Abstract
The kinase inhibitor imatinib mesylate targeting the oncoprotein Bcr-Abl has revolutionized the treatment of chronic myeloid leukemia (CML). However, even though imatinib successfully controls the leukemia in chronic phase, it seems not to be able to cure the disease, potentially necessitating lifelong treatment with the inhibitor under constant risk of relapse. On a molecular level, the cause of disease persistence is not well understood. Initial studies implied that innate features of primitive progenitor cancer stem cells may be responsible for the phenomenon. Here, we describe an assay using retroviral insertional mutagenesis (RIM) to identify genes contributing to disease persistence in vivo. We transplanted mice with bone marrow cells retrovirally infected with the Bcr-Abl oncogene and subsequently treated the animals with imatinib to select for leukemic cells in which the proviral integration had affected genes modulating the imatinib response. Southern blot analysis demonstrated clonal outgrowth of cells carrying similar integration sites. Candidate genes located near the proviral insertion sites were identified, among them the transcription factor RUNX3. Proviral integration near the RUNX3 promoter induced RUNX3 expression, and Bcr-Abl-positive cell lines with stable or inducible expression of RUNX1 or RUNX3 were protected from imatinib-induced apoptosis. Furthermore, imatinib treatment selected for RUNX1-expressing cells in vitro and in vivo after infection of primary bone marrow cells with Bcr-Abl and RUNX1. Our results demonstrate the utility of RIM for probing molecular modulators of targeted therapies and suggest a role for members of the RUNX transcription factor family in disease persistence in CML patients.
The Bcr-Abl fusion protein arising from the t(9,22) translocation plays a decisive role in the pathogenesis of chronic myeloid leukemia (CML) and a subset of Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL) (1, 2). With the introduction of imatinib, a 2-aminophenylpyrimidine inhibitor of the Abl tyrosine kinase, a potent new therapy for the treatment of Bcr-Abl-expressing leukemias, has become available (3). Treatment with imatinib alone has been shown to induce hematologic remissions in most patients with chronic-phase CML, and >80% of these patients achieve a complete cytogenetic response (CCR) (3). However, even though patients with chronic-phase CML respond well with durable remissions, imatinib treatment seems not to be able to eradicate the disease. Evidence for disease persistence in CML patients on imatinib treatment comes from reports demonstrating that Bcr-Abl mRNA can still be detected in patients in CCR and that molecular remissions are rare in CML patients treated with imatinib (3). Furthermore, case reports on patients who had to stop imatinib treatment for different reasons indicated a high incidence of relapse (4). Impaired drug action or Bcr-Abl-independent growth, either because of intrinsic or acquired properties of residual Ph+ stem cells, has been implicated in disease persistence. Studies performed on Ph+ early hematopoietic progenitor cells suggested that imatinib treatment limits proliferation but does not induce apoptosis in these cells (5, 6). The mechanisms underlying the insensitivity of the CML progenitor cells toward imatinib are not yet well understood. cDNA microarray analyses comparing CML and normal stem cells have revealed a host of data on differentially expressed genes, but they are limited in their ability to identify functionally important candidate genes from the complex genetic networks interacting in imatinib-resistant cells (7). Retroviral insertional mutagenesis (RIM) as a functional genetic screen may be able to overcome some of these limitations. During the retroviral life cycle, viral RNA is reverse transcribed into DNA, which then stably integrates into the host genome (8). The insertion of proviral DNA near oncogenes or tumor suppressor genes can lead to a dysregulated expression of these genes and promote cellular transformation (9). As a negative consequence, RIM is believed to be responsible for ALL development in patients treated in a gene therapy trial aiming to correct the common cytokine γ-chain deficiency in SCID-X1 syndrome (10). Advanced PCR techniques and the availability of near-complete sequences for several vertebrate genomes, including the mouse, have facilitated the recovery of the proviral flanking regions and the assignment of candidate genes potentially affected by retroviral integration (9). Insertional mutagenesis mediated by DNA-integrating viruses or retrotransposons and subsequent identification of genes affected by vector integration thus represents a powerful tool for the rapid analysis of cooperating oncogenes (9). Furthermore, it has also been used in cell culture screens for the analysis of the development of drug resistance to conventional chemotherapy (11). In this work, we used a RIM screen with a replication-defective retrovirus carrying the Bcr-Abl oncogene to identify candidate genes modulating the cellular imatinib response in a murine model of CML/ALL.
