Abstract
Myxococcus xanthus cells coordinate cellular motility, biofilm formation, and development through the use of cell signaling pathways. In an effort to understand the mechanisms underlying these processes, the inner membrane (IM) and outer membrane (OM) of strain DK1622 were fractionated to examine protein localization. Membranes were enriched from spheroplasts of vegetative cells and then separated into three peaks on a three-step sucrose gradient. The high-density fraction corresponded to the putative IM, the medium-density fraction corresponded to a putative hybrid membrane (HM), and the low-density fraction corresponded to the putative OM. Each fraction was subjected to further separation on discontinuous sucrose gradients, which resulted in discrete protein peaks for each major fraction. The purity and origin of each peak were assessed by using succinate dehydrogenase (SDH) activity as the IM marker and reactivities to lipopolysaccharide core and O-antigen monoclonal antibodies as the OM markers. As previously reported, the OM markers localized to the low-density membrane fractions, while SDH localized to high-density fractions. Immunoblotting was used to localize important motility and signaling proteins within the protein peaks. CsgA, the C-signal-producing protein, and FibA, a fibril-associated protease, were localized in the IM (density, 1.17 to 1.24 g cm−3). Tgl and Cgl lipoproteins were localized in the OM, which contained areas of high buoyant density (1.21 to 1.24 g cm−3) and low buoyant density (1.169 to 1.171 g cm−3). FrzCD, a methyl-accepting chemotaxis protein, was predominantly located in the IM, although smaller amounts were found in the OM. The HM peaks showed twofold enrichment for the type IV pilin protein PilA, suggesting that this fraction contained cell poles. Two-dimensional polyacrylamide gel electrophoresis revealed the presence of proteins that were unique to the IM and OM. Characterization of proteins in an unusually low-density membrane peak (1.072 to 1.094 g cm−3) showed the presence of Ta-1 polyketide synthetase, which synthesizes the antibiotic myxovirescin A.
The cell envelope defines the border of living cells, mediates interactions with the environment, and is the arena for numerous processes important for cellular survival. In gram-negative bacteria, membranes aid in the assembly of cellular appendages, regulation of a motility motor(s), and coordination of cell division. Membranes are also involved in the biosynthesis of cell envelope components, such as lipopolysaccharide (LPS), peptidoglycan, and phospholipids, and the secretion of extracellular matrix components, such as enzymes, polysaccharides, and secondary metabolites. Membranes of Myxococcus xanthus support two motility motors and enable biogenesis of two different types of appendages, type IV pili (TFP) (19) and fibrils (3, 4). In addition, membranes facilitate cell-to-cell exchange of motility proteins (16) and specific signals required for development (15).
M. xanthus cells move by translocation along the long axis accompanied by periodic reversals in direction (32). This type of movement involves coordination of the social (S) and adventurous (A) motility motors, which govern group and individual cell movement, respectively (16). The A motility motor has not been identified yet, although it has been suggested that cells secrete a propellant (60). S motility is similar to twitching of Neisseria gonorrhoeae and Pseudomonas aeruginosa and requires TFP, which are polar appendages made of PilA pilin subunits. TFP generate movement by extension, attachment, and retraction that pull the cell forward like a grappling hook (35, 51, 53). The TFP gene cluster of M. xanthus contains at least 14 homologues of P. aeruginosa TFP genes but also requires unique genes encoding an ATP transporter and the lipoprotein Tgl (32, 45, 63). S motility requires appendages known as fibrils, a polysaccharide core decorated with proteins that constitutes the extracellular matrix, and LPS O-antigen (2, 7).
Under starvation conditions, M. xanthus cells exchange signals that orchestrate a developmental program for the production of fruiting bodies containing dormant, asexual spores (48). Cells incapacitated in the production of one of the signals can be extracellularly complemented for development in the presence of wild-type cells (15). The predominant developmental signal is the C-signal, which acts as a timer to sequentially induce each stage of fruiting body morphogenesis (22, 26, 29). C-signaling requires CsgA, a membrane-associated protein that activates gene expression (25, 49, 50). CsgA also modifies cell motility by increasing cellular speed and decreasing cellular reversal (17). CsgA is present in two forms, a 25-kDa protein with homology to short-chain alcohol dehydrogenases (28) and a processed 17-kDa form (23, 26). It is unclear whether the 17-kDa form of CsgA acts as a protein signal or if the signal is an unidentified enzymatic product of the 25-kDa CsgA.
In this study inner membranes (IM) and outer membranes (OM) of vegetative M. xanthus DK1622 cells were separated, and the locations of several motility and signaling proteins were examined by Western blotting. The Tgl and Cgl motility proteins, which can be transferred between cells by contact, were localized to the OM. However the 25-kDa form of CsgA was found in the IM. Finally, a polyketide synthetase (PKS) that synthesizes the antibiotic myxovirescin, also known as TA, was also found to be associated with the membrane.
MATERIALS AND METHODS
Bacterial strains and reagents.
Wild-type M. xanthus strain DK1622 was used in all experiments. DK1622 cells were grown in Casitone yeast extract medium containing 1% Casitone (Difco), 0.5% yeast extract (Difco), 10 mM 4-morpholinepropanesulfonic acid (MOPS) buffer (pH 7.6), and 0.1% MgSO4. Sucrose solutions were made in 20 mM HEPES with 5 mM EDTA (pH 7.6) (HE5 buffer) or in 20 mM HEPES (pH 7.6) buffer (HE buffer). Membrane enrichments were diluted in 20 mM HEPES-0.1 mM EDTA (pH 7.6) (HE0.1 buffer). The density of each sucrose solution was determined at room temperature with a refractometer. Sucrose solutions were stored at 4°C and mixed before use.
Membrane separation.
