Abstract
Attainment of a brown adipocyte cell phenotype in white adipocytes, with their abundant mitochondria and increased energy expenditure potential, is a legitimate strategy for combating obesity. The unique transcriptional regulators of the primary brown adipocyte phenotype are unknown, limiting our ability to promote brown adipogenesis over white. In the present work, we used microarray analysis strategies to study primary preadipocytes, and we made the striking discovery that brown preadipocytes demonstrate a myogenic transcriptional signature, whereas both brown and white primary preadipocytes demonstrate signatures distinct from those found in immortalized adipogenic models. We found a plausible SIRT1-related transcriptional signature during brown adipocyte differentiation that may contribute to silencing the myogenic signature. In contrast to brown preadipocytes or skeletal muscle cells, white preadipocytes express Tcf21, a transcription factor that has been shown to suppress myogenesis and nuclear receptor activity. In addition, we identified a number of developmental genes that are differentially expressed between brown and white preadipocytes and that have recently been implicated in human obesity. The interlinkage between the myocyte and the brown preadipocyte confirms the distinct origin for brown versus white adipose tissue and also represents a plausible explanation as to why brown adipocytes ultimately specialize in lipid catabolism rather than storage, much like oxidative skeletal muscle tissue.
Keywords: microarray, myocyte, principal component analysis, differentiation, transcriptome
At the onset of obesity, mitochondrial function is attenuated in white adipocytes (1), exemplifying a negative interaction between obesity and mitochondrial biogenesis, a process that potentially contributes to diabetes (2–4). In contrast to white adipocytes, brown adipocytes have an abundance of mitochondria and are able to contribute positively to energy expenditure through the specific expression of the mitochondrial uncoupling protein, UCP1 (5). By using this narrow criterion, primary white adipocytes can also obtain a brown adipocyte-like phenotype (6, 7) through overexpression of peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC1α). PPARγ has been found to be essential for adipogenesis per se (8, 9), whereas PPARγ agonists, such as rosiglitazone, promote reversal of mitochondrial dysfunction in adipocytes (1) and skeletal muscle (3), but unfortunately they also promote adiposity and weight gain, potentially limiting their clinical effectiveness (5).
There is support for the possibility that unique precursor cell types give rise to brown versus white preadipocyte pools because these distinct adipose tissue depots appear at different developmental stages (10). Identification of the transcriptional strategies that characterize the distinct brown adipocyte differentiation process may help identify safe mechanisms to promote mitochondrial biogenesis or increase energy expenditure in white adipocytes. However, the factors that determine primary adipocyte differentiation remain ill defined (9, 11, 12). We would argue that utilization of a primary cell model is critical to address this research aim because it will faithfully document the endogenous process through which “preadipocytes” pass to form more mature adipocytes. In contrast, immortalized cell models are a priori committed to an adipocytic lineage in a manner that may have limited bearing on endogenous differentiation (9), thus potentially limiting their usefulness. In the present study, we contrasted the transcriptional regulation of brown and white primary preadipocytes and robustly demonstrated that brown preadipocytes uniquely share an overlapping transcriptional program with muscle cells, whereas a number of developmental genes further help to distinguish the two preadipocyte cell types.
Results
Discovery of a Myogenic Signature in Brown Preadipocytes.
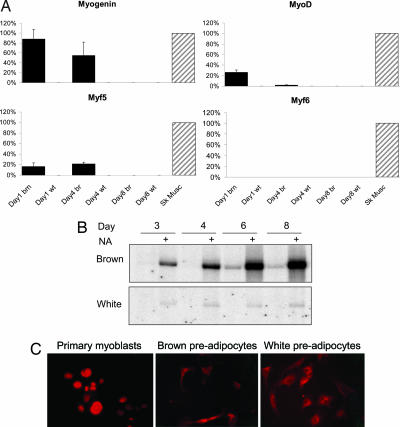
By using a variety of array analysis methodologies we provide here an extensive analysis of primary brown and white preadipocytes early during their respective differentiation programs. Using the Affymetrix U74A v2 chip, we characterized the abundance of ≈8,000 transcripts (12,000 probe sets) in undifferentiated primary brown and white preadipocytes (n = 6–8) cultured for 4 days (13). At this stage, the preadipocytes are morphologically undifferentiated yet committed to their final phenotype (see below). By using significance analysis of microarrays (14) we identified ≈300 genes demonstrating at least 2-fold differential expression between brown and white undifferentiated preadipocytes (false discovery rate, <1%) [supporting information (SI) Data Set 1]. The most differentially expressed of these genes are listed in SI Table 1. As can be seen, within the brown preadipocytes “transcriptome” we found a substantial enrichment of genes belonging to the “skeletal muscle” gene ontology (GO) analysis class (15). Remarkably, these genes included the previously claimed “muscle-specific” basic helix–loop–helix (bHLH) myogenic regulator myogenin, which was expressed in brown preadipocytes at a level comparable with that found in differentiating confluent C2C12 myoblasts (16). To confirm the expression of myogenic genes in brown preadipocytes, we used an independent set of primary cell cultures in which we harvested RNA after 1, 4, and 8 days in culture and performed fluorescence-based real-time quantitative PCR (RT-qPCR). Convincingly, in brown preadipocytes (but not in white), myogenin was expressed at day 1 (at a level similar to that in young adult skeletal muscle), with expression diminishing during differentiation (Fig. 1A). Although MyoD and Myf5 were detectable in brown preadipocytes after 1 and 4 days of culture, Myf6 was detectable only in skeletal muscle tissue, demonstrating that brown preadipocytes do not express the full repertoire of myogenic regulatory factors. By using the brown adipocyte marker gene UCP1 (Fig. 1B), we confirmed that despite identical culture conditions, our primary brown cell cultures were phenotypically distinct from our white preadipocytes: increasing norepinephrine-induced ucp1 induction was only observed in the maturating brown cultures.
