Abstract
Salmonella spp. are enteropathogenic gram-negative bacteria that use a large array of virulence factors to colonize the host, manipulate host cells, and resist the host's defense mechanisms. Even closely related Salmonella strains have different repertoires of virulence factors. Bacteriophages contribute substantially to this diversity. There is increasing evidence that the reassortment of virulence factor repertoires by converting phages like the GIFSY phages and SopEΦ may represent an important mechanism in the adaptation of Salmonella spp. to specific hosts and to the emergence of new epidemic strains. Here, we have analyzed in more detail SopEΦ, a P2-like phage from Salmonella enterica serovar Typhimurium DT204 that encodes the virulence factor SopE. We have cloned and characterized the attachment site (att) of SopEΦ and found that its 47-bp core sequence overlaps the 3′ terminus of the ssrA gene of serovar Typhimurium. Furthermore, we have demonstrated integration of SopEΦ into the cloned attB site of serovar Typhimurium A36. Sequence analysis of the plasmid-borne prophage revealed that SopEΦ is closely related to (60 to 100% identity over 80% of the genome) but clearly distinct from the Fels-2 prophage of serovar Typhimurium LT2 and from P2-like phages in the serovar Typhi CT18 genome. Our results demonstrate that there is considerable variation among the P2-like phages present in closely related Salmonella spp.
Bacteria of the genus Salmonella enterica can cause diseases ranging from self-limiting enterocolitis to systemic infection (typhoid fever). It is well established that most infections in warm-blooded animals are caused by S. enterica strains belonging to subspecies 1 and that different S. enterica subspecies 1 serovars are often characterized by different host ranges, disease, and epidemic fitness. Interestingly, even different strains belonging to the same S. enterica subspecies 1 serovar (i.e., Typhimurium) can have different virulence potentials. It is thought that these strain-specific differences are (at least partially) attributable to additional genes present in some strains but absent in others (10, 13, 14, 24, 25, 26, 28, 30, 34).
Work done in recent years has revealed that temperate phages, including the lambdoid GIFSY-1, -2, -3 phages and phages e, P22, Fels-1, and SopEΦ, are important vehicles for horizontal gene transfer between different S. enterica strains (4). These phages encode extra gene cassettes (morons) that are not essential for the phage life cycle but enhance the proliferation of the prophage by improving the fitness and/or virulence of the lysogen. The moron-encoded functions include superoxide dismutases, enzymes for lipopolysaccharide modification, and toxins (effector proteins) that are injected by the bacteria into cells of the host animal by specialized type III secretion systems (17).
The P2-like phage SopEΦ has been identified in serovar Typhimurium strain DT49/DT204, which caused an epidemic in the United Kingdom and the former East Germany during the 1970s and 1980s. In the tail fiber region of SopEΦ, we had identified a moron encoding the type III effector protein SopE (27, 28). After injection into animal cells, SopE modulates host cellular signaling and leads to dramatic responses like membrane ruffling and invasion of host cells (12, 36). Disruption of the sopE gene reduces the invasiveness of the natural SopEΦ lysogen serovar Typhimurium SL1344 (27, 33, 40). Also, lysogenic conversion of virulent serovar Typhimurium strain ATCC 14028 (normally SopEΦ−) with SopEΦ enhances its invasiveness in cultured cells (S. Mirold and W.-D. Hardt, unpublished data) and its enteropathogenesis in a bovine infection model (39). These observations indicate that lysogenic conversion by SopEΦ has been an important step in the emergence of epidemic serovar Typhimurium strain DT49/DT204. For this reason, it was of interest to characterize the integration site (att) and the sequence of SopEΦ in more detail.
Here we describe the identification of the attachment site of SopEΦ. In addition, we have “captured” a plasmid-borne version of SopEΦ by using a vector that harbors the bacterial attachment site (attB). This allowed purification of the prophage DNA, sequence analysis, and comparison of SopEΦ with P2 and P2-like phages present in the genomes of serovar Typhimurium and serovar Typhi.
MATERIALS AND METHODS
Bacterial strains.
Serovar Typhimurium reference strain A36 (1) is from the Robert Koch Institut, Wernigerode, Germany. Serovar Typhimurium strain DT204 isolate 3351/78 (a natural SopEΦ lysogen) and its isogenic derivative M106 (3351/78, a SopEΦsopE::aphT lysogen) have been described recently (28). M824 is a SopEΦsopE::aphT lysogen of A36.
Recombinant DNA techniques and sequencing.
pM36 has been described previously and harbors a 4.5-kb EcoRV/NcoI fragment of serovar Typhimurium M106 (3351/78, SopEΦsopE::aphT) comprising the right end of the SopEΦsopE::aphT prophage, attR, and the flanking bacterial sequences (28). This vector was used to sequence the attR region of the SopEΦsopE::aphT prophage.
pM47 harbors the attB core sequence of serovar Typhimurium A36. It was isolated via colony hybridization with the insert of pM36 as a probe from a library of 1- to 1.6-kb chromosomal EcoRI/EcoRV DNA fragments from serovar Typhimurium A36 cloned into the pBluescript SKII+ vector (Stratagene).
pM800 (Cmr) is a pACYC184 derivative harboring a 1.2-kb BamHI/HindIII fragment of pM47 that harbors the attB site.
pM802 (Cmr Kanr) was obtained by integration of SopEΦsopE::aphT into the attB site of pM800.
For Southern blot analysis, DNA was isolated (QIAamp DNA Mini Kit) and digested with endonucleases and the DNA fragments were resolved through 1% agarose gels. DNA was transferred to ZETA-Probe BT blotting membrane with a vacuum blotter (Bio-Rad Laboratories). Southern hybridization was performed overnight at 58°C. Fluorescein-modified probes and detection were prepared in accordance with the protocols of the manufacturer of the random prime labeling system, version II (Amersham Biosciences, Amersham, England).