The technique used for membrane separation was a combination of methods developed by Orndorff and Dworkin (39) and Kotarski and Salyers (24). Cells were grown in a 2-liter flask containing 1,200 ml of Casitone yeast extract medium to a final cell density of 2 ×108 cells ml−1. The culture was split into 300-ml aliquots; each aliquot was mixed with 100 ml of ice-cold double-distilled water and harvested by centrifugation at 10,000 × g for 10 min at 4°C. The cells were washed with 100 ml of ice-cold distilled water and centrifuged at 10,000 × g for 10 min. Each washed cell pellet was resuspended in 10 volumes (approximately 40 to 50 ml) of 23.5% sucrose in HE buffer. Freshly prepared chicken egg white lysozyme (0.12 mg ml−1 in water; Sigma Chemical Co., St. Louis, Mo.) and 100 mM EDTA in water (pH 7.6) were added to final concentrations of 300 μg ml−1 and 1 mM, respectively. The cell suspension was incubated at 4°C with gentle stirring for 14 to 16 h. The lysozyme-EDTA-treated cells were distributed into four 50-ml tubes and collected by centrifugation (27,000 × g, 15 min, 4°C). The pellets were resuspended in 3 volumes of ice-cold double-distilled water (approximately 6 ml) with vigorous pipetting to induce spheroplast formation and stirred for 30 min at 4°C. Conversion of osmotically shocked cells into circular spheroplasts was monitored microscopically.
Spheroplasts were collected by centrifugation (12,000 × g, 10 min, 4°C). The supernatant was collected, and the pellet was resuspended in 3 volumes of 5 mM EDTA (pH 7.6). One tablet of complete EDTA-free protease inhibitor (Roche, Indianapolis, Ind.) was dissolved, and the suspension was stirred for 1 h. The supernatant was added back to the suspension and stirred for an additional 30 min. Finally, 1 ml of RNase A and 1 ml of DNase type II (each at a concentration of 10 mg ml−1 in distilled water; Sigma Chemical Co.) were added, and the preparation was stirred for another 30 min. The suspension was placed in two 50-ml Falcon tubes and diluted fourfold with HE0.1 buffer, and intact cells were separated from the spheroplasts by repeated centrifugation (5,000 × g, 10 min, 4°C).
The final suspension was divided into aliquots in 12 tubes, and the membranes were collected by ultracentrifugation (100,000 × g, 2 h, 4°C) in a 70.1 Ti rotor (Beckman Coulter, Fullerton, Calif.). Each membrane pellet was gradually resuspended in 0.5-ml aliquots of 23.5% sucrose in HE5 buffer by using a Dual 21 tissue homogenizer (Kimble Kontes, Vineland, N.J.). The membranes were ground by using vertical and circular strokes with minimal introduction of air bubbles. The membrane suspension was incubated overnight with gentle stirring. The next day, 1 tablet of complete EDTA-free protease inhibitor was added, and 0.4 ml of the membrane suspension was stored at 4°C as a control for immunoblotting and enzyme analysis. The remaining 12 ml was divided and loaded on top of four three-step gradients, each consisting of 10 ml of 60% sucrose, 10 ml of 48% sucrose, and 10 ml of 35% sucrose in HE5 buffer. The membranes were harvested by ultracentrifugation (120,000 × g, 3 h, 4°C) in an SW28 rotor. Unless stated otherwise, all remaining steps in the isolation procedure were performed at 4°C by using an SW28 rotor.
The material that migrated to the middle of the 35% sucrose layer (light membrane enrichment) was transferred to a new 38-ml centrifuge tube and diluted with enough HE0.1 buffer to nearly fill the tube. Then 1 ml of 30% sucrose was placed in the bottom of the tube to create a cushion. The material that migrated to the interface of the 35 and 48% sucrose layers (medium-density membrane enrichment) was transferred to another tube, diluted with HE0.1 buffer as described above, and cushioned with 1 ml of 40% sucrose. The material that migrated at the interface of the 48 and 60% sucrose layers (heavy membrane enrichment) was diluted with HE0.1 buffer and cushioned with 1 ml of 50% sucrose. Heavy and medium-density membrane enrichments were collected by ultracentrifugation (120,000 × g, 3 h), while the light membrane enrichment was collected by ultracentrifugation for 6 h. Concentrated enrichments were diluted with 1 to 3 ml of HE0.1 buffer to obtain a final sucrose concentration of 20% (heavy and medium-density membrane enrichments) or 15% (light membrane enrichment), and a protease inhibitor was added.
Each membrane enrichment was layered on top of a different discontinuous gradient. The heavy membrane gradient consisted of 4 ml of a 70% sucrose solution, 4 ml of a 60% sucrose solution, 15 ml of a 55% sucrose solution, 3 ml of a 40% sucrose solution, and 3 ml of a 30% sucrose solution in HE5 buffer. The intermediate membrane enrichment gradient contained 3 ml of a 70% sucrose solution, 4 ml of a 60% sucrose solution, 4 ml of a 55% sucrose solution, 15 ml of a 46% sucrose solution, 3 ml of a 40% sucrose solution, and 3 ml of a 30% sucrose solution in HE5 buffer. The light membrane enrichment gradient contained 4 ml of a 70% sucrose solution, 4 ml of a 60% sucrose solution, 4 ml of a 55% sucrose solution, 15 ml of a 37.5% sucrose solution, 3 ml of a 30% sucrose solution, and 3 ml of a 20% sucrose solution in HE5 buffer. The gradients were ultracentrifuged (120,000 × g, 16 h), and 1-ml aliquots were collected with a gradient fractionator (Hoefer Scientific, San Francisco, Calif.). The density and relative protein content (A280) were determined for every other fraction by using a refractometer (Bausch and Lomb, Rochester, N.Y.) and an UV-VIS spectrophotometer (Beckman Coulter), respectively. Peak fractions containing protein were pooled, diluted to 35 ml with HE0.1 buffer, and consolidated by centrifugation in a Beckman SW28 rotor (120,000 × g, 12 to 16 h). This washing step was repeated to ensure sucrose removal. Membrane pellets were resuspended in 1 ml of HE0.1 buffer by using a tissue homogenizer, and samples were stored at −20°C. Quantification of succinate dehydrogenase (SDH) and lipid A was performed within 48 h. During this time 50-μl aliquots of each sample were kept at 4°C, while the remaining samples were frozen at −20°C.
Preparation of fraction VII.