Results
CML Mice Respond Rapidly to Imatinib Treatment but Eventually Develop Imatinib-Resistant ALL or CML.
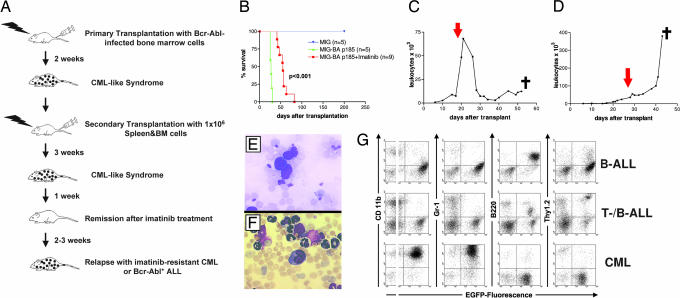
Biological systems can be powerful tools for the selection of gain-of-function mutations in the appropriate settings. Using imatinib as the selective agent on leukemic cells in a murine retroviral infection/transplantation model of CML, we aimed to recover genes induced by RIM affecting the cellular imatinib response. A CML-like syndrome was induced in primary animals by high-titer (>106 viral particles per ml) infection of bone marrow (BM) cells with an MIG-p185 Bcr-Abl-expressing retroviral construct [supporting information (SI) Fig. 5]. To increase disease latency and the probability of recovering recurrent integrations, leukemic cells from diseased CML mice were serially transplanted to 15 sublethally irradiated mice (Fig. 1A and SI Table 1). After establishment of CML disease in the secondary transplanted mice, imatinib treatment was initiated, extending the overall survival of the treated mice compared with an untreated control group (Fig. 1 A and B). Seven of nine treated mice displayed a quick hematologic response with normalization of the white blood cell counts (WBC) (Fig. 1C), whereas in two mice the WBC fell briefly after initiation of imatinib treatment but then continued to rise despite imatinib treatment with an increased dose of 100 mg/kg bid (Fig. 1D and SI Fig. 5C). The seven mice responding with a hematologic remission subsequently developed a rapidly fatal ALL resistant to imatinib (Fig. 1E). Two mice with short-lived responses succumbed to an imatinib-resistant CML-like disease (Fig. 1F). The ALL cells from resistant mice exhibited either a B or a mixed B/T cell immunophenotype in the flow cytometric analysis, whereas the resistant CML mice showed mostly CD11b (Mac-1)- and Gr-1-positive granulocytes in the spleen (Fig. 1G). Thus, all diseased mice treated with imatinib eventually developed resistance to imatinib with progressive ALL or CML disease despite continuing imatinib treatment (SI Table 1).
Fig. 1.
Imatinib-treated mice relapse with resistant CML or ALL disease. (A) Outline of the experimental setup. (B) Kaplan–Meier survival curves of mice serially transplanted with Bcr-Abl-infected BM cells. The curves represent mice transplanted with empty vector infected cells (blue), untreated Bcr-Abl-transplanted mice (green), and imatinib-treated Bcr-Abl-transplanted mice (red). (C) WBC of a mouse with imatinib-resistant ALL. The red arrow indicates the start of imatinib treatment. (D) WBC of a mouse with imatinib-resistant CML disease. (E) Hematoxylin/eosin stain of peripheral blood from a mouse with imatinib-resistant ALL. (Magnification, ×600.) (F) Hematoxylin/eosin stain of a peripheral blood smear from a mouse with imatinib-resistant CML disease. (Magnification, ×600.) (G) Flow cytometric analysis of the different resistant leukemic phenotypes. The expression of lineage-specific antigens versus EGFP is shown in two-parameter dot-plots of spleen-derived leukemic cells.
Evidence for Clonal Selection of Leukemic Cells Refractory to Imatinib.