The attB site of serovar Typhimurium A36 was amplified with primers 48F (5′-GATCGCTGGTCCGTTTCTTCT-3′) and 36R (5′-CGGGGATCAAGAGAGGTCAAAC-3′). The attL site of SopEΦ in M824 was amplified with primers attBF (5′-TCTGTGCTACGGGGCTTCGC-3′) and 1R (5′-CATCCCCTTCAGCCAACGTC-5′), and the attR site was amplified with primers 36R and 36F (5′-CGGAAGCAAAACAGGGTGATTA-3′). The attP site of SopEΦ was amplified with primers 2R (5′-GTTCTGCAGTTTGCTGGAGAGAGATGAATAGGC-3′) and 36F. The initial denaturation step (94°C, 7 min) was followed by 35 cycles of denaturing, annealing, and extension and one final extension step. The annealing and extension temperatures were set according to the primers used (Metabion, Martinsried, Germany, and Microsynth GmbH, Balgach, Switzerland). PCR amplification products were subcloned into the 2.1-Topo vector (Invitrogen) in accordance with the manufacturer's protocol and sequenced with primers M13 and T7, which anneal inside the vector. All of the phage sequences described in the present report were retrieved from the database except for that of SopEΦsopE::aphT (this study). Shotgun-cloned fragments of the captured phage (pM802) were sequenced by AGOWA (Berlin, Germany). The sequence was assembled with the DNASTAR/Seqman 5.05 program (DNASTAR Inc., Madison, Wis.), and open reading frames (ORFs; start codons ATG and GTG) were predicted with the DNASTAR/GeneQuest 5.05 program. Assigned ORFs were compared to those in the database with the National Center for Biotechnology Information blastx program. To compare phage genomes, we aligned the deduced amino acid sequences by using blastx. The graphic representation of the alignments was prepared with PowerPoint XP software.
Lytic induction and propagation of phages.
Lysates of SopEΦsopE::aphT were produced as described recently (28). Briefly, overnight cultures of the lysogens M106 (3351/78, SopEΦsopE::aphT) were diluted 1:10 into fresh Luria-Bertani medium (LB; 1.5 ml) supplemented with 2 μg of mitomycin C (Sigma) per ml and grown for 6 h at 37°C. Lysates were processed by centrifugation (10,000 × g, 5 min, 4°C) and 0.45-μm-pore-size membrane filtration. Purification, determination of phage titers, and preparation of plate lysates on serovar Typhimurium A36 were performed in accordance with standard protocols for phage λ (31). Plate lysates typically had titers of about 107 PFU/ml. For the definition of the SopEΦ-attP site, 7 ml of an overnight LB culture of strain M824 (carrying the phage integrated into the chromosome) was incubated with 2 μg of mitomycin C per ml. After 6 h of incubation, the cells were centrifuged. The pellet was resuspended in 200 μl of S1 (Qiagen protocol) buffer, and then 200 μl of S2 and S3 buffer was added. After centrifugation (10 min, 10,000 × g), the supernatant and 400 μl of isopropanol were centrifuged for an additional 30 min. Afterward, the pellet was washed (70% ethanol), dried, and resuspended in 140 μl of double-distilled H2O. Five microliters of this DNA preparation was used as the template for PCR amplification (50 μl) of the attP region.
Lysogenic conversion of bacterial strains.
Preparation of lysogens was performed as previously described (28). We incubated 106 PFU of SopEΦsopE::aphT for 15 min at 21°C with 4 × 107 bacteria (kanamycin sensitive) in a total volume of 100 μl of λ buffer. The mixture was plated on LB (50 μg of kanamycin per ml), and kanamycin-resistant clones were purified by streaking on selective agar plates.
Nucleotide sequence accession numbers.
The nucleotide sequences of pM36, pM47, and SopEΦ have been deposited in the GenBank database and assigned the following accession numbers: pM36 (right terminal region of the SopEΦ prophage), AY319520; pM47 (chromosomal region of serovar Typhimurium A36 harboring the SopEΦ attachment site), AY316313; SopEΦsopE::aphT prophage, AY319521.
RESULTS
Subcloning and sequence analysis of the attB site.
Bacteriophage P2 integrates into the host chromosome via site-specific recombination between the attB site of the bacterial chromosome and the attP site of the circularized P2 DNA. This leads to the “recombinant” attR and attL sites at either end of the integrated prophage. All four types of P2 attachment site (attB, attP, attL, and attR) have a 27-nucleotide (nt) conserved core sequence that serves as a key recognition site for the P2 enzymes catalyzing integration and excision. Even between related phages, attachment sites differ significantly (2, 16, 37, 38).
We wanted to identify the attachment site of P2-like phage SopEΦ. In previous work, the right end of the SopEΦ prophage (pM36) from the natural lysogen serovar Typhimurium DT204 isolate 3351/78 had been subcloned (28). pM36 comprises the attR site of the SopEΦ prophage, which is a hybrid attachment site with one part originating from the unoccupied attB site of the bacterial chromosome (Fig. 1). For this reason, the pM36 insert could be used as a probe to identify the attB site from serovar Typhimurium A36. A36 does not naturally harbor a SopEΦ prophage but readily yields lysogens. A library of EcoRI/EcoRV-restricted chromosomal DNA fragments from serovar Typhimurium A36 was screened by colony hybridization, and a 1.3-kb DNA insert (pM47) harboring the attB region was retrieved (see Materials and Methods).
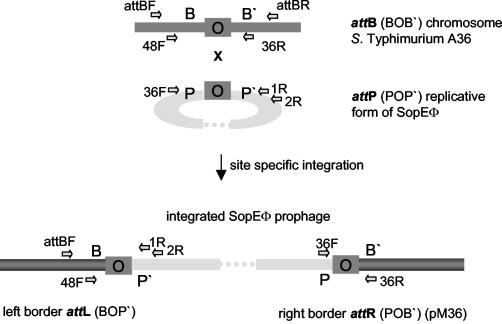
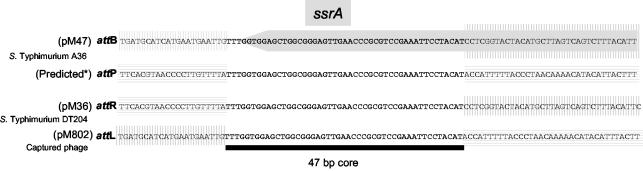
FIG. 1.