Fig. 1.
Phenotypic and myogenic factors in brown and white primary preadipocytes. Cell cultures were generated, harvested at the time points indicated, and examined for expression of genes by using RT-qPCR (n = 3 per cell type and time point) or by immunohistochemistry. (A) Myogenin, MyoD, Myf5, and Myf6 mRNA expression was determined by using RT-qPCR. Data (mean ± SE) are expressed as a percentage of the positive control value (hindlimb muscle obtained from 4-week-old mice). (B) A Northern blot was carried out to demonstrate the ucp1 expression status in primary white and brown preadipocytes at the time intervals shown. Norepinephrine (0.1 μM for 4 h) was used to induce ucp1 expression to demonstrate that the brown preadipocytes were committed to the brown cell lineage and distinct from the white preadipocytes. (C) Given the striking findings from the array analysis we aimed to rule out the possibility of any contamination from skeletal muscle cells within the brown preadipocyte culture. Immunohistochemistry was carried out to detect MyoD expression in day 2 preadipocytes (day 2 cultures have enough cells to ensure that a reasonable image can be obtained). Primary mouse myoblasts (Left) demonstrated nuclear staining (red) for MyoD, whereas at the most both brown and white preadipocytes demonstrated diffuse cytosolic staining resulting from the secondary antibody (see SI Fig. 4). At no time did we find any evidence that a distinct subpopulation of cells stained strongly for MyoD, and thus the brown preadipocytes cultures are not contaminated with myoblasts.
Gene expression profiling is a sensitive and powerful tool to study the cellular differentiation process (17), but the issue may be raised that contaminating myoblasts could have influenced the analysis outcome. However, several lines of experimental data and calculation demonstrate that this problem does not exist. For example, we carefully examined for nuclear staining of MyoD (Fig. 1C) and myogenin (data not shown) in primary myoblasts (18) and contrasted them with HeLa cells artificially contaminated with myoblasts, and with our two primary preadipocyte cell types (white and brown; SI Fig. 4 A and B). Primary myoblasts demonstrated clear nuclear staining for MyoD, whereas both brown and white preadipocytes demonstrated only cytosolic staining (a nonspecific action of the secondary antibody; Fig. 1C), a pattern distinct from the artificial “contamination profile” obtained when mixing primary myoblasts with HeLa cells (representative images can be found in SI Fig. 4). The presence in brown preadipocytes of myogenic transcription factor (TF) mRNA is supported by recent findings of Atit et al. (19), who published findings clearly demonstrating that the myogenin-expressing central dermomyotome gives rise to dermis, muscle, and brown adipose tissue but not white adipose tissue, data that wholeheartedly support the present analysis. Thus, the lack of MyoD or myogenin protein expression in brown preadipocyte cultures is consistent with the lack of sensitivity of the protein detection methodology and, as one might now expect, remnant myogenic transcription factor (TF) activity reflecting the common developmental origin of brown preadipocytes and striated muscle.
Contrasting Differentiation Processes.