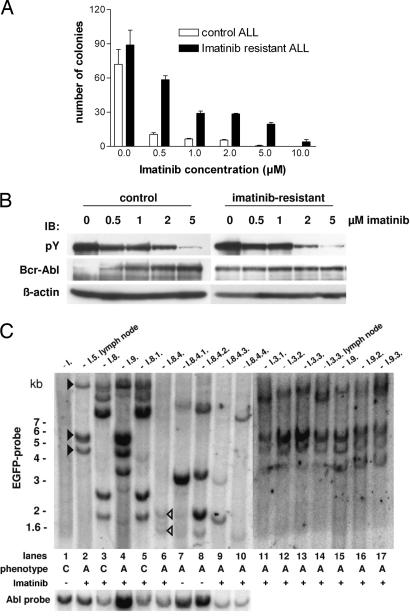
We further analyzed the leukemic cells ex vivo to delineate the mechanisms underlying the decreased imatinib responsiveness. Cobblestone area-forming cell (CAFC)-like assays were performed with leukemic cells from imatinib-resistant and naïve control mice. Imatinib treatment effectively suppressed colony formation in control cells, whereas the growth of ALL cells from imatinib-resistant mice could not be completely blocked even at concentrations of 10 μM imatinib (Fig. 2A). Direct sequencing of a 1-kb stretch spanning the Abl kinase domain in all nine resistant mice did not reveal Bcr-Abl kinase mutations (data not shown). Furthermore, Bcr-Abl autophosphorylation was efficiently inhibited in imatinib-resistant and previously untreated control ALL cells (Fig. 2B).
Fig. 2.
ALL cells from resistant mice are also refractory toward imatinib treatment ex vivo, but the Bcr-Abl kinase is still inhibited by imatinib. Southern blot analysis shows recurrent proviral integration patterns in different resistant mice. (A) Cobblestone area-forming cell (CAFC)-like assay of imatinib-resistant and control ALL cells on MS-5 stromal feeder cells in the presence of different concentrations of imatinib. (B) Analysis of Bcr-Abl autophosphorylation in resistant and control ALL cells cultured ex vivo in the presence of different concentrations of imatinib. Blots were sequentially probed with the indicated antibodies. (C) Southern blot analysis of the proviral integration pattern in leukemic cells from imatinib-resistant mice. Genomic DNA was cut with EcoRI and probed with EGFP sequences for the analysis of proviral integration distribution or with Abl sequences to check the integrity of the provirus. The leukemic phenotype is specified below the graph by C for CML and A for ALL disease. Filled arrows indicate recurrent proviral integrations in resistant ALL clones; open arrows highlight proviral insertions in resistant clones that are overgrown by other leukemic clones after imatinib withdrawal (see “Results”).
Next, we investigated whether recurrent proviral integration patterns indicating clonal selection could be found in the different diseased mice. The initial CML-like disease from the primary transplantation was polyclonal (Fig. 2C, lane 1), whereas all imatinib-resistant mice showed an oligoclonal insertion pattern. Several mice with imatinib-resistant B-ALL (Fig. 2C, lanes 2, 4, and 11–17) displayed similar integration patterns (filled black arrowheads). The transplantation of spleen cells from a mouse with imatinib-resistant CML led to a similar resistant disease with the identical insertion profile in a tertiary recipient mouse (Fig. 2C, lanes 3 and 5). Because resistant disease in the mice with B-ALL could be caused either by a single clone containing multiple retroviral integrations or several clones containing one retroviral integration each, quantitation of the number of retroviral integration events per cell was performed by real-time PCR, indicating that the predominant clone contained three to four retroviral insertions in these mice (data not shown).
Four mice were serially transplanted with cells from a mouse with imatinib-resistant mixed B-/T-ALL phenotype disease. Two mice were treated with imatinib, and the other two mice did not receive the inhibitor (Fig. 2C, lanes 6–10). Interestingly, the treated mice again developed a mixed ALL, whereas the untreated mice succumbed to a T-ALL. Accordingly, besides the integrations in the imatinib-resistant primary mouse (Fig. 2C, lane 6, open arrowheads), other clones appeared in the serially transplanted untreated mice (Fig. 2C, lanes 7 and 8) but only to a lesser extent in the treated animals (Fig. 2C, lanes 9 and 10). Taken together, the results of the analysis indicated that imatinib treatment led to a clonal selection of leukemic cells carrying similar retroviral integration patterns in the diseased mice.