Map of the chromosomal SopEΦ attachment site (attB), the circular replicative form of SopEΦ (harboring attP), and the left (attL) and right (attR) sites of the integrated SopEΦ prophage. Site-specific integration of the phage into the bacterial chromosome occurs at the core (“O”) part of attB (BOB′). The integration leads to the duplication of the core at either end of the prophage and to formation of the hybrid attL (BOP′) and attR (POB′) sites. att core sites are marked as gray boxes. Horizontal arrows indicate the positions of the primers used in this study.
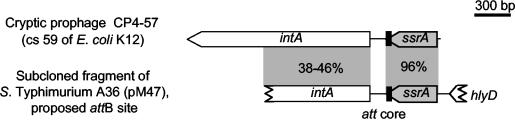
Sequence analysis revealed that pM47 bears part of an integrase-like gene (intA) that is similar to integrases of P4-like phages (i.e., CP4-57), as well as the ssrA gene (Fig. 2). The latter gene encodes the 363-nt stable tmRNA (= ssrA, 10Sa RNA), which has tRNA and mRNA functions, is required for recovery of stalled ribosomes from degraded mRNAs, and tags partially synthesized proteins for degradation (20, 21, 23). Genes in this region are induced in vivo, and deletions have been found to attenuate Salmonella virulence (8, 18). ssrA has been identified as an integration site for a cryptic P4-like phage (CP4-57) (22) from Escherichia coli, a chromosomal virulence-associated region of Dichelobacter nodosus (15), the Fels-2 phage from serovar Typhimurium LT2 (accession no. NC_003197), and VPIΦ from Vibrio cholerae, a filamentous phage encoding the Tcp adhesin (19). This suggested that the subcloned A36 fragment did indeed represent the attB site for SopEΦ. Furthermore, the similarity to CP4-57 from E. coli (Fig. 2) suggested that this attB site is the remnant of a P4-like prophage.
FIG. 2.
Alignment of the subcloned fragment of serovar Typhimurium A36 with the attachment site from the cryptic E. coli prophage CP4-57 The proposed attB site of serovar Typhimurium A36 encodes a P4-like integrase (white box), the ssrA gene, and the 3′-terminal part of hlyD (55% identical to export protein HlyD from Xylella fastidiosa).
Integration of SopEΦsopE-aphT into the plasmid-borne attB site of serovar Typhimurium A36.
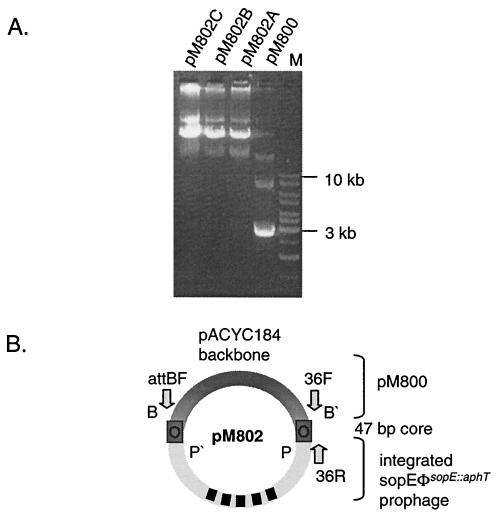
In the next step, it was necessary to analyze whether the cloned serovar Typhimurium A36 fragment of pM47 can function as the attB site for SopEΦ. For this purpose, the insert of pM47 was subcloned into pACYC184 and the resulting plasmid, pM800 (Cmr; see Materials and Methods), was transferred into serovar Typhimurium A36 (originally Cms Kms). We infected 107 CFU of serovar Typhimurium A36 (pM800) with SopEΦsopE::aphT (107 PFU/ml) and selected lysogens by plating on LB agar plates supplemented with kanamycin (50 μg/ml) and chloramphenicol (20 μg/ml). Plasmid (pM802) DNA was isolated from three independent clones (A, B, and C) and analyzed by agarose gel electrophoresis. All three pM802 clones had the same restriction pattern (data not shown) and were significantly larger than pM800, which suggested that SopEΦsopE::aphT had integrated into the attB site of pM800 (Fig. 3A).
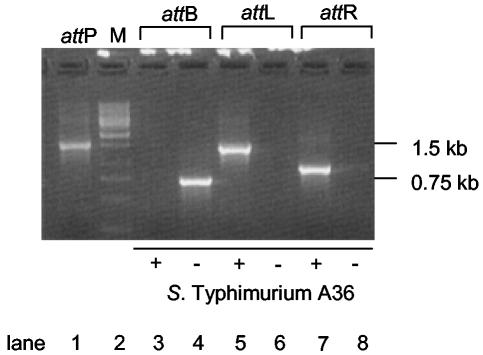
FIG. 3.
Plasmid preparation from serovar Typhimurium A36 (pM800) after infection with a SopEΦsopE::aphT lysate. (A) Plasmids of three independent clones (pM802A, -B, and -C) that were selected on LB plates supplemented with kanamycin and chloramphenicol were isolated. The larger size of pM802 plasmids suggested that SopEΦsopEΦ::aphT had integrated into the proposed attB site of pM800. (B) Schematic drawing of pM802. Dark gray, pM800 part; light gray, SopEΦsopE::aphT. att core sites are marked as gray boxes. Arrows indicate the positions of the primers used in this study.