Low yields of fraction VII were obtained on the discontinuous sucrose gradient, so a quicker method for isolation of this fraction was developed. Cells were grown and converted to spheroplasts as described above. Total membranes were collected by ultracentrifugation for 2 h at 100,000 × g in a 70.1 Ti rotor, after which the membranes were disrupted with a tissue homogenizer. Following addition of 1 tablet of protease inhibitor, the membranes were loaded on a linear sucrose gradient consisting of 5 ml of a 60% sucrose solution, 5 ml of a 50% sucrose solution, 5 ml of a 45% sucrose solution, 5 ml of a 40% sucrose solution, 5 ml of a 35% sucrose solution, 5 ml of a 30% sucrose solution, and 5 ml of a 25% sucrose solution. Gradient ultracentrifugation and fractionation were performed as described above.
Protein (25 μg) was separated on a 7.5% polyacrylamide gel and stained with Coomassie blue R250 for 2 h. The five most intense protein bands were excised and frozen in 1% acetic acid. The protein in gel slices was digested with trypsin, and fragment masses were determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Since fingerprinting data did not yield strong hits with any protein in the public database, the fingerprinting data were sent to Cereon LLT to identify the matching gene in the M. xanthus genome database. Only one gene, MYX12U 3797, was predicted to encode the protein that matched the fingerprint. A portion of this gene was reported to encode PKS Ta-1 (47). The GenBank accession numbers for the complete gene encoding Ta-1 are MYX0C1270 and AY282522.
Gel electrophoresis and immunoblotting.
Protein concentrations were determined by the bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, Ill.). Proteins were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-PAGE) (27). Low-range prestained SDS-PAGE standards (Bio-Rad Laboratories, Hercules, Calif.) were used for SDS-PAGE gels. Proteins were transferred onto a 45-μm-pore-size nitrocellulose membrane (Bio-Rad Laboratories) in a Bio-Rad Trans Blot cell (80 mA for 5 h) by using Towbin buffer (54). Most membranes were blocked in Tris-buffered saline containing 5% powdered milk; the exception was FrzCD, which was blocked in the presence of 10% powdered milk (34). For Tgl (44), CglB (43), PilA (62), CsgA (26), and FrzCD (34) immunoblots, 4 to 20 μg of protein was loaded per lane and reacted with the corresponding polyclonal antibodies at dilutions of 1:2,000, 1:10,000, 1:2,000, 1:5,000, and 1:500, respectively. Secondary anti-rabbit antibody conjugated with horseradish peroxidase (Cell Signaling) was added at a 1:3,000 dilution. For lipid A, O-antigen, and FibA immunoblots, 4 to 10 μg of protein was loaded per lane and reacted with monoclonal antibodies 2254, 783, and 2105 at dilutions of 1:500, 1:1,000, and 1:500, respectively (10, 12, 13). Secondary goat anti-mouse antibody was used at a 1:10,000 dilution. Reactive bands were visualized by using the ECL reagents and were recorded on Hyperfilm (Amersham Biosciences, Piscataway, N.J.). Reactive bands were quantified by using Scion Image software (Scion, Frederick, Md.).
SDH assay.
SDH was assayed as described by Kasahara and Anraku (20), with the following modifications. The assays were performed by using 15 μg of protein, and Sarkosyl was added to a final concentration of 0.25%. The reaction mixture was equilibrated for 2 min before addition of membrane protein. After 3 min of incubation, 2,6-dichlorophenolindophenol (Fluka, Buchs, Switzerland) and phenazine methosulfate were added. The decrease in absorption at 600 nm was recorded after 3 min. The overall level of recovery of SDH was 74% ± 9.0%, and the overall level of protein recovery was 51% ± 4.6%.
Two-dimensional SDS-PAGE analysis.
Two-dimensional electrophoresis was performed by the method of O'Farrell et al. (38) by Kendrick Labs, Inc. (Madison, Wis.). Isoelectric focusing was carried out in 2.0-mm-inside-diameter glass tubes by using 2% pH 3.5 to 10 ampholines (Gallard Schlesinger Industries, Inc., Garden City, N.Y.) for 9,600 V-h. One hundred micrograms of protein was equilibrated in 10% glycerol-50 mM dithiothreitol-2.3% SDS-62.5 mM Tris (pH 6.8) for 10 min. Carbamylated carbonic anhydrase markers with known isoelectric points were added to each sample. Each tube was sealed to the top of a stacking gel. SDS-PAGE was carried out with a 10% polyacrylamide gel (thickness, 0.75 mm) for 4 h at 15 mA. The gels were stained with Sypro Ruby stain (Bio-Rad Laboratories), and images were obtained by using 2D 2920 Master Imager (Amersham Biosciences) at 410 nm.
RESULTS
The M. xanthus cell envelope facilitates the exchange of proteins and chemical signals involved in motility and development. Many of the genes involved in cell signaling and motility stimulation have been identified by traditional genetic approaches. What remains is to understand the mechanisms by which the proteins and signals are exchanged and used to activate the recipient cell. Accomplishing this goal will require a thorough understanding of the membrane architecture. To begin this task, we devised a method for separating IM and OM and used this approach to localize several signaling proteins by Western blotting.