Retroviral Insertion Site Analysis Identifies Proviral Integrations Near Transcription Factor Genes.
The previous results were compatible with a role for RIM in the induction of the imatinib-resistant phenotype. To recover the genes influenced by the retroviral integration, we used a linker-mediated (LM) PCR method (12) to extract the proviral flanking sequence and subsequently screen a public murine genomic database (www.ensembl.org) to identify the retroviral integration sites (RIS) (SI Table 2). Mice serially transplanted with spleen cells from the same donor mouse (Fig. 2C, mice I.3.1–I.3.3.) with a similar integration pattern in the Southern blot had identical RIS near the RUNX3 and Irf2 gene. Overall, the identified flanking sequences demonstrated retroviral integration in the vicinity of transcription factors (RUNX3, Nfix1, Irf2), other DNA-binding proteins (histone genes), kinases (Stk24), and genes involved in DNA-repair mechanisms (Rad51L1) (SI Table 2). Two of the RIS identified correspond to known common integration sites (RUNX3 and Kif13a), which have been described to cooperate with c-myc overexpression and p16INK4a/p19ARF deficiency to induce lymphomas and leukemias in mice (13, 14). Interestingly, the integration site near the RUNX3 locus was to be identical to the previously characterized Dsi1 locus, a RIS found in murine and rat lymphomas (15, 16).
Multicolor FISH (M-FISH) Reveals a Normal Karyotype, and FISH Corroborates the Results of the Proviral Flanking Region Analysis.
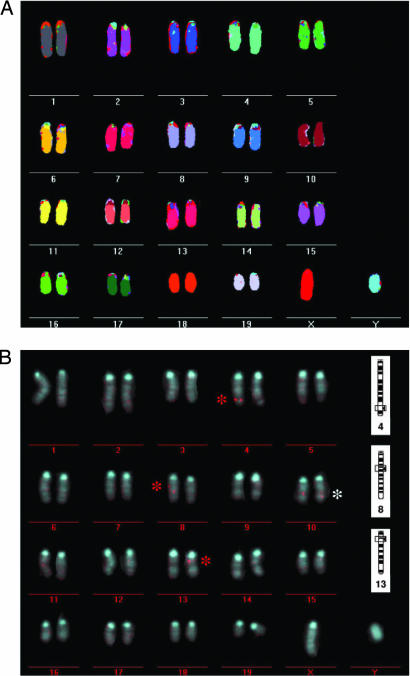
The M-FISH analysis on leukemic cells from three imatinib-resistant and untreated control mice with ALL demonstrated the absence of major structural abnormalities and a normal male karyotype (2n = 40) in all analyzed metaphases (Fig. 3A). The results from the Southern blot analysis and the identification of the proviral integration sites had shown that the predominant leukemic clone from the mice I.3, I.5, and I.9 contained proviral integrations near the genes Irf2, RUNX3, and the histone gene cluster on chromosome 13. Ten metaphase spreads of resistant and control ALL cells were analyzed by FISH with a probe matching human Bcr sequences. The predominant leukemic clone in mouse I.3 contained three proviral integrations that located to the telomeric part of chromosome 4 (RUNX3 locus), more centromeric on chromosome 8 (Irf2 locus) and also centromeric on chromosome 13 (histone gene locus) (Fig. 3B). Furthermore, the probe hybridized to the endogenous mouse Bcr locus on both alleles on chromosome 10. Interestingly, whereas the hybridization signals on chromosomes 8 and 13 were monoallelic in all metaphase spreads, we found biallelic hybridization in some metaphases on chromosome 4 (SI Fig. 6A), suggesting that a further amplification of the integrated provirus and loss of heterozygosity of the wild-type RUNX3 allele had taken place in some subclones. This finding strengthened the case for a functional role of the proviral integration at the RUNX3 locus for imatinib resistance.
Fig. 3.