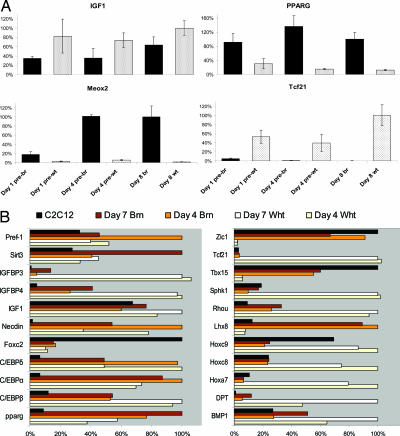
We used both correlation and principal component analysis (PCA) (20, 21) to define the differentiation processes in the brown and white adipocyte cultures. Day 4 undifferentiated brown preadipocytes are more similar to maturing brown adipocytes than either to day 4 white preadipocytes or to C2C12 myoblasts (16) (SI Fig. 5 A–C). The undifferentiated white preadipocyte transcriptome was specifically enriched with extracellular matrix genes (SI Data Set 1), consistent with a recent cDNA array study examining human preadipocytes (22). There were also evident similarities between the two adipocyte cultures; e.g., Igf1 and Pparg mRNA differed modestly between the cell types (Fig. 2A). However, to identify potential “unique” markers for brown and white preadipocytes, we examined the array hybridization data by using a simple threshold filter approach (Fig. 2B). Several genes from this analysis appeared interesting, and two were validated further by using RT-qPCR: Meox2 and Tcf21. The vascular developmental gene Meox2 (mesenchyme homeobox 2) was expressed markedly in brown preadipocytes (Fig. 2A), although it was essentially absent in white adipocytes (Fig. 2A) and in skeletal muscle cells (data not shown). During active thermogenesis, brown adipose tissue becomes highly vascularized (5), and interestingly, Meox2 can promote VEGF-driven vessel maturation (23), a process central to brown adipose tissue angiogenesis and one that may be distinct (24) from the hypoxia-driven process in skeletal muscle (25, 26). Intriguingly, the bHLH “antimyogenic” transcription factor Tcf21 (27) (also known as Pod-1) appeared distinctly expressed in white preadipocytes (Fig. 2 and SI Data Set 1).
Fig. 2.
Expression of established and novel adipocyte marker genes in brown (br) and white (wt) preadipocytes. Primary cultures were produced, and cells were harvested at the time points indicated. (A) RT-qPCR validation of Pparg and Igf1 expression patterns in white (pre-wt) and brown (pre-br) preadipocytes (n = 3 independent cultures per cell type). In addition, verification of the microarray data for novel markers for brown (Meox2) and white (Tcf21) preadipocytes was carried out. (B) Affymetrix hybridization data for literature (Left) and novel (Right) markers of brown and white preadipocytes. Data from C2C12 (see SI Methods) are provided as a reference and further illustrate the difference in profile among brown, white, and muscle cells. The analysis is illustrative of genes that are expressed differentially across these cell types. Values are normalized to the cell type with the greatest signal; values much <20% are indicative of an “absent” call within the MAS5.0 algorithm. Of the “literature” genes, only Igfbp3 displays a qualitative difference in expression pattern; among the novel genes, Lhx8 (and Zic1) are characteristic of brown adipocytes, and Tcf21, Sphk1, Hoxa7, and DPT are characteristic of white adipocytes.
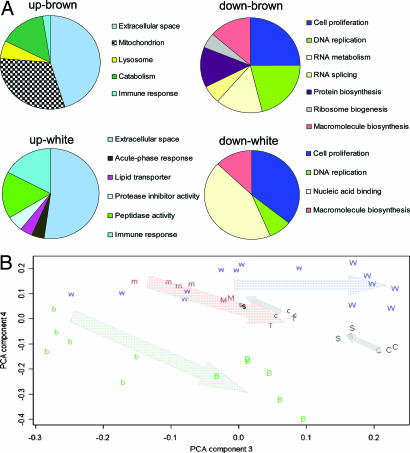
We identified the biological processes that were altered during differentiation by examining the genes expressed differentially (28) at day 8 versus day 4 in culture. We applied gene ontology analysis (15) to the ≈1,400 modulated genes in brown preadipocytes and the ≈450 modulated in white preadipocytes during maturation (SI Data Set 2). Because both cell types become lipid-laden during the differentiation process, it was not surprising that 72% of the genes up-regulated in white preadipocytes were also up-regulated in brown preadipocytes and that 52% of the genes down-regulated in white preadipocytes were also down-regulated in brown preadipocytes (Fig. 3A and SI Data Set 3). In both brown and white preadipocytes, extracellular matrix modeling genes were abundantly up-regulated during differentiation, giving rise to the highest ranked enriched group in the gene ontology analysis (Fig. 3A and SI Data Set 2). Down-regulated processes included cell cycle and nucleic acid biosynthesis genes (Fig. 3A and SI Data Set 2), as expected during growth arrest, a process known to be essential for adipocyte differentiation (29).
Fig. 3.
