenesis. Development 129:5377-5388. [DOI] [PubMed] [Google Scholar]
- 3.Baonza, A., and M. Freeman. 2002. Control of Drosophila eye specification by Wingless signalling. Development 129:5313-5322. [DOI] [PubMed] [Google Scholar]
- 4.Bessa, J., B. Gebelein, F. Pichaud, F. Casares, and R. S. Mann. 2002. Combinatorial control of Drosophila eye development by Eyeless, Homothorax, and Teashirt. Genes Dev. 16:2415-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonini, N. M., Q. T. Bui, G. L. Gray-Board, and J. M. Warrick. 1997. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124:4819-4826. [DOI] [PubMed] [Google Scholar]
- 6.Bonini, N. M., W. M. Leiserson, and S. Benzer. 1998. Multiple roles of the eyes absent gene in Drosophila. Dev. Biol. 196:42-57. [DOI] [PubMed] [Google Scholar]
- 7.Brand, A., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 8.Bui, Q. T., J. E. Zimmerman, H. Liu, and N. M. Bonini. 2000. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155:709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R., M. Amoui, Z. Zhang, and G. Mardon. 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91:893-903. [DOI] [PubMed] [Google Scholar]
- 10.Chen, R., G. Halder, Z. Zhang, and G. Mardon. 1999. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development 126:935-943. [DOI] [PubMed] [Google Scholar]
- 11.Cheyette, B. N., P. J. Green, K. Martin, H. Garren, V. Hartenstein, and S. L. Zipursky. 1994. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12:977-996. [DOI] [PubMed] [Google Scholar]
- 12.Curtiss, J., and M. Mlodzik. 2000. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127:1325-1336. [DOI] [PubMed] [Google Scholar]
- 13.Czerny, T., G. Halder, U. Kloter, A. Souabni, W. J. Gehring, and M. Busslinger. 1999. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3:297-307. [DOI] [PubMed] [Google Scholar]
- 14.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 15.Flores, G. V., H. Duan, H. Yan, R. Nagaraj, W. Fu, Y. Zou, M. Noll, and U. Banerjee. 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103:75-85. [DOI] [PubMed] [Google Scholar]
- 16.Galindo, M. I., S. A. Bishop, S. Greig, and J. P. Couso. 2002. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science 297:256-259. [DOI] [PubMed] [Google Scholar]
- 17.Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter, and W. J. Gehring. 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125:2181-2191. [DOI] [PubMed] [Google Scholar]
- 18.Halfon, M. S., A. Carmena, S. Gisselbrecht, C. M. Sackerson, F. Jimenez, M. K. Baylies, and A. M. Michelson. 2000. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103:63-74. [DOI] [PubMed] [Google Scholar]
- 19.Heanue, T. A., R. Reshef, R. J. Davis, G. Mardon, G. Oliver, S. Tomarev, A. B. Lassar, and C. J. Tabin. 1999. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13:3231-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao, F. C., A. Williams, E. L. Davies, and I. Rebay. 2001. Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev. Cell 1:51-61. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, K., Y. Watanabe, H. Ohto, and K. Kawakami. 2002. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell. Biol. 22:6759-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami, K., H. Ohto, K. Ikeda, and R. G. Roeder. 1996. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 24:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami, K., S. Sato, H. Ozaki, and K. Ikeda. 2000. Six family genes—structure and function as transcription factors and their roles in development. BioEssays 22:616-626. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S. S., R. Zhang, S. E. Braunstein, A. Joachimiak, A. Cvekl, and R. S. Hegde. 2002. Structure of the retinal determination protein dachshund reveals a DNA binding motif. Structure (Cambridge) 10:787-795. [DOI] [PubMed] [Google Scholar]