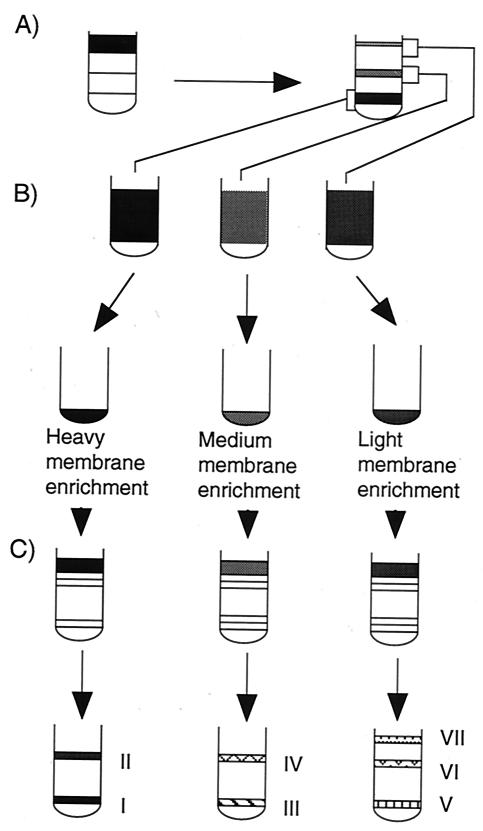
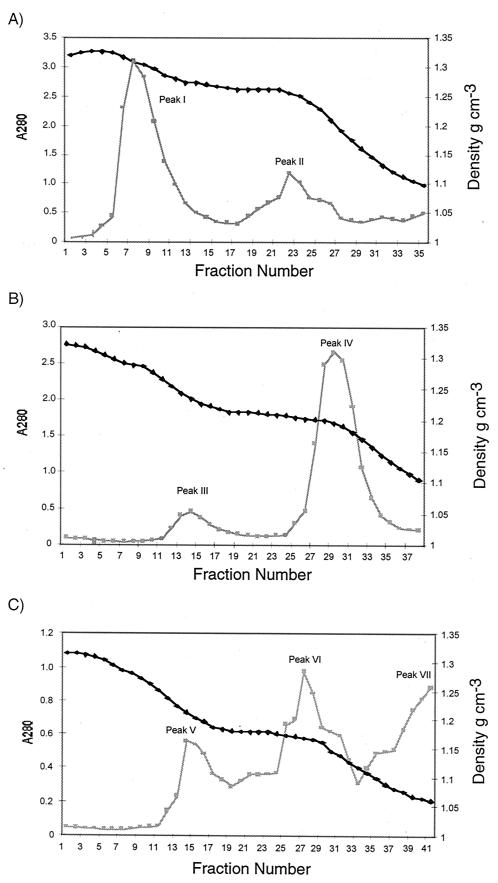
Vegetative DK1622 cells were osmotically shocked to form spheroplasts, subjected to mechanical grinding with a tissue homogenizer, and then applied to a three-step sucrose gradient (Fig. 1A). Three bands were observed, corresponding to light, medium-density, and heavy membrane fractions. Each fraction was concentrated (Fig. 1B) and then separated on a discontinuous sucrose gradient, which yielded seven membrane peaks (Fig. 1C). The heavy membrane enrichment yielded two peaks (Fig. 2). One of these peaks, peak I, accounted for 72% of the protein in the heavy membrane fraction and was later discovered to be IM. The medium-density membrane enrichment separated into two peaks (Fig. 2). The major peak, peak IV, accounted for about 90% of the total protein in this enrichment and was later found to be hybrid membrane (HM). The light membrane enrichment fractionated into three peaks; peaks V and VI were derived from the OM.
FIG. 1.
Scheme for separating membranes of M. xanthus DK1622 vegetative cells. Total membranes were separated with a three-step sucrose gradient by centrifugation for 3 h at 100,000 × g (A). Each membrane enrichment was concentrated (B) and separated with separate discontinuous sucrose gradients (C). The peaks were designated peaks I through VII.
FIG. 2.
Protein contents of the heavy membrane (A), medium-density membrane (B), and light membrane peaks (C) from discontinuous sucrose gradients as a function of absorbance at 280 nm (A280) (squares) and buoyant density (diamonds).
Biochemical characterization of the isolated membrane fractions.
Biochemical markers were used to confirm the identities of the isolated membrane fractions. SDH activity was used as the IM marker (20). The lipid A and O-antigen portions of LPS were used as the OM markers and were quantified by Western blotting (10).
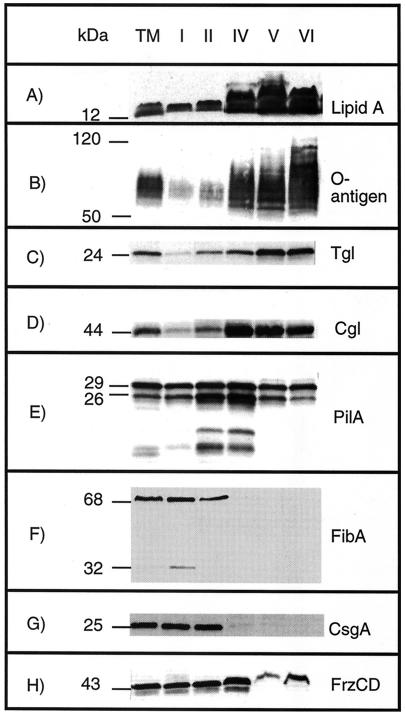
Peaks II and IV, which together accounted for 48% of the total protein, appeared to be HM peaks (Fig. 2) since they contained 73% of the SDH and 56% of the lipid A (data not shown). Large HM peaks are not unusual with the technique used (39). A small amount of peak III was isolated, and this peak was not characterized (Fig. 2B). Peaks V and VI together contained 10% of the total protein, 2% of the total SDH activity (data not shown), and 27% of the lipid A (data not shown). These fractions were highly enriched in lipid A (Fig. 3A) and O-antigen (Fig. 3B) relative to protein and were OM peaks. Interestingly they varied widely in density; peak V had a density of 1.213 to 1.24 g cm−3, while peak VI had a density of 1.169 to 1.171 g cm−3. Peak I, with a density of 1.292 to 1.310 g cm−3 (Fig. 2), contained 42% of the total membrane protein, 25% of the total SDH activity (data not shown), and 16% of the lipid A, suggesting that this peak was derived from the IM. The IM contained more lipid A than the OM contained SDH, which was expected as lipid A is synthesized in the IM. The IM and OM fractions appeared to be relatively pure based on these biochemical assays.
FIG. 3.
Immunoblots of isolated membrane fractions containing LPS lipid A (A), LPS O-antigen (B), Tgl (a lipoprotein required for assembly of TFP) (C), CglB (a lipoprotein required for adventurous gliding) (D), PilA (the structural pilin protein for TFP) (E), FibA (an extracellular matrix-associated zinc metalloprotease) (F), CsgA (a protein required for C-signaling) (G), and FrzCD (a methylated chemotaxis protein) (H). Total membrane (lane TM), IM peak I (lane I), HM peak II (lane II), HM peak IV (lane IV), OM peak V (lane V), and OM peak VI (lane VI) fractions were separated by SDS-PAGE for Western blotting. The numbers on the left indicate molecular masses of protein standards (in kilodaltons).
Immunoblot analysis of membrane fractions.
Membrane peaks were probed with antibodies against well-known antigens (Fig. 3). Identical amounts of total protein from the different peaks were subjected to Western analysis.