GO analysis of differentiating brown and white primary preadipocytes compared with C2C12 myoblasts and PCA. (A) Diagrammatic representation of the GO analysis carried out by using EASE. GO subgroups were collapsed into related groups to facilitate visualization of the data. For the complete breakdown of significantly enriched gene ontology groups, see SI Data Set 3. All GO groups demonstrated enhanced statistical representation (based on an EASE score and a Bonferroni-adjusted value of P < 0.05). (B) We used a PCA-based approach to compare the C2C12 differentiation data set, the SIRT1 differentiation data set (where SIRT1 overexpression prevents myoblast differentiation) with the differentiating pre-brown adipocytes and pre-white adipocytes. The data set includes day 4 undifferentiated brown (b) and white (w) preadipocytes and day 7/8 differentiating brown (B) and white (W) preadipocytes. C2C12 myoblasts are shown as m, and progressively differentiating myoblasts are plotted as M and T (http://pepr.cnmcresearch.org/). Sirt1-overexpressing cells (16) (s for 12-h differentiation and time control c, whereas S is 36-h differentiation with SIRT1 viral overexpression and time control C). After global normalization, we examined the top 100 genes that contributed to components 3 to 5 and used GO analysis to identify the putative SIRT1-regulated genes (see Methods). Components 3 and 4 are plotted. The arrows are used to illustrate the general direction of cell differentiation. It should be noted that SIRT1 (s and S) overexpression opposes the differentiation process.
However, there were also substantial differences in the expression programs of the two cell types. For example, only brown preadipocytes substantially switched on an oxidative phosphorylation program, where ≈100 mitochondria-related genes were up-regulated (SI Data Set 2). Gene ontology analysis demonstrated that this characteristic was unique among brown preadipocytes during differentiation and verified that mitochondrial genes were indeed statistically overrepresented (P < 0.00001, Bonferroni-adjusted; SI Data Set 3). The most down-regulated genes in brown preadipocytes were myogenin and several other skeletal muscle-related genes (although they represented only a small proportion of the modulated gene list and thus were not detected by gene ontology analysis).
Contrasting Transcriptional Profiles in Primary Preadipocytes and Immortalized Cell Line Models.
Studies of an immortalized brown adipocyte cell line have implied that insulin receptor substrate 1 (IRS-1) deficiency (and ensuing necdin overexpression) prevent differentiation of “brown-like” preadipocytes (11). We used the multiple-array study comparison approach (30), comparing our data set and the IRS-1-knockout immortalized brown adipocyte cell line data set (11), to examine consistency between the two models for brown adipocyte differentiation (i.e., we wanted to identify a number of inversely related gene expression responses across these “differentiation” transcriptomes) (SI Data Set 4). However, with a 2-fold differential expression data set, we found that only four genes were modulated consistently in the IRS-1 model and our brown adipocyte differentiation model, one of them being Dkk3, a wnt signaling “antagonist” that was 2-fold overexpressed in IRS1−/− adipocytes cell lines and 2-fold down-regulated during differentiation of brown preadipocytes. Given our ability to establish a large common transcriptional profile even between primary brown and white preadipocytes, the lack of commonality between the IRS1−/− adipocyte cell line and brown preadipocytes is perplexing but apparently robust.
SIRT1 as a Plausible Brown Adipocyte Differentiation Regulator.
Mitochondrial abundance is important to both brown adipocyte (5) and skeletal muscle differentiation and function (31, 32). The Sir2 ortholog SIRT1 would represent an excellent candidate for carrying out the dual role of suppressing the myogenic signature because it prevents myoblast differentiation (16) as well as promoting mitochondrial biogenesis as it can modulate PGC1α action through its class III histone deacetylase activity (33). The role of SIRT1 in brown adipocytes has not been studied, and thus we used the transcriptome changes generated after SIRT1 overexpression in myoblasts (16) to evaluate the possible role of SIRT1 during brown preadipocyte differentiation. A total of 40 genes significantly overlapped between SIRT1 down-regulated genes and genes down-regulated during brown preadipocyte maturation (SI Data Set 5). This overlapping gene list was very specific, being highly enriched with muscle development genes (P < 0.00003 after Bonferroni correction) and thus was clearly nonrandom. Further, we found that 59 genes were common to SIRT1 up-regulated genes and genes up-regulated during brown preadipocyte maturation; this list was highly enriched for extracellular matrix genes and antigen-responsive genes (P < 0.0003 after Bonferroni correction).
The potential importance of the SIRT1-related overlapping gene lists was also assessed by using an approach to PCA (20, 21), where we combined PCA with GO analysis to give the top 100 ranking genes within a component some biological context. During differentiation of all cell types studied (or inhibition of differentiation in the case of SIRT1 overexpression in myoblasts), the first few components reflected modulation of ribosomal, cell cycle, and RNA regulation genes (e.g., Fig. 3B, component 2, and SI Data Set 6, component 3). Compellingly, component 4 (Fig. 3B) captured a variance category that described the maturation of brown preadipocyte and C2C12 muscle cells and the negative impact of SIRT1 overexpression (16) on differentiation, whereas this component was neutral for white adipocyte differentiation (SI Data Set 6). Consistent with our hypothesized role for SIRT1 in brown preadipocyte differentiation, component 4 represented mitochondrial and metabolism genes (P < 0.00001 after Bonferroni correction; SI Data Set 6). Component 5 (SI Data Set 6) was statistically enriched for muscle development genes where both SIRT1 overexpression and brown preadipocyte differentiation are associated with the down-regulation of this common group of muscle-specific genes, also supporting our hypothesis. These substantial and biologically distinct overlapping transcriptional signatures were in stark contrast to the lack of similarity between the primary brown adipocytes and the IRS1−/− brown adipocytes, suggesting that SIRT1 would be a valid and interesting target for further biochemical studies.