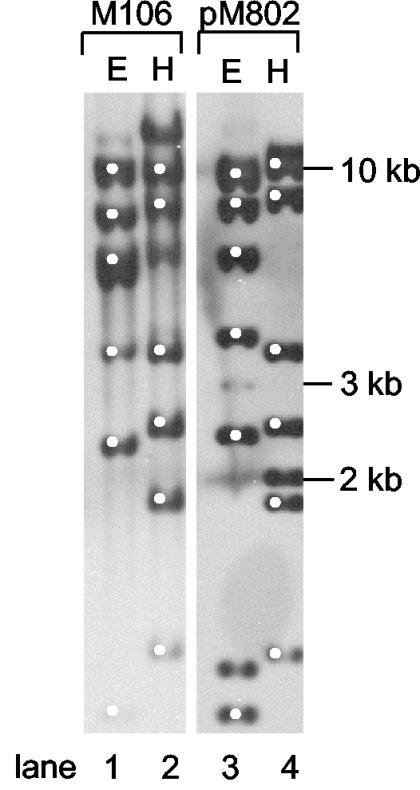
To verify that pM802 indeed harbors the SopEΦsopE::aphT prophage, Southern blot analysis was performed. Chromosomal DNA of strain M106 (the original SopEΦsopE::aphT lysogen) (28) and plasmid pM802A were digested with the enzymes EcoRV (Fig. 4, lanes 1 and 3) and HindIII. (Fig. 4, lanes 2 and 4). Hybridization was performed with the entire pM802A plasmid as a probe. The results shown in Fig. 4 confirm that pM802 harbors the SopEΦsopE::aphT prophage. Extra bands (e.g., >10 kb in lanes 2 and 4) are thought to represent the left and right junctions between the prophage and the chromosome (lane 2) or the plasmid (lane 4).
FIG. 4.
Southern blot analysis of M106 and pM802. The chromosomal DNA of M106 (SopEΦsopE::aphT lysogen) and plasmid pM802 were digested with EcoRV (E) and HindIII (H) and hybridized with the entire pM802 plasmid as a probe. White dots indicate DNA fragments identical in size in M106 and pM802.
Identification of the attB, attL, attR, and attP core sequences of SopEΦsopE::aphT.
Next, we defined the core sequence of the SopEΦ attachment site. Sequence alignment of the inserts of pM36 (attR region of the SopEΦ prophage in serovar Typhimurium DT204 3351/78) and pM47 (attB region from serovar Typhimurium A36) suggested that the “right” end of the att core sequence maps to the 3′ nucleotides of the ssrA gene. To identify the left att core sequence, we have determined the nucleotide sequence of the attL region from the SopEΦ prophage present in pM802. The primer used for this purpose, attBF (Fig. 1 and 3B), was designed on the basis of the sequenced attB region present in pM36.
Alignment of the sequence obtained (the putative attL region) with the attB and attR region sequences indicated that the conserved core sequence of the SopEΦ attachment site is 47 bp long and overlaps 45 nt of the ssrA 3′ terminus (Fig. 5). This is in line with results of the sequence comparison of the integration sites of SopEΦ-like prophages (35). Because of the duplication of these nucleotides during the integration process, the presence of SopEΦ at this position is not expected to disrupt the function of the ssrA gene. On the basis of these sequences, the attP region of the circular, replicative SopEΦ form could be predicted (Fig. 5).
FIG. 5.
Alignment of the sequenced DNA fragments corresponding to the attR, attL, attB, and attP sites. Horizontal lines mark phage DNA, and vertical lines mark bacterial DNA. The black bar indicates the 47-bp core sequence present in all four att sites. The gray arrow marks the 3′ terminus of the ssrA gene. *, later, the attP site was confirmed by PCR and sequencing with primers 2R and 36F (see Fig. 6).
In order to confirm these results, PCR analyses of the putative attB region (primers 48F and 36R) and the proposed attL (primers attBF and 1R) and attR (primers 36R and 36F) regions in wild-type serovar Typhimurium A36 and a freshly prepared serovar Typhimurium A36 SopEΦsopE::aphT lysogen (M824; see Materials and Methods) were performed. All PCR products were analyzed by agarose gel electrophoresis, cloning into the 2.1-Topo vector, and sequencing (Fig. 6 and data not shown). The PCR product corresponding to the attB sequence could be detected in the wild-type serovar Typhimurium A36 strain (Fig. 6, lane 4) but not in its lysogen (Fig. 6, lane 3). Conversely, the PCR products corresponding to the attL and attR regions could only be detected in the lysogen M824 (Fig. 6, lanes 5 and 7) but not in the parental strain (serovar Typhimurium A36; Fig. 6, lanes 6 and 8).
FIG. 6.
PCR amplification of the predicted att sites. Serovar Typhimurium A36 was infected with SopEΦsopE::aphT lysate, and lysogens were selected on kanamycin-containing LB plates. PCRs for the defined att sites were performed with primers 36R and 36F for attR, attBF and 1R for attL, and 48F and 36R for attB. attP was amplified from a plasmid preparation of mitomycin C-induced M824 with primers 36F and 2R. −, serovar Typhimurium A36; +, serovar Typhimurium A36 harboring SopEΦsopE::aphT (= M824).
In order to detect the attP site, the serovar Typhimurium lysogen M824 was treated with mitomycin C to induce the prophage (see Materials and Methods). PCR analysis of plasmid DNA prepared from this culture yielded a 1.5-kb PCR product (primers 2R and 36F). Sequencing of this PCR product verified it to be identical with the predicted attP core of the circular replicative phage (Fig. 5). These data confirmed that the attP region of the replicative form of SopEΦsopE::aphT, the attB region of serovar Typhimurium A36, and the attL/attR regions for the SopEΦsopE::aphT prophage were assigned correctly as depicted in Fig. 5.
SopEΦsopE::aphT att sequences in the published S. enterica genomes.
Database searches revealed that two copies of the 47-bp SopEΦ att core sequence are present in both completely sequenced S. enterica genomes (serovar Typhimurium strain LT2 and serovar Typhi strain CT18; Fig. 7A to D).