Certain genetic defects in both the A and S motility systems can be restored through contact with wild-type cells (16). Contact stimulation is transient phenotypic rescue of a genetic defect that is due to acquisition of the wild-type protein by a mutant (16, 44, 45). For example, mutations in tgl eliminate pilus assembly (57), which can be restored by contact with tgl+ cells (59). Similarly, defects in cglB, which eliminate A motility, can be transiently corrected through contact with cglB+ cells (43). Tgl is a lipoprotein and contains a conserved 19-amino-acid N-terminal signal sequence (44). Tgl antibody reacted strongly with OM peaks V and VI, while 20-fold- and 10-fold-lower levels of Tgl were detected in peaks I and IV, respectively (Fig. 3C). CglB, another lipoprotein, was localized in OM peaks V and VI, but unlike Tgl, CglB was equally abundant in HM peak IV (Fig. 3D). The predominance of these proteins in the OM is consistent with their exchange by contact stimulation.
PilA is the structural protein for TFP, which are found exclusively at the cell poles (19, 61). Prepilin monomers should reside in the IM and become assembled after N-terminal signal cleavage and methylation via a type II secretion mechanism (31). Accordingly, prepilin and pilin should yield 25- and 23-kDa proteins based on the predicted amino acid sequence. However, mature pilin is observed as 29- and 26-kDa proteins when it is sheared from intact cells (59). While the presence of prepilin could not be detected, 29- and 26-kDa proteins were observed in all membrane fractions (Fig. 3E). P. aeruginosa 1244 PilA is glycosylated with O-antigen, which may account for the observation that M. xanthus PilA is slightly larger than the predicted molecular weight (8). In addition, two smaller bands were present at amounts proportional to the 29- and 26- kDa antigens and most likely were pilin degradation products. Approximately similar levels of PilA were found in IM peak I and OM peaks V and VI, while HM peaks II and IV showed twofold enrichment.
IM vesicles, and to a lesser degree HM peak II vesicles, agglutinated rapidly in a manner reminiscent of the behavior of intact cells during a cohesion assay (47). Cell cohesion is a consequence of the adhesive nature of fibrils (2). FibA is predicted to be a member of the M4 zinc-dependent metalloprotease family (21), whose production and processing are associated with fibril formation (4, 21). FibA is present in growing cells, as well as in the extracellular matrix of nutrient-depleted cells. FibA was found exclusively in IM peak I and to a lesser degree in HM peak II (Fig. 3F).
CsgA is the C-signal-producing protein and is essential for fruiting body development (22, 26, 29). CsgA is present in two forms during development: a full-length protein with homology to short-chain alcohol dehydrogenases (25-kDa form) (28) and a processed 17-kDa form (23, 26). The nature of the C-signal is controversial as it is not clear whether the 17-kDa form of CsgA acts as a protein signal or if the signal is an unknown enzymatic product of the 25-kDa CsgA form. Whereas the first model envisions CsgA cleavage and export to the OM, the second model predicts that CsgA is part of the IM. The full-length protein localizes to the IM, while a small amount of full-length protein is present in HM peak IV (Fig. 3G). The processed form, which was present at 10-fold-lower levels, could not be detected in vegetative cells. Localization of CsgA to the IM supports the prediction that CsgA acts as an enzyme and adds to the intrigue regarding this signaling pathway, but it does not disprove the hypothesis that the small form is a signal. Extracellular CsgA has been detected by colloidal gold scanning electron microscopy, although it is still not clear whether the extracellular antigen is the product of cell lysis or the product of a developmental secretion system (50). To critically evaluate the two C-signaling models, the proteolytic product needs to be localized during development. Despite the fact that CsgA colocalized with FibA, CsgA does not appear to be a fibril-associated protein as fibrilless, dsp cells enable development of csgA cells by extracellular complementation (30).
FrzCD is a homologue of E. coli methyl-accepting chemotaxis proteins (33). It regulates cell reversals and is essential for fruiting body morphogenesis (5, 65). Generally, methyl-accepting chemotaxis proteins have transmembrane topologies with a characteristic periplasmic domain for ligand binding. Despite lacking predicted membrane-spanning domains, FrzCD is membrane associated (33). FrzCD was distributed in the IM and HM peaks (Fig. 3H). The amounts present in peak IV were 1.5- to 2-fold higher than the amounts in the IM. FrzCD was also detected in OM peaks V and VI.
Two-dimensional analysis of isolated fractions.
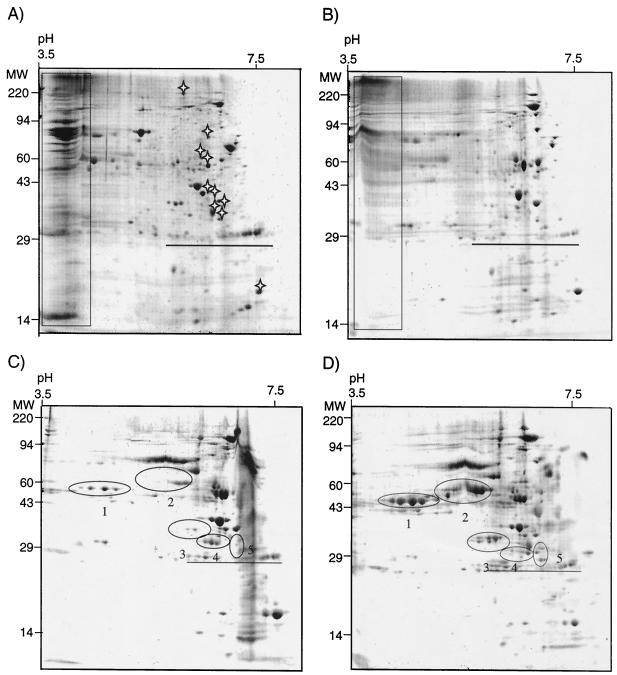
The proteomes of the four most abundant membrane fractions were separated by two-dimensional SDS-PAGE. IM peak I contained many acidic proteins of various sizes that did not resolve well during isoelectric focusing. The high intensities of the streaks suggested that they contained major components of the IM proteome (Fig. 4A). Manual alignment of IM peak I and HM peak IV revealed a high level of similarity. In addition to the acidic proteins (Fig. 4A and B), the two membrane peaks shared at least 37 protein spots. For comparison, alignment of IM peak I and OM peak VI gels revealed only 10 spots in common (Fig. 4A and D). Comparison of HM peak IV and OM peak V revealed 21 protein spots that were common to the two peaks (Fig. 4B and C). In contrast to IM peak I and HM peak IV, neither OM peak contained large quantities of acidic proteins. The two OM proteomes appeared to be similar, yet they differed greatly in the levels of several groups of proteins (Fig. 4C and D). Proteins in clusters 1, 2, 3, and 5 were less abundant in OM peak V than in OM peak VI. Only cluster 4 proteins appeared to be more abundant in OM peak V than in OM peak VI.