TF Analysis.
Although our analysis has provided a unique insight into primary preadipocyte maturation, identification of the TFs potentially responsible for these coordinated gene expression programs would further enhance our understanding. To address this aim, we carried out in silico prediction analysis (SI Fig. 6) of TF-binding sites within the upstream regions of all genes on the Affymetrix U74A v2 array and then compared them with the frequency of occurrence within each of the modulated gene lists from brown and white preadipocytes during differentiation (SI Data Set 2). Robust prediction of TF sites in eukaryotic genes is challenging because of the degenerative nature of binding sites (34) that yields a high rate of false-positive predictions. We therefore used phylogenetic footprinting (35), using human sequences for comparison, and restricted our analysis to only the 1,000 bp upstream. ClustalW (36) alignments between upstream mouse and corresponding human sequences were scanned for TF-binding sites by using the TFBS software (37) and JASPAR database (38). Between 12 and 39% of all TF-binding sites within the JASPAR database (111 sites) were significantly underrepresented within each modulated gene list. Only one TF-binding site, that for E2F, was overrepresented (SI Data Set 7), and it was in the list of genes down-regulated during brown adipocyte differentiation. It is unclear why the modulated gene lists were specifically underenriched for TF-binding sites, but the general outcome most likely indicates that the repertoire of the JASPAR database is still limited.
Discussion
Although both adipocytes and myocytes can differentiate from a common mesenchymal precursor cell (39), our analysis indicates that brown adipocytes endogenously share a common early transcriptional program with skeletal muscle cells, which is then suppressed early during differentiation. There was no Tcf21 expression in brown adipocytes, which would suggest that Tcf21 may be the first useful positive selection marker for white preadipocytes. Tcf21 positively regulates bone morphogenetic protein 4 (bmp4) (40) expression, and bmp4 can commit pluripotent mesenchymal cells to form white adipocytes, suggesting that Tcf21 also has the potential to play an important role in adipogenesis. Conversely, we found that Meox2 was expressed predominantly in brown adipocytes (and not in white adipocytes or skeletal muscle), and we speculate that it may contribute to the unique angiogenesis process in brown adipose tissue (24).
Skeletal muscle development relies on the expression of four bHLH transcription factors (myogenin, MyoD, Myf5, and Myf6), expressed in a coordinated manner (17). However, our observation of expression of three of these myogenic factors in brown adipocytes clearly indicates that they can no longer be considered unique myogenic markers. In skeletal muscle, MyoD regulates myogenin expression (17), and hence early loss of MyoD expression after day 1 could explain the diminished expression of myogenin (Fig. 1A), leading to the down-regulation of the skeletal muscle gene expression pattern from day 4 to day 8 in culture. Importantly, several lines of experimental data demonstrate that our preadipocyte cultures lacked any contaminating myoblasts, indicating that the myogenic expression signature is a genuine characteristic of brown preadipocytes. First, the myogenic program is successively “switched off” as the sparsely distributed cells become more confluent, directly contrasting with the response of myoblasts as they approach confluence (17). Second, smooth muscle cells specifically express myf6 and lack myf5 (41), which contrasts with the brown preadipocytes profile (Fig. 1A). Third, if a few percent of cells in the brown preadipocyte culture were indeed myoblasts, the relative expression of MyoD (and myogenin) in these cells would have to approach the level of ribosomal RNA to produce the average levels detected in the total RNA obtained from brown preadipocyte cultures, and we feel that this scenario is implausible. Finally, the recent discovery that the central dermomyotome gives rise to both muscle and brown adipose tissue but not white adipose tissue (19) clearly supports the validity of the present analysis.
Although it has been shown previously that modulating the cellular fate between myoblast and adipocyte is possible, through artificially modulating PPARγ or Rho GTPase (8, 39, 42), our analysis would suggest that an endogenous mechanism should exist to shut down the myogenic program in brown adipocytes during maturation. Clearly, mitochondriogenesis is important for both brown adipocyte (5) and skeletal muscle differentiation (31, 32). We evaluated SIRT1 as a potential candidate for both suppressing the myogenic signature (16) and regulating genes involved in mitochondrial biogenesis (33). The PCA/GO analysis provides clear evidence (based on a large statistically enriched transcript signature) that SIRT1 may indeed regulate mitochondrial gene expression in brown adipocytes, perhaps by direct impact on PGC-1α activity (43). Given data demonstrating that SIRT1 deacetylates MyoD, thus terminating myogenic gene expression (16), together with our PCA analysis, it is also plausible that SIRT1 activity terminates the myogenic gene expression signature during brown adipocyte maturation (Fig. 3B). Overall, our approach, combining PCA and GO analysis, illustrates the value of using multiple-array-derived transcriptome signatures for investigating novel biological scenarios.