M-FISH analysis of resistant ALL cells shows a normal male murine karyotype (2n = 40). FISH analysis with a human Bcr probe on the same cells confirms the loci of proviral integration as determined by LM-PCR cloning of proviral flanking regions. (A) M-FISH analysis was performed on chromosomal metaphase spreads of an imatinib-resistant ALL clone. Leukemic clones from three different resistant mice were analyzed with identical results. Chromosome numbers are indicated below each chromosome. (B) To locate the proviral integrations of the MIG-Bcr-Ablp185 vector, chromosome metaphase spreads of resistant ALL cells were analyzed by FISH employing a human Bcr-Probe. The red and the white asterisks highlight FISH signals from proviral integrations and from the endogenous murine Bcr-locus on chromosome 10, respectively. On the right side of the graph the proviral integration loci as determined by LM-PCR are shown for comparison.
Increased RUNX3 and RUNX1 Expression Protects Bcr-Abl-Transformed Cells from Imatinib-Mediated Apoptosis.
To determine the effects of the proviral integration near the RUNX3 promoter, we analyzed RUNX3 expression in leukemic cells with a proviral integration at the RUNX3 locus on chromosome 4. In line with a previous report (16), we could detect increased RUNX3 mRNA and protein expression by Northern blotting, real-time PCR, and Western blotting in these cells compared with controls (SI Fig. 6 B–D). Direct sequencing of the RUNX3 cDNA amplified from two leukemic samples with a proviral integration near the RUNX3 locus did not reveal any mutations in the RUNX3 sequence (data not shown).
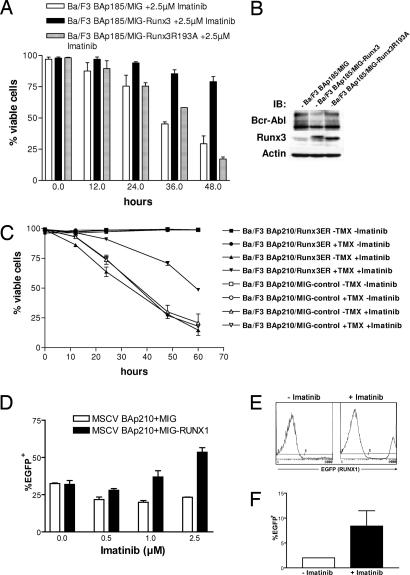
To study the effects of increased RUNX expression on the response to imatinib treatment, we compared the effects of RUNX3 expression on survival of Ba/F3-Bcr-Abl cells treated with imatinib by measuring cell viability by propidium iodide (PI) exclusion. As a control, we used Ba/F3-Bcr-Abl cells infected either with empty vector or with a murine RUNX3R193A mutant (corresponding to human RUNX1R174A), which has a reduced DNA-binding capacity (17). Expression of RUNX3wt cDNA significantly decreased imatinib-mediated apoptosis compared with the empty vector control (Fig. 4A). The RUNX3R193A mutant, although equally well expressed, was unable to protect the cells from imatinib effects, implicating that RUNX3 DNA-binding activity is required for the observed effect (Fig. 4 A and B).
Fig. 4.
Exogenous expression of RUNX3 or RUNX1 protects Bcr-Abl-transformed cells from imatinib-induced apoptosis. (A) Imatinib response of Bcr-Abl-transformed Ba/F3 cells coexpressing RUNX3wt or a RUNX3R193A mutant deficient in DNA binding. Dead cells were determined by PI exclusion. All experiments were done in triplicate and repeated three times with identical results. (B) Western blot analysis of the cells from A. The blot was probed with the indicated antibodies. (C) Effect of conditionally active RUNX3ER on the imatinib response of Bcr-Abl-transformed Ba/F3 cells. TAM (200 nM) was added 48 h before exposition to 2.5 μM imatinib. Dead cells were measured by flow cytometry at the indicated time points after PI staining. All cells were plated in triplicate and analyzed in three independent experiments. (D) Analysis of primary BM cells coinfected with Bcr-Abl (EGFP−) and RUNX1 or empty vector control (EGFP+) after plating in methylcellulose. One × 104 cells were plated in duplicate in the presence of indicated concentrations of imatinib without growth factors. The bars represent percentage of EGFP+ cells per well after 6 days. (E) EGFP histograms of BM cells from either an untreated (Left) or imatinib-treated (Right) mouse transplanted with RUNX1 (EGFP+) and Bcr-Abl-coinfected cells. (F) Average percentage of EGFP+ cells in the BM of RUNX1/Bcr-Abl-coinfected mice without (n = 1) and with (n = 3) imatinib treatment.