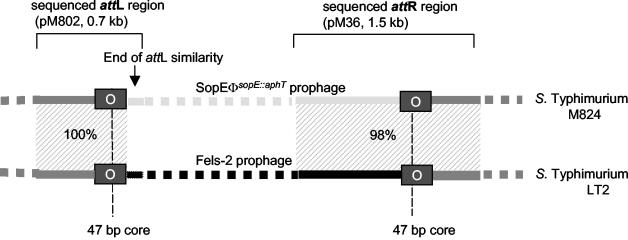
FIG. 7.
Presence of the 47-bp attachment core site of SopEΦ in M824, serovar Typhi CT18, and serovar Typhimurium LT2. Two perfectly conserved 47-bp direct repeats flank the SopEΦsopE::aphT prophage in M824 and the Fels-2 prophage in serovar Typhimurium LT2. The serovar Typhi CT18 prophage at cs 93 has the 47-bp core sequence (with one mismatch) at its right end but only a remnant of 8 nt at its left end. The cryptic phage (CT18, cs 57) harbors the 47-bp sequence at its right border. The left part of this phage is absent. Whereas the right 47-bp direct repeat of SopEΦ, Fels-2, and the cryptic phage are part of the ssrA gene that is followed by hlyD, the direct repeat of the CT18 prophage at cs 93 is located in the samA gene.
In serovar Typhi CT18 (accession no. NC_003198), one copy of the 47-bp sequence is located at centisome (cs) 93 and one is located at cs 57. The sequence at cs 93 has one mismatch (a→g) and is located at the right end of a P2-like prophage that is inserted into the samAB operon (Fig. 7C). This 47-nt sequence is part of a truncated ssrA gene, and the left end of the prophage bore an 8-nt fragment of the 47-nt sequence. This suggested that the attL site of the cs 93 prophage had been destroyed. This may be ascribed to illegitimate recombination that occurred during the insertion into the 3′ end of samA.
The cs 57 region of serovar Typhi CT18, which harbors the second 47-bp sequence, is remarkably similar to the attR region of the SopEΦsopE::aphT prophage in M824. It overlaps the 3′ terminus of the ssrA gene and is located at the right end of the ca. 9-kb tail fragment of a P2-like prophage. No significant match for the attL site could be found in the cs 57 region of the serovar Typhi CT18 genome (Fig. 7D).
In serovar Typhimurium LT2, both copies of the 47-bp att core-like sequence are located 33.7 kb apart from each other in the cs 59 region of the chromosome (Fig. 7B). They represent the attL and attR sites of the P2-like prophage Fels-2. Interestingly, there are even more similarities between both prophages. (i) The chromosomal regions flanking the attR and attL core sequences are 98 to 100% identical between the SopEΦsopE::aphT prophage in pM802 and M824 and the Fels-2 prophage from serovar Typhimurium LT2 (Fig. 8). (ii) The right end of the SopEΦ prophage directly adjacent to the att core sequence is 98% identical between the two prophages (Fig. 8). However, there are also differences between the two prophages: Fels-2 does not carry the sopE gene cassette of SopEΦ, and the sequences at the left end of both prophages (phage part of attL) are entirely different (Fig. 8). Nevertheless, SopEΦ and Fels-2 are clearly similar. This mosaic of similarity and clear differences prompted us to sequence SopEΦsopE::aphT and to compare the two phages in more detail.
FIG. 8.
Sequence comparison with attR and attL of the SopEΦsopE::aphT prophage in M824 and the Fels-2 prophage in the genome of serovar Typhimurium LT2. The chromosomal regions of M824 flanking the attR and attL core of the integrated SopEΦsopE::aphT prophage and serovar Typhimurium LT2 flanking the Fels-2 prophage are 98 and 100% identical. The right end of the SopEΦ prophage directly adjacent to the att core sequence is 98% identical between the two prophages. In contrast, the sequences at the left end of both prophages (phage part of attL) are entirely different. The values in the shaded areas indicate percent DNA sequence identity.
Sequence analysis of SopEΦsopE::aphT.
As the titers of SopEΦ lysates were generally too low (106 to 107 PFU/ml) for isolation of an amount of DNA sufficient for sequence analysis, the plasmid-borne SopEΦsopE::aphT prophage (pM802) was used to purify phage DNA. The DNA sequence of pM802 was determined as described in Materials and Methods. The sequence of wild-type SopEΦ was deduced by replacing the inserted aphT cassette with the original sequence of the sopE region from serovar Typhimurium SL1344 (accession no. AF043239). We found that the SopEΦ prophage was 36 kb in size and encoded 45 ORFs (Fig. 9; Table 1). In line with our earlier observations (14, 28), the phage is similar to the P2 phage in sequence (i.e., 40 to 67% amino acid identity in the tail fiber region) and genome organization (Fig. 9A). The similarity to P2-like prophage Fels-2 is even more striking. The two phages have 68 to 100% sequence identity over large parts of the genome (Fig. 9A). This also pertains to the orf1 to -8 region of SopEΦ, which is similar to Fels-2 but not to P2 (Fig. 9A). Nevertheless, three regions of SopEΦ are clearly different from Fels-2.
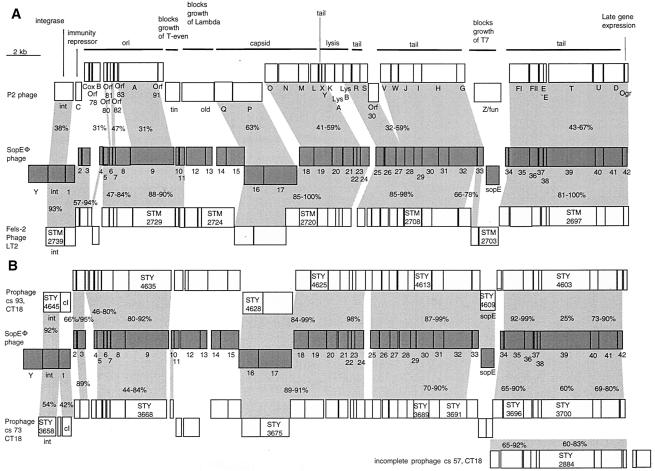
FIG. 9.