The transcriptional regulators and developmental genes presented in Fig. 2B provide some interesting contrasts with the expression patterns determined in immortalized cell models of adipogenesis. For example, a general model has been presented, inferring a central role for pRB in regulating adipocyte differentiation, including interactions with E2F. Interestingly, we demonstrated that E2F-binding sites were 1.8-fold more frequent (P < 0.01) in the genes down-regulated during brown preadipocyte differentiation than in the population of genes present on the chip. The 43 “E2F” genes down-regulated included hdac3 (histone deacetylase 3) and rbbp4 (retinoblastoma protein-binding protein 4). Active suppression of E2F activity may influence PPARγ function during brown differentiation (29). We found that pRB expression was increased during brown preadipocyte differentiation, whereas neither foxc2 nor RIα expression was modulated (Fig. 2 Lower and data not shown). Pref-1 and necdin have been described as being potential regulators of brown preadipocyte differentiation (11). Necdin has been shown to promote differentiation of skeletal muscle (44) and to be modulated during adipocyte differentiation (45, 46). Specifically, necdin down-regulation appears to promote adipogenesis in immortalized brown preadipocytes (11), whereas overexpression in 3T3-L1 white preadipocytes prevents differentiation (45). However, necdin expression increases in the 3T3-L1 adipocyte cell model (45) during differentiation (but is concurrently associated with inhibition of differentiation when expressed at artificially high levels; ref. 45). We observed that both pref-1 (dlk1) and necdin are down-regulated in both brown and white primary cells. Given the lack of consistency between the different models, we can conclude that there may be substantial differences between the early differentiation transcriptional program characterized in artificially immortalized cell lines (11, 45) and the primary brown preadipocyte differentiation program investigated in the present work. In addition, our data contrast with general pRB-E2F brown adipocyte differentiation model (47) developed by using mouse embryo fibroblasts.
We also noted some distinctive expression patterns for several developmental genes, including Tbx15, Hoxc8, and Hoxa7 (Fig. 2 Lower), between brown and white preadipocytes, further supporting the argument that these mesenchymal stem cells have distinct origins. We originally postulated that our analysis may provide insight into potential strategies for targeting the transdifferentiation of white adipocytes, in obese patients. During the review of our article, Gesta et al. (48) discovered that Tbx15, Hoxc8, and Hoxc9 were expressed differentially between s.c. and visceral preadipocyte fractions, and, critically, that this expression pattern was sustained during in vitro culture of the cells. Furthermore, when profiled across patients with a large range in body mass index (as a measure of obesity), they were able to demonstrate that Tbx15 was profoundly suppressed as body mass index increased above normal levels. Although clearly requiring further investigation, this finding suggests that genes preferentially expressed in brown adipocytes may have relevance for human obesity (Tbx15 expression was far more substantial in brown adipocytes).
In conclusion, we present a detailed transcriptome analysis of primary brown versus white preadipocyte maturation. Superficially, white preadipocytes share an early differentiation program with brown, yet striking and novel differences in transcription factor expression were found. A multilevel approach, supported by direct gene and protein expression analysis, demonstrates that the origin of brown preadipocytes is distinct from white preadipocytes. Although brown preadipocytes express previously defined muscle-specific bHLH transcription factors, consistent with their embryological origins (19), white preadipocytes express the bHLH transcription factor Tcf21, a transcription factor that suppresses the antiadipogenic androgen nuclear receptor (49) and the promyogenic bHLH factor MyoD (27). Our discovery of a myogenic signature represents a logical explanation as to how brown adipocytes do ultimately specialize in lipid catabolism, much like highly oxidative skeletal muscle tissue.
Methods
Primary Cell Culture and Differentiation.
Male NMRI mice (age 3–4 weeks; B&K, Stockholm, Sweden) were killed by CO2, and brown adipose tissue (from the interscapular, cervical, and axillary depots) and epididymal white adipose tissue were isolated as described previously (13). The cells were cultured in 10-cm2-well plates (Corning, Corning, NY), culture medium was DMEM with 10% (vol/vol) newborn calf serum (Invitrogen, Carlsbad, CA)/2.4 nM insulin/25 μg/ml sodium ascorbate/10 mM Hepes, pH 7.4/4 mM glutamine/50 units/ml penicillin/50 μg/ml streptomycin. To verify the cell phenotype (using ucp1 expression as a marker), cells were treated with either vehicle or 0.1 μM norepinephrine for 4 h, all as described previously (5).