FIG. 4.
Two-dimensional SDS-polyacrylamide gel separated in the first dimension by pH 3.5 to 10 and in the second dimension on 10% polyacrylamide. (A) IM peak I; (B) HM peak IV; (C) OM peak V; (D) OM peak VI. Molecular masses (MW) (in kilodaltons) are indicated on the left. A series of internal, carbamylated carbonic anhydrase markers ranging from pH 5 to 7.3 are underlined on each gel. The rectangles indicate acidic proteins common to IM peak I and HM peak IV which did not resolve well. The stars in panel A indicate protein spots common to IM peak I and HM peak VI. The numbers in panels C and D indicate clusters of proteins that are differentially expressed in the two OM peaks.
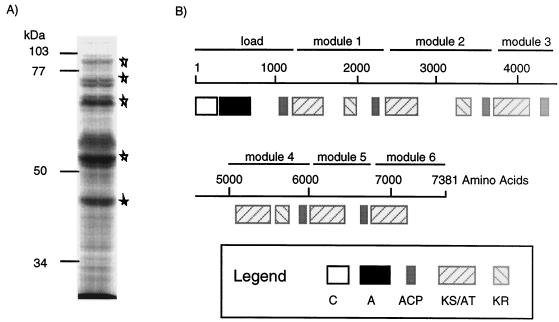
Peak VII contained Ta-1 PKS.
Peak VII had an unusually low buoyant density (1.094 to 1.072 g cm−3) (Fig. 2 C), low SDH activity, and lipid A levels comparable to those found in the total membrane (data not shown), which made it difficult to determine the cellular location. Despite high absorbance at 280 nm, the protein yields of peak VII were small compared to those of the other fractions. However, when the experimental procedure was shortened and the spheroplast membrane material was loaded directly onto a linear sucrose gradient, larger amounts were recovered. Peak VII produced a small set of very intense bands upon SDS-PAGE (Fig. 5A). Five peptides (90, 86, 71, 55, and 45 kDa) were fingerprinted following trypsin digestion by MALDI-TOF MS. Comparison with the M. xanthus genome revealed that all five proteins were processed products of polypeptide MYX12U-3797, which is predicted to be 7,831 amino acids long. A portion of MYX12U-3797 was found in the GenBank database as M. xanthus Ta-1 PKS, which exhibited 99% identity over 2,392 amino acids (40).
FIG. 5.
Fraction VII contains a PKS. (A) Polyacrylamide (7.5%) gel of fraction VII stained with Coomassie blue. The stars indicate protein bands analyzed by MALDI-TOF MS. (B) Predicted organization of the 7,831-amino-acid Ta-1 PKS. The conserved domains are as follows: C domain, A domain, ACP, KS/AT, and KR. The domains are organized in modules, which are basic units responsible for incorporation of extender units. The scale bar indicates the amino acid numbers in the primary structure of Ta-1.
Ta-1 has combined structural elements of a nonribosomal peptide synthetase and a type I PKS (40) (Fig. 5 B). While nonribosomal peptide synthetases assemble products by covalently linking amino acids (64), type I PKS assemble products through incorporation of acetate or propionate. Ta-1 exhibits modular organization similar to that of type I PKS (52). Each module in a type I PKS is responsible for covalent addition, and sometimes reduction, of an acetate or propionate unit in the elongating chain. The Ta-1 gene is a member of a large cluster of PKS genes originally discovered in M. xanthus strain TA isolated in Tel Aviv. The TA cluster was reported to encode the antibiotic TA (46), also known as myxovirescin A (55), whose structure in Myxococcus virescens has been elucidated.
Analysis of Ta-1 conserved domains by using the conserved domain search program (1) indicated that the first 1,000 amino acids in the amino terminus comprise a loading domain, which consists of a condensation domain (C domain), an amino acid activation domain (A domain), and an acyl carrier protein (ACP) group (Fig. 5B). The C domain is responsible for peptide bond formation in nonribosomal peptide biosynthesis. Downstream of the C domain is the A domain, also known as the adenylation domain, which is responsible for amino acid activation as an adenylate intermediate at the expense of ATP. The activated amino acid-acyl extender unit is sequentially transferred to the thiol residue of a peptydyl-ACP domain. Peptydyl-ACP transfers the thiol derivative to the β-keto-synthase (KS) domain of the first chain extension module. The A domain of Ta-1 contains all five conserved motifs (64), with the following sequence conservation: motif A (L-AG-AYVP), motif B (Y-SGTTGXPKGV), motif C (GEL-IGGXGXARGYL), motif D (YXTGD), and motif E (VK-RGXRIE-GEIE), where X indicates any amino acid and hyphens indicate mismatches with the published motifs. Motifs B, C, D, and E are engaged in ATP binding and formation of amino acid adenylate (14, 41, 42, 56). Analysis of other peptide synthetases has indicated that each activation domain is responsible for activation of a single amino acid.
Modules 1, 2, and 4 are organized as typical type I PKS modules consisting of β-KS/acyltransferase (AT) and ACP chain elongation units and β-keto-reductase (KR) reducing units. Modules 3 and 5 lack KR units, which is not unusual and contributes to the structural variation in natural products (52). Finally, the sixth module consists of only the AT domain. There is likely to be a frameshifting error in module 7 which when corrected adds KR and ACP domains.