Alignment of genetic maps of SopEΦ with the P2 phage and prophages present in the sequenced genomes of serovar Typhi CT18 and serovar Typhimurium LT2. Boxes indicate the predicted ORFs starting with the phage integrase gene. Numbers in boxes refer to the chromosomal sequences of serovar Typhi CT18 and serovar Typhimurium LT2. (A) Alignment of SopEΦ (center), Fels-2 prophage (serovar Typhimurium LT2; bottom), and the P2 phage (accession no. NC_001895; top). For P2, gene functions are annotated. (B) Alignment of SopEΦ (center), the serovar Typhi CT18 prophage at cs 93 (top), the serovar Typhi CT18 prophage at cs 73, and the serovar Typhi CT18 incomplete prophage at cs 57 (bottom). The values in the shaded areas indicate percent amino acid sequence identity.
TABLE 1.
ORFs of the SopEΦ phage
| ORF | Size (kb) | % Identity (blastx) | Presencea or function in P2 phage |
|---|---|---|---|
| y | 1.21 rev.b | 27, hypothetical protein YopC of Bacillus subtilis phage SPBc2 | — |
| int | 1.0 rev. | 53, int (phage 186) | Integrase |
| 1 | 0.63 rev. | 30, Cl protein (phage 186) | Immunity repression |
| 2 | 0.25 | 31, Apl (phage 186) | — |
| 3 | 0.51 | 89, hypothetical 18.1-kDa protein of Escherichia coli retron Ec67 | — |
| 4 | 0.2 | 82, Fil (phage 186) | — |
| 5 | 0.34 | 84, hypothetical 12.8-kDa protein of Escherichia coli retron Ec67 | — |
| 6 | 0.23 | 79, hypothetical protein 79 (phage 186); 31, Orf80 (phage P2) | — |
| 7 | 0.23 | 60, Orf80 (phage 186); 47, Orf82 (phage P2) | Origin of replication |
| 8 | 0.85 | 73, retron EC67 DNA adenine methylase | — |
| 9 | 2.2 | 35, A protein (phage 186); 31, gpA (phage P2) | Origin of replication |
| 10 | 0.19 | 90, hypothetical protein of Fels-2 prophage (serovar Typhimurium LT2) | — |
| 11 | 0.23 | 42, Tum95.5 (phage 186) | — |
| 12 | 1.1 | 29, unknown (Methanothermobacter, wolfeii prophage psiM100) | — |
| 13 | 0.38 | 39, unknown (Methanothermobacter, wolfeii prophage psiM100) | — |
| 14 | 0.58 | 22, hypothetical protein (Pyrococcus furiosus DSM 3638) | — |
| 15 | 0.98 | 34, hypothetical protein (Microbulbifer degradans 2-40) | — |
| 16 | 1.0 rev. | 64, gpQ (phage P2); 63, capsid portal protein (phage 186) | Capsid |
| 17 | 1.78 rev. | 63, gpP (phage P2); 63, terminase subUnit (phage 186) | Capsid |
| 18 | 0.83 | 55, gene V protein (phage 186); 44, gpO (phage P2) | Capsid |
| 19 | 1.0 | 55, gpN (phage P2); 55, major capsid protein (phage 186) | Capsid |
| 20 | 0.65 | 42, R protein (phage 186); 41, gpM (phage P2) | Capsid |
| 21 | 0.53 | 47, Q protein (phage 186); 40, gpL (phage P2) | Capsid |
| 22 | 0.2 | 59, gpX (phage P2) | Lysis |
| 23 | 0.21 | 100, phage-holin analog protein of Fels-2 prophage (serovar Typhimunum LT2) | — |
| 24 | 0.51 | 37, gp17 lysozyme (phage P1) | — |
| 25 | 0.43 | 32, LysB (phage P2) | Lysis |
| 26 | 0.5 | 49, N protein (phage 186); 54, gpR (phage P2) | Tail |
| 27 | 0.45 | 43, O protein (phage 186); 34, gpS (phage P2) | Tail |
| 28 | 0.58 | 40, gpV (phage P2) | Tail |
| 29 | 0.36 | 59, M protein (phage 186); 55, gpW (phage P2) | Tail |
| 30 | 0.9 | 60, P2 J (phage 186); 59, gpJ (phage P2) | Tail |
| 31 | 0.6 | 58, gpI (phage P2); 57, Orf38 (phage 186) | Tail |
| 32 | 1.6 | 42, gpH (phage P2) | Tail |
| 33 | 0.4 | 50, gpG (phage P2); 46, unnamed protein product (phage 186) | Tail |
| sopE | 0.72rev. | sopE (exchanged for a kanamycin cassette in SopEΦsopE::aphT) invasion-associated secreted protein | — |
| 34 | 0.2 | 69, DNA invertase from lambdoid prophage e14 | — |
| 35 | 1.18 | 67, gpFI (phage P2); 69, (phage 186) | Tail |
| 36 | 0.51 | 50, gpFII (phage P2); 55, (phage 186) | Tail |
| 37 | 0.3 | 43, gpE (phage P2) | Tail |
| 38 | 0.12 | 57, tail protein, probably produced by a translational frameshift from orf51 into orf52 (phage 186); 55, gpE + E′ (phage P2) | Tail |
| 39 | 2.8 | 26, G protein (phage 186) | Tail |
| 40 | 0.5 | 50, gpU (phage P2); 45, F protein (phage 186) | Tail |
| 41 | 1.1 | 50, gpD (phage P2); 50, D protein (phage 186) | Late gene expression |
| 42 | 0.26 | 63, B protein (phage 186); 60, Ogr (phage P2) | Late gene expression |
—, not present.
rev., reverse.
(i) An additional 1.2-kb ORF (orfY) was located at the left end of the SopEΦ prophage (Fig. 9A). It had limited similarity (27% identical on the amino acid level) to a gene of unknown function (yopC) from the Bacillus subtilis phage SPBc2. orfY was located between the integrase gene int and the attL site. In Fels-2 and P2, int is located directly adjacent to the att sites.