Microarray Analysis and RT-qPCR and Northern Blot Analysis.
We used the Affymetrix (Santa Clara, CA) U74A v2 array platform, and all array data were normalized through implementations of the MAS5 algorithm and then subjected to a multilevel gene array analysis strategy. Full details can be found in SI Methods.
For RT-qPCR, total RNA was isolated by the use of Ultraspec (Biotecx, Houston, TX) followed by DNase treatment with the RNeasy mini kit column procedure (Qiagen, Valencia, CA). RNA (750 ng) was reverse-transcribed by reverse transcription reagents (Applied Biosystems, Foster City, CA) with random hexamer primers, in a total volume of 40 μl. RT-qPCR aliquots contained 2–5 μl of the sample cDNA, 2× TaqMan universal PCR master mix, and an optimized concentration of each primer, added to a final volume of 24–26 μl and were measured in triplicate from three independent cell cultures. For the muscle-specific factors (MyoD, myogenin, Myf5 and Myf6), hindlimb skeletal muscle was obtained from 4-week-old animals to act as a positive control. In this situation, expression was normalized first to 18S and then to expression levels found in skeletal muscle. For all other genes, data were normalized to 18S and then expressed as a percentage of the sample with the highest expression (shown as 100% in Fig. 2). Northern blotting was carried out as described in SI Methods.
Immunohistochemistry.
Primary muscle cells were plated in 6-well plates containing collagen-coated coverslips. The cells were rinsed in PBS and before fixing in 4% paraformaldehyde for 20 min at room temperature. Fixed cells were washed in PBS containing 0.2% fish skin gelatin, permeabilized in 0.2% Triton X-100 for 4 min, and then blocked in goat serum for 10 min. The coverslips were then incubated in primary antibody (1:50) for 20 min at room temperature and washed in 0.2% fish skin gelatin. Primary antibody binding was detected after incubation with Cy3-labeled secondary antibodies (1:500) and mounting in Vectashield (Vector Laboratories, Burlingame, CA). Adipocytes were grown on coverslips in 6-well plates and were washed in 37°C PBS and fixed in 3.7% formaldehyde in PBS, then permeabilized in 0.1% Triton X-100 in PBS for 2 min at room temperature and processed as above.
Supplementary Material
Acknowledgments
We thank Birgitta Leksell for technical assistance. This work was supported by Swedish Science Research Council Grant 621-2004-3858 (to B.C. and J.N.), Swedish Cancer Foundation Grant 05-0291 (to J.N.), Swedish Diabetes Society Grant DIA2005-004 (to J.T.), VINNOVA Grant P25770-1 (to C.W.), and funding from Heriot–Watt University Grant L6004 (to J.T.).
Abbreviations
- bHLH
basic helix–loop–helix
- GO
gene ontology
- IRS-1
insulin receptor substrate 1
- PCA
principal component analysis
- PGC1α
peroxisome proliferator-activated receptor γ coactivator 1α
- PPARγ
peroxisome proliferator-activated receptor γ
- RT-qPCR
real-time quantitative PCR
- TF
transcription factor.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE 7032).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610615104/DC1.
References
- 1.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 3.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmons JA, Norrbom J, Scheele C, Thonberg H, Wahlestedt C, Tesch P. Genomics. 2006;87:165–172. doi: 10.1016/j.ygeno.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Cannon B, Nedergaard J. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 6.Orci L, Cook WS, Ravazzola M, Wang MY, Park BH, Montesano R, Unger RH. Proc Natl Acad Sci USA. 2004;101:2058–2063. doi: 10.1073/pnas.0308258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D. J Biol Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 8.Hu E, Tontonoz P, Spiegelman BM. Proc Natl Acad Sci USA. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen ED. Prostaglandins Leukotrienes Essent Fatty Acids. 2005;73:31–34. doi: 10.1016/j.plefa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Moulin K, Truel N, Andre M, Arnauld E, Nibbelink M, Cousin B, Dani C, Penicaud L, Casteilla L. Biochem J. 2001;356:659–664. doi: 10.1042/0264-6021:3560659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, Cypess AM, Niinobe M, Yoshikawa K, Patti ME, Kahn CR. Nat Cell Biol. 2005;7:601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 12.Nedergaard J, Petrovic N, Lindgren EM, Jacobsson A, Cannon B. Biochim Biophys Acta. 2005;1740:293–304. doi: 10.1016/j.bbadis.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Nechad M, Nedergaard J, Cannon B. Am J Physiol. 1987;253:C889–C894. doi: 10.1152/ajpcell.1987.253.6.C889. [DOI] [PubMed] [Google Scholar]