DISCUSSION
This is the first study in which characterization of the IM and OM of wild-type M. xanthus DK1622 cells is reported. The procedure used for initial preparation of cells was similar to the procedure that Orndorff and Dworkin used for mutant MD-2 (39). In a comparison with the previous work on MD-2, the density of OM peak VI was similar to the density of the single OM peak of MD-2 (1.170 and 1.166 g cm−3, respectively). The OM of strain MD-2 eluted as a single peak, whereas that of DK1622 eluted as two peaks. HM peak IV appeared to be a heavier counterpart of the MD-2 HM (densities, 1.197 and 1.185 g cm−3, respectively), while the difference between the buoyant densities of the IM of strains DK1622 and MD-2 is larger (densities, 1.30 and 1.22 g cm−3, respectively). Strain MD-2, which has two point mutations in the OM channel gene pilQ (58), is hypopiliated (19) and hypofibrilated (9). However, the differences appear not to be due to the presence of pili and fibrils. Membrane fractions of MD-2 separated by this procedure had the same densities as the membrane fractions of the wild-type strain and similar SDH specific activities (data not shown). These results suggest that the differences in the buoyant densities of IM, as well as the differences in the measured SDH activities obtained in the two studies, are probably due to differences in the physical treatment of membranes (i.e., syringe versus a tissue homogenizer).
The reproducibility of the separation technique depended on three factors: the volume of water used for spheroplast preparation, the volume of 5 mM EDTA added during subsequent incubation of spheroplasts, and the manner in which shearing was performed. To provide adequate and uniform shearing, the total membrane was centrifuged in 7- to 8-ml increments in a 70.1 Ti rotor to produce smaller pellets that were then gradually resuspended and ground in 0.5-ml aliquots of 23% sucrose. Collection of membranes with the SW28 rotor produced larger pellets that could not be uniformly homogenized, which resulted in unpredictable protein elution profiles from the discontinuous gradients. Introduction of a three-step gradient as the initial step in separation was essential to provide physical separation of the OM peak from HM peak IV even though these two fractions had almost identical densities (1.197 and 1.220 g cm−3, respectively). A factor that had a deleterious effect on membrane separation was the presence of calcium cations, which caused clumping of cells. Problems with calcium were due to residual detergent and were eliminated by extensive rinsing of glassware with distilled water.
HM peak IV had a threefold-higher content of SDH than IM peak I. Even though large HM peaks, such as peak IV, are not unusual for this type of procedure (6, 24) and could be an artifact of the separation procedure, peak IV might also be an area of tight juncture between the IM and OM. Western analysis revealed twofold enrichment of PilA compared to the IM. Higher levels of PilA suggest that HM fraction IV is enriched in cell poles, where the pilus motor is located. Tighter coupling between the two membranes might be necessary for export of TFP to the surface and retraction of pili by PilT, the proposed pilus retraction motor (18).
The IM and OM proteomes appear to cluster in specific pI ranges. The IM proteome has a bimodal distribution of proteins as a function of pI, with the greatest abundance in the pI 3.5 to 4.0 and pI 5.0 to 7.0 ranges. The OM proteome has a unimodal protein distribution in the pI 5.0 to 7.0 range. In the few previously described two-dimensional gel analyses of OM proteomes, the OM of E. coli exhibited the same trend (36), while Caulobacter crescentus OM proteins were found predominantly in the pI 8.0 to 11 range (37). Additional proteome analyses should reveal if this is a general trend and if it has any biological repercussions for pathways for protein transport and localization.
Myxovirescin A is produced during exponential growth and has been purified from culture supernatants (11). The membrane association of PKS probably facilitates safe export of the final product. Myxovirescin A inhibits peptidoglycan synthesis in a wide range of gram-negative cells, including cells of the producing organism. M. virescens displays twofold-greater sensitivity to myxovirescin A than E. coli (11). In contrast, 60-fold more myxovirescin A was required to inhibit cell growth of P. aeruginosa. It is possible that the myxovirescin A biosynthetic machinery is associated with phospholipid to protect the peptidoglycan. This hypothesis is consistent with experiments indicating that myxovirescin A activity is inhibited by phospholipids (11).
In conclusion, the technique described here enables separation of IM and OM on a scale suitable for proteomics. Given the many signaling and protein export-import pathways observed for M. xanthus, one would predict extensive protein and signal traffic across the OM (48). Systematic identification of OM proteins should be facilitated by the MALDI-TOF MS approach and the availability of the genome sequence.
Acknowledgments
We thank James Carroll for performing MALDI-TOF MS, Barry Goldman for analysis of MALDI-TOF MS fingerprint data, and Tracy Andacht for two-dimensional gel imaging. We thank Dale Kaiser, Marty Dworkin, Lotte Sogaard-Andersen, Alfred Spormann, and David Zusman for providing antibodies. The genome sequence was generously provided by Monsanto Company prior to public release. We thank Pam Bonner and Patrick Curtis for critical reading of the manuscript.