(ii) orf12 to -15, which probably represent the immunity region of SopEΦ, show no similarity to Fels-2 and P2. At the equivalent position, P2 carries the tin and old genes, which block the growth of T-even and lambda phages. However, orf12 to -15 are similar to ORFs from Methanothermobacter wolfeii phages (29 and 39% identical on the amino acid level) and ORFs from Pyrococcus furiosus (gene 14; 22%) and Microbulbifer degradans (gene 15; 34%). Additional work is required to determine the function (i.e., phage exclusion, serovar Typhimurium pathogenesis) of orf12 to -15 of SopEΦ.
(iii) As described earlier, the sopE gene is located in the tail fiber region of SopEΦ at a hot spot for the insertion of additional gene cassettes (14, 17). In Fels-2 and P2, this position is occupied by orfSTM2703 and Z/fun (Fig. 9A).
In conclusion, these observations demonstrate that SopEΦ from serovar Typhimurium DT204 is a close relative of but clearly distinct from the Fels-2 phage of serovar Typhimurium LT2.
Serovar Typhi CT18 prophages related to SopEΦ.
In the serovar Typhi CT18 genome, we detected two prophages and one phage remnant with similarity to SopEΦ (Fig. 9B). Of these, the cs 93 prophage was most similar to SopEΦ. Most ORFs displayed 80 to 100% sequence identity (Fig. 9B). Moreover, the sopE moron was present and located at the same position in both phages. However, no similarity to the orf10 to -15 region of SopEΦ was detected and the cs 93 phage lacked a gene with similarity to orfY between the attL site and int (Fig. 9B). As discussed above, this phage may be defective in excision from the chromosome (Fig. 7).
The cs 73 prophage displayed extensive but somewhat less sequence similarity to SopEΦ (60 to 90% identity; Fig. 9B). It lacks genes similar to the orf11 to -15 region and to orfY of SopEΦ and carries the ORFs (STY3693 and STY3694) in place of the sopE moron. The cs 73 prophage is integrated into the yifB gene, and we did not detect sequences with similarity to the att core sequences of SopEΦ at either end of the prophage.
At cs 57, serovar Typhi CT18 carries a phage remnant similar to the tail region of SopEΦ (Fig. 9B). As discussed above, the right end of this phage remnant harbors the 47-nt SopEΦ att core sequence. However, in contrast to SopEΦ, it harbors two additional ORFs between the orf42/ogr homolog and the 47-nt SopEΦ att core sequence (Fig. 9B).
These data show that the sequence and genomic organization of SopEΦ are similar but not identical to those of prophages and phage remnants present in serovar Typhi CT18.
DISCUSSION
Work done in the last few years has established that horizontal gene transfer can modify the virulence of serovar Typhimurium strains and contribute to the emergence of new epidemic clones (29). It has been found that temperate bacteriophages are involved in this process (4, 9, 10) Therefore, it was of interest to analyze phage SopEΦ in more detail. This P2-like phage was isolated from an epidemic serovar Typhimurium strain and carries a moron encoding the virulence factor SopE (27, 28). Here, we have characterized the attachment site of SopEΦ in the serovar Typhimurium chromosome, determined the sequence of the SopEΦ prophage, and compared it to other P2-like phages form serovars Typhimurium and Typhi.
We have mapped the attachment site attB for SopEΦ to the 3′ terminus of ssrA (Fig. 7), which encodes the small stable tmRNA integration of SopEΦ. The ssrA sequence is not disrupted by the integration of SopEΦ. This may be important, since earlier work has shown that deletions in the ssrA region attenuate Salmonella virulence (18). Indeed, it has been reported that the virulence of a serovar Typhimurium ATCC 14028 SopEΦsopE::aphT lysogen is not attenuated in the bovine infection model (39). ssrA also serves as an integration site for several other phages, including VPIΦ, a filamentous phage from V. cholerae that encodes the Tcp adhesin (19), and the P4-like cryptic prophage CP4-57 from E. coli (22). Moreover, ssrA has been identified as a hot spot of integration for various genetic elements. Among enterobacteria, at least four different integrase subfamilies mediate integration into the ssrA locus (35).
This lends further support to the notion that ssrA serves (very much like tRNA genes) as an integration site for pathogenicity islands and phages (7, 11, 35).
Interestingly, the chromosomal region of serovar Typhimurium A36 that harbors the SopEΦ attachment site is similar to the P4-like cryptic prophage from E. coli (Fig. 2) (22). In both cases, a P4-like integrase is located downstream of the ssrA gene. This indicates that SopEΦ may actually use the remnant of a P4-like cryptic prophage as an attachment site in the serovar Typhimurium A36 chromosome.
SopEΦ had been isolated from epidemic serovar Typhimurium strain DT204 (28). The present work revealed that it is highly similar to the P2-like Fels-2 prophage from serovar Typhimurium LT2. The structural genes, the ori regions, and the integrases of both phages are 75 to 100% identical at the amino acid level, and the two phages seem to occupy the same chromosomal attachment sites (Fig. 7 and 9A). Nevertheless, the Fels-2 prophage encodes STM2703 in place of the sopE moron, the genes in the immunity region are different from those of SopEΦ, and Fels-2 does not harbor an orfY-like gene at the attL site (Fig. 9A). This shows that SopEΦ is a close relative of Fels-2 and indicates that the P2-like phages are diverse even between closely related S. enterica strains.