- 25.Knust, E., K. A. Bremer, H. Vassin, A. Ziemer, U. Tepass, and J. A. Campos-Ortega. 1987. The enhancer of split locus and neurogenesis in Drosophila melanogaster. Dev. Biol. 122:262-273. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, M., K. Nishikawa, T. Suzuki, and M. Yamamoto. 2001. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 232:315-326. [DOI] [PubMed] [Google Scholar]
- 27.Kronhamn, J., E. Frei, M. Daube, R. Jiao, Y. Shi, M. Noll, and A. Rasmuson-Lestander. 2002. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development 129:1015-1026. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, P., and K. Moses. 2001. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell 104:687-697. [DOI] [PubMed] [Google Scholar]
- 29.Kurusu, M., T. Nagao, U. Walldorf, S. Flister, W. J. Gehring, and K. Furukubo-Tokunaga. 2000. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the Eyeless, Twin of eyeless, and Dachshund genes. Proc. Natl. Acad. Sci. USA 97:2140-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X., V. Perissi, F. Liu, D. W. Rose, and M. G. Rosenfeld. 2002. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297:1180-1183. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Rios, J., K. Tessmar, F. Loosli, J. Wittbrodt, and P. Bovolenta. 2003. Six3 and Six6 activity is modulated by members of the groucho family. Development 130:185-195. [DOI] [PubMed] [Google Scholar]
- 32.Mardon, G., N. M. Solomon, and G. M. Rubin. 1994. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120:3473-3486. [DOI] [PubMed] [Google Scholar]
- 33.Ohto, H., S. Kamada, K. Tago, S.-I. Tominaga, H. Ozaki, S. Sato, and K. Kawakami. 1999. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137-147. [DOI] [PubMed] [Google Scholar]
- 35.Ozaki, H., Y. Watanabe, K. Ikeda, and K. Kawakami. 2002. Impaired interactions between mouse Eyal harboring mutations found in patients with branchio-oto-renal syndrome and Six, Dach, and G proteins. J. Hum. Genet. 47:107-116. [DOI] [PubMed] [Google Scholar]
- 36.Pascal, E., and R. Tjian. 1991. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 5:1646-1656. [DOI] [PubMed] [Google Scholar]
- 37.Pignoni, F., B. Hu, K. H. Zavitz, J. Xiao, P. A. Garrity, and S. L. Zipursky. 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91:881-891. [DOI] [PubMed] [Google Scholar]
- 38.Pignoni, F., and S. L. Zipursky. 1997. Induction of Drosophila eye development by decapentaplegic. Development 124:271-278. [DOI] [PubMed] [Google Scholar]
- 39.Rebay, I., R. G. Fehon, and S. Artavanis-Tsakonas. 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74:319-329. [DOI] [PubMed] [Google Scholar]
- 40.Seo, H. C., J. Curtiss, M. Mlodzik, and A. Fjose. 1999. Six class homeobox genes in Drosophila belong to three distinct families and are involved in head development. Mech. Dev. 83:127-139. [DOI] [PubMed] [Google Scholar]
- 41.Shen, W., and G. Mardon. 1997. Ectopic eye development in Drosophila induced by directed dachshund expression. Development 124:45-52. [DOI] [PubMed] [Google Scholar]
- 42.Treisman, J. E., and U. Heberlein. 1998. Eye development in Drosophila: formation of the eye field and control of differentiation. Curr. Top. Dev. Biol. 39:119-158. [DOI] [PubMed] [Google Scholar]
- 43.Xu, C., R. C. Kauffmann, J. Zhang, S. Kladny, and R. W. Carthew. 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103:87-97. [DOI] [PubMed] [Google Scholar]
- 44.Xu, P. X., J. Cheng, J. A. Epstein, and R. L. Maas. 1997. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc. Natl. Acad. Sci. USA 94:11974-11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, P. X., I. Woo, H. Her, D. R. Beier, and R. L. Maas. 1997. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development 124:219-231. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, C. C., M. A. Dyer, M. Uchikawa, H. Kondoh, O. V. Lagutin, and G. Oliver. 2002. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129:2835-2849. [DOI] [PubMed] [Google Scholar]
- 47.Zimmerman, J. E., Q. T. Bui, E. Steingrimsson, D. L. Nagle, W. Fu, A. Genin, N. B. Spinner, N. G. Copeland, N. A. Jenkins, M. Bucan, and N. M. Bonini. 1997. Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res. 7:128-141. [DOI] [PubMed] [Google Scholar]