This study was supported by research grant MCB-0090946 from the National Science Foundation.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behmlander, R. M., and M. Dworkin. 1991. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J. Bacteriol. 173:7810-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackhart, B. D., and D. R. Zusman. 1985. Frizzy genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 82:8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bledsoe, H. A., J. A. Carroll, T. R. Whelchel, M. A. Farmer, D. W. Dorward, and F. C. Gherardini. 1994. Isolation and characterization of Borrelia burgorferi inner and outer membranes by using iospycnic centrifugation. J. Bacteriol. 176:7447-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowden, M. G., and H. B. Kaplan. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30:275-284. [DOI] [PubMed] [Google Scholar]
- 8.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 9.Dana, J. R., and L. J. Shimkets. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 175:3636-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fink, J. M., and J. F. Zissler. 1989. Characterization of lipopolysaccharide from Myxococcus xanthus by use of monoclonal antibodies. J. Bacteriol. 171:2028-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerth, K., H. Irschik, H. Reichenbach, and W. Trowitzsch. 1982. The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J. Antibiot. 35:1454-1459. [DOI] [PubMed] [Google Scholar]
- 12.Gill, J., E. Stellwag, and M. Dworkin. 1985. Monoclonal antibodies against cell-surface antigens of developing cells of Myxococcus xanthus. Ann. Inst. Pasteur Microbiol. 136A:11-18. [DOI] [PubMed] [Google Scholar]
- 13.Gill, J. S., and M. Dworkin. 1986. Cell surface antigens during submerged development of Myxococcus xanthus, as examined with monoclonal antibody probes. J. Bacteriol. 168:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gocht, M., and M. A. Marahiel. 1994. Analysis of core sequences in the d-Phe activating domain of the multi-functional peptide synthetase TycA by site-directed mutagenesis. J. Bacteriol. 176:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 17.Jelsbak, L., and L. Sogaard-Andersen. 1999. The cell surface-associated intercellular C-signal induces behavioral changes in individual Myxococcus xanthus cells during fruiting body morphogenesis. Proc. Natl. Acad. Sci. USA 96:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasahara, M., and Y. Anraku. 1974. Succinate dehydrogenase of Escherichia coli membrane vesicles. Activation and properties of the enzyme. J. Biochem (Tokyo) 76:959-966. [PubMed] [Google Scholar]
- 21.Kearns, D. B., P. J. Bonner, D. R. Smith, and L. J. Shimkets. 2002. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 184:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, S. K., and D. Kaiser. 1990. Purification and properties of Myxococcus xanthus C-factor, an intercellular signalling protein. Proc. Natl. Acad. Sci. USA 87:3635-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotarski, S. F., and A. A. Salyers. 1984. Isolation and characterization of outer membranes of Bacterioides thetaiotaomicron grown on different carbohydrates. J. Bacteriol. 158:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroos, L., P. Hartzell, K. Stephens, and D. Kaiser. 1988. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 2:1677-1685. [DOI] [PubMed] [Google Scholar]
- 26.Kruse, T., S. Lobedanz, N. M. S. Berthelsen, and L. Sogaard-Andersen. 2001. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 40:156-168. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B.-U., K. Lee, J. Mendez, and L. J. Shimkets. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 9:2964-2973. [DOI] [PubMed] [Google Scholar]
- 29.Li, S., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 30.Li, S. F., and L. J. Shimkets. 1993. Effect of dsp mutations on the cell-to-cell transmission of CsgA in Myxococcus xanthus. J. Bacteriol. 175:3648-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147-155. [DOI] [PubMed] [Google Scholar]
- 32.McBride, M. 2001. Bacterial gliding motility: multiple mechanisms for cell movement. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 33.McBride, M. J., R. A. Weinberg, and D. R. Zusman. 1989. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc. Natl. Acad. Sci. USA 86:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCleary, W., M. McBride, and D. Zusman. 1990. Developmental sensory transduction in Myxococcusxanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 172:4877-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merz, A., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-101. [DOI] [PubMed] [Google Scholar]
- 36.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267. [DOI] [PubMed]
- 37.Molloy, M. P., N. D. Phadke, H. Chen, R. Tyldesley, D. E. Garfin, J. R. Maddock, and P. C. Andrews. 2002. Profiling the alkaline membrane proteome of Caulobacter crescentus with two-dimensional electrophoresis and mass spectroscopy. Proteomics 2:899-910. [DOI] [PubMed] [Google Scholar]
- 38.O'Farrell, P. Z., H. M. Goodman, and P. H. O'Farrell. 1977. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 2:1133-1142. [DOI] [PubMed] [Google Scholar]
- 39.Orndorff, P. E., and M. Dworkin. 1980. Separation and properties of the cytoplasmic and outer membranes of vegetative cells of Myxococcus xanthus. J. Bacteriol. 141:914-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paitan, Y., G. Alon, E. Orr, E. Z. Ron, and E. Rosenberg. 1998. The first gene in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus codes for a unique PKS module coupled to a peptide synthetase. J. Mol. Biol. 286:465-474. [DOI] [PubMed] [Google Scholar]
- 41.Pavela-Vrancic, M., E. Pfeifer, W. Schroder, H. von Dohren, and H. Kleinkauf. 1994. Identification of ATP-binding site in tyrocidine synthetase I by selective modification with fluorescein 5′-isothiocynate. J. Biol. Chem. 269:14962-14966. [PubMed] [Google Scholar]
- 42.Pavela-Vrancic, M., E. Pfeifer, H. van Liempt, H. J. Schafer, H. von Dohren, and H. Kleinkauf. 1994. ATP-binding in peptide synthatases: determination of contact sites of the adenine moiety by photoaffinity labelling of tyrocidine synthetase I with 2-azido-adenosine triphosphate. Biochemistry 33:6276-6283. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez, A. M., and A. M. Spormann. 1999. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 181:4381-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Soto, J. P., and D. Kaiser. 1997. Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J. Bacteriol. 179:4372-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Soto, J. P., and D. Kaiser. 1997. The tgl gene: social motility and stimulation in Myxococcus xanthus. J. Bacteriol. 179:4361-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg, E., S. Fytlovitch, S. Carmeli, and Y. Kashman. 1982. Chemical properties of Myxococcus xanthus antibiotic TA. J. Antibiot. 35:788-793. [DOI] [PubMed] [Google Scholar]
- 47.Shimkets, L. J. 1986. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J. Bacteriol. 166:837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimkets, L. J. 1999. Intercellular signaling during fruiting body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 49.Shimkets, L. J., R. E. Gill, and D. Kaiser. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. USA 80:1406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimkets, L. J., and H. Rafiee. 1990. CsgA, an extracellular protein essential for Myxococcus xanthus development. J. Bacteriol. 172:5299-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauton, J., and B. Wilkinson. 1997. Biosynthesis of erythromycin and rapamycin. Chem. Rev. 97:2611-2629. [DOI] [PubMed] [Google Scholar]
- 53.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 54.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trowitzsch, W., V. Wray, K. Gerth, and G. Hofle. 1982. Structure of myxovirescen A, a new macrocyclic antibiotic from gliding bacteria. J. Chem. Soc. Chem. Commun. 1982:1340-1342. [Google Scholar]
- 56.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wall, D., and D. Kaiser. 1998. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc. Natl. Acad. Sci. USA 95:3054-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wall, D., P. E. Kolenbrander, and D. Kaiser. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wall, D., S. S. Wu, and D. Kaiser. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 61.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. 18:547-558. [DOI] [PubMed]
- 62.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene of Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, S. S., J. Wu, Y. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]
- 64.Zocher, R., and U. Keller. 1997. Thiol template peptide synthesis systems in bacteria and fungi. Adv. Microb. Physiol. 38:85-131. [DOI] [PubMed] [Google Scholar]
- 65.Zusman, D. R. 1982. Frizzy mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J. Bacteriol. 150:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]