In the serovar Typhi CT18 genome, there are two prophages and one phage remnant with similarity to the P2 family. In line with the current “modular” model of phage evolution (3, 5, 6, 17, 32), different blocks of genes show different degrees of sequence similarity to SopEΦ. Nevertheless, the majority of ORFs of SopEΦ and the P2-like elements in serovar Typhi CT18 are 60 to 90% identical at the protein level (Fig. 9B). Again, the differences include the immunity region (orf10 to -15 of SopEΦ) and the absence of orfY at the attL region. The sopE moron is absent from the cs 73 prophage and the phage remnant at cs 57. In contrast, the tail region of the cs 93 prophage harbors the sopE moron and is up to 99% identical on the amino acid level to the equivalent region of SopEΦ (Fig. 9B). Taken together, these observations show that SopEΦ is distinct from but closely related to the cs 93 phage from serovar Typhi.
It remains unclear whether the two phages have diverged from a single sopE-positive ancestor. Alternatively, the high sequence similarity in the tail fiber regions including the sopE moron suggests that this region was recently transferred en bloc between or into both phages and prophages.
Are the P2-like prophages at cs 93 and 73 of serovar Typhi functional? The cs 93 prophage is inserted into the samAB UV repair operon (Fig. 7). Its attL and attR sites seem to be disrupted and show signs of illegitimate recombination. This suggests that this prophage has lost the ability to excise. Alternatively, the 8-bp repeat of the samA sequence found at the two ends of the cs 93 prophage may present a secondary attachment site for this phage (35). It remains to be determined whether the “defective” att sequences prohibit excision from the chromosome or not.
It is interesting to speculate whether the disruption of prophage attachment sites may serve the stabilization of favorable phage-encoded genes in the bacterial chromosome or the avoidance of lytic induction.
In conclusion, our data show that SopEΦ is highly similar but not identical to several P2-like prophages from Salmonella spp. There are even clear differences between SopEΦ and the cs 93 prophage from serovar Typhi CT18, which both carry the sopE moron. Earlier work on sopE+ Salmonella spp. has provided evidence for the transfer of the sopE moron between a chromosomal site and the tail fiber regions of λ- and P2-like phages (27). The data presented here show that there is a diverse population of P2-like phages even in closely related Salmonella strains and suggest that the sopE moron has been transferred between different P2-like phages. The transfer of functional phage modules between chromosomal sites and phages, their transfer between different phage families, and their transfer between different members of the same phage family are well documented (17). However, to our knowledge, the Salmonella sopE moron provides the first example of a bacterial virulence factor being transferred to a similar extent as functional phage modules. Several other virulence factors also have a diverse distribution between different Salmonella strains, and some are carried by prophages (4, 10, 24). Evidence is accumulating that some of these virulence factors have also been transferred between chromosomal sites and phages (9, 10). It is tempting to speculate that the shuffling of virulence factors between different phages may enhance the exchange of virulence factors between different Salmonella spp. and play a role in adaptation to new animal hosts and the emergence of new epidemic strains.
Acknowledgments
We thank Wolfgang Rabsch and Helmut Tschäpe for helpful discussion. We are grateful to Alexander Rakin, Andrea Friebel, and Kristin Ehrbar for comments on the manuscript and to J. Heesemann and J. E. Galán for support and scientific advice.
This work was funded by a grant from the Kompetenznetzwerk “Genomforschung an pathogenen Bakterien”—Verbund 3 to W.-D. Hardt.
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreiro, V., and E. Haggard-Ljungquist. 1992. Attachment sites for bacteriophage P2 on the Escherichia coli chromosome: DNA sequences, localization on the physical map, and detection of a P2-like remnant in E. coli K-12 derivatives. J. Bacteriol. 174:4086-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 5.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-223. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397. [Google Scholar]
- 7.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201-208. [DOI] [PubMed] [Google Scholar]
- 8.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 10.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-32. In J. Hacker and J. B. Kaper (ed.), Pathogenicity islands and other mobile virulence elements. American Society for Microbiology, Washington, D.C.
- 12.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galán. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 13.Hardt, W. D., and J. E. Galán. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94:9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardt, W. D., H. Urlaub, and J. E. Galán. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haring, V., S. J. Billington, C. L. Wright, A. S. Huggins, M. E. Katz, and J. I. Rood. 1995. Delineation of the virulence-related locus (vrl) of Dichelobacter nodosus. Microbiology 141:2081-2089. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., H. Matsumoto, M. Ohnishi, and Y. Terawaki. 1993. Molecular analysis of a cytotoxin-converting phage, φCTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol. Microbiol. 7:657-667. [DOI] [PubMed] [Google Scholar]
- 17.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 18.Julio, S. M., D. M. Heithoff, and M. J. Mahan. 2000. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol. 182:1558-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaolis, D. K., S. Somara, D. R. Maneval, J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 20.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 21.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 22.Kirby, J. E., J. E. Trempy, and S. Gottesman. 1994. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol. 176:2068-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komine, Y., M. Kitabatake, T. Yokogawa, K. Nishikawa, and H. Inokuchi. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:9223-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao, E. A., and S. I. Miller. 1999. Bacteriophages in the evolution of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 96:9452-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 27.Mirold, S., K. Ehrbar, A. Weissmüller, R. Prager, H. Tschäpe, H. Rüssmann, and W. D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 183:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschäpe, H. Rüssmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knüppel, W. Rabsch, W. D. Hardt, and H. Tschäpe. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schmieger, H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 34.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, K. P. 2003. Traffic at the tmRNA gene. J. Bacteriol. 185:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood, M., W. R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1999. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 37.Yu, A., L. E. Bertani, and E. Haggard-Ljungquist. 1989. Control of prophage integration and excision in bacteriophage P2: nucleotide sequences of the int gene and att sites. Gene 80:1-11. [DOI] [PubMed] [Google Scholar]
- 38.Yu, A., and E. Haggard-Ljungquist. 1993. Characterization of the binding sites of two proteins involved in the bacteriophage P2 site-specific recombination system. J. Bacteriol. 175:1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, S., R. L. Santos, R. M. Tsolis, S. Mirold, W. D. Hardt, L. G. Adams, and A. J. Bäumler. 2002. Phage mediated horizontal transfer of the sopE gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for calves. FEMS Microbiol. Lett. 17:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galán. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-260. [DOI] [PubMed] [Google Scholar]