- 14.Larsson O, Wahlestedt C, Timmons JA. BMC Bioinformatics. 2005;6:129. doi: 10.1186/1471-2105-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 17.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang YC, Dennis RG, Baar K. Am J Physiol. 2006;291:C11–C17. doi: 10.1152/ajpcell.00366.2005. [DOI] [PubMed] [Google Scholar]
- 19.Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raychaudhuri S, Stuart JM, Altman RB. Pac Symp Biocomput. 2000:455–66. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urs S, Smith C, Campbell B, Saxton AM, Taylor J, Zhang B, Snoddy J, Jones Voy B, Moustaid-Moussa N. J Nutr. 2004;134:762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 23.Wu Z, Guo H, Chow N, Sallstrom J, Bell RD, Deane R, Brooks AI, Kanagala S, Rubio A, Sagare A, et al. Nat Med. 2005;11:959–965. doi: 10.1038/nm1287. [DOI] [PubMed] [Google Scholar]
- 24.Fredriksson JM, Nikami H, Nedergaard J. FEBS Lett. 2005;579:5680–5684. doi: 10.1016/j.febslet.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 25.Timmons JA, Jansson E, Fischer H, Gustafsson T, Greenhaff PL, Ridden J, Rachman J, Sundberg CJ. BMC Biol. 2005;3:19. doi: 10.1186/1741-7007-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hang J, Kong L, Gu JW, Adair TH. Am J Physiol. 1995;269:H1827–H1831. doi: 10.1152/ajpheart.1995.269.5.H1827. [DOI] [PubMed] [Google Scholar]
- 27.Funato N, Ohyama K, Kuroda T, Nakamura M. J Biol Chem. 2003;278:7486–7493. doi: 10.1074/jbc.M212248200. [DOI] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 30.Timmons JA, Larsson O, Jansson E, Fischer H, Gustafsson T, Greenhaff PL, Ridden J, Rachman J, Peyrard-Janvid M, Wahlestedt C, Sundberg CJ. FASEB J. 2005;19:750–760. doi: 10.1096/fj.04-1980com. [DOI] [PubMed] [Google Scholar]
- 31.Rochard P, Rodier A, Casas F, Cassar-Malek I, Marchal-Victorion S, Daury L, Wrutniak C, Cabello G. J Biol Chem. 2000;275:2733–2744. doi: 10.1074/jbc.275.4.2733. [DOI] [PubMed] [Google Scholar]
- 32.Timmons JA, Gustafsson T, Sundberg CJ, Jansson E, Hultman E, Kaijser L, Chwalbinska-Moneta J, Constantin-Teodosiu D, Macdonald IA, Greenhaff PL. J Clin Invest. 1998;101:79–85. doi: 10.1172/JCI1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 34.Wasserman WW, Sandelin A. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 35.Lenhard B, Sandelin A, Mendoza L, Engstrom P, Jareborg N, Wasserman WW. J Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenhard B, Wasserman WW. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 38.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sordella R, Jiang W, Chen GC, Curto M, Settleman J. Cell. 2003;113:147–158. doi: 10.1016/s0092-8674(03)00271-x. [DOI] [PubMed] [Google Scholar]
- 40.Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. Development (Cambridge, UK) 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 41.Graves DC, Yablonka-Reuveni Z. J Histochem Cytochem. 2000;48:1173–1193. doi: 10.1177/002215540004800902. [DOI] [PubMed] [Google Scholar]
- 42.Taylor-Jones JM, McGehee RE, Rando TA, Lecka-Czernik B, Lipschitz DA, Peterson CA. Mech Ageing Dev. 2002;123:649–661. doi: 10.1016/s0047-6374(01)00411-0. [DOI] [PubMed] [Google Scholar]
- 43.Nemoto S, Fergusson MM, Finkel T. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 44.Kuwajima T, Taniura H, Nishimura I, Yoshikawa K. J Biol Chem. 2004;279:40484–40493. doi: 10.1074/jbc.M404143200. [DOI] [PubMed] [Google Scholar]
- 45.Goldfine AB, Crunkhorn S, Costello M, Gami H, Landaker EJ, Niinobe M, Yoshikawa K, Lo D, Warren A, Jimenez-Chillaron J, Patti ME. Diabetes. 2006;55:640–650. doi: 10.2337/diabetes.55.03.06.db05-1015. [DOI] [PubMed] [Google Scholar]
- 46.Boeuf S, Klingenspor M, Van Hal NL, Schneider T, Keijer J, Klaus S. Physiol Genomics. 2001;7:15–25. doi: 10.1152/physiolgenomics.00048.2001. [DOI] [PubMed] [Google Scholar]
- 47.Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, Petrovic N, Enerback S, Nedergaard J, Cinti S, et al. Proc Natl Acad Sci USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Proc Natl Acad Sci USA. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong CY, Gong EY, Kim K, Suh JH, Ko HM, Lee HJ, Choi HS, Lee K. Mol Endocrinol. 2005;19:2245–2257. doi: 10.1210/me.2004-0400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.