We then established a conditional RUNX3 expression construct by fusing the RUNX3 cDNA to parts of a modified murine estrogen receptor (18). This construct enabled inducible activation of RUNX3 by addition of 4-OH-tamoxifen (TAM) (SI Fig. 7 A and B). Ba/F3-Bcr-Abl cells infected with the RUNX3ER construct were treated with imatinib, and cell viability was measured by PI exclusion in TAM induced and uninduced cells, demonstrating that RUNX3 activation by the addition of TAM led to a significantly higher proportion of cells surviving imatinib treatment (Fig. 4C). Interestingly, a TAM-inducible RUNX1ER-construct analogous to RUNX3ER (SI Fig. 7 C and D) was equally able to rescue Ba/F3-Bcr-Abl cells from imatinib-mediated apoptosis (SI Fig. 8A). The results were further confirmed in two different approaches measuring imatinib-induced apoptosis in Ba/F3-Bcr-Abl cells (SI Fig. 8 B and C). In contrast, RUNX1/3 expression did not have an effect on the response of Bcr-Abl-transformed Ba/F3 cells toward two other conventional chemotherapeutic agents commonly used in the treatment of leukemia, cytarabine and etoposide (SI Fig. 8 D and E and data not shown). RUNX3 expression also did not protect Ba/F3 cells without Bcr-Abl from apoptosis induced by growth factor withdrawal (SI Fig. 8F).
To analyze the effects of increased RUNX gene expression on imatinib response in primary cells, we coinfected murine BM cells with a p210 Bcr-Abl construct without EGFP and either a RUNX1-IRES-EGFP or a IRES-EGFP control construct and plated the cells in duplicates in methylcellulose in the presence of 0–2.5 μM imatinib or transplanted them into lethally irradiated recipient mice with three mice per group. The control/Bcr-Abl and the RUNX1/Bcr-Abl group received 1 mg of imatinib twice daily by oral gavage.
The methylcelluose plates were evaluated after 6 days in culture by flow cytometric analysis of the pooled colonies of each well for EGFP/RUNX1 expression, showing an increased number of RUNX1/EGFP-expressing cells in the presence of imatinib (Fig. 4D). The untreated RUNX1/Bcr-Abl mice died within 14 days, whereas the treated mice survived for >30 days, also displaying an enrichment of RUNX1/EGFP-expressing cells in the BM after 1 month of treatment (Fig. 4 E and F). These results implicated that imatinib treatment selected for primary RUNX1/Bcr-Abl-coexpressing cells in vivo.
Discussion
Disease persistence and drug resistance are central problems in molecular targeted therapy with imatinib (3). The mechanisms of resistance to imatinib have been studied extensively, implicating point mutations in the Bcr-Abl kinase region that prevent imatinib binding, gene amplification, and clonal evolution as the clinically most important causes of resistance development (19, 20). However, even though a range of studies suggested that the cause of persistence resides in the CML stem cell, which seems to be refractory to imatinib treatment, the molecular events contributing to disease persistence are not known so far (5, 6). To identify genes influencing the cellular responses against imatinib, we used a RIM screen in a murine model of imatinib resistance.
Imatinib Resistance Is Mediated by Clonal Leukemic Cells Carrying Recurrent Integration Sites.
By infecting BM cells with a high-titer replication-incompetent retrovirus, we achieved leukemia induction by Bcr-Abl expression and insertional mutagenesis by multiple-copy retroviral integration at the same time. In diseased mice, imatinib treatment led to initial hematologic responses, but all animals subsequently relapsed despite continuing treatment. Further analysis ruled out point mutations in the Abl kinase region or Bcr-Abl amplification as a cause of the resistant phenotype. Proviral integration analysis showed an oligoclonal integration pattern in leukemic cells from resistant mice. The reduction from polyclonal to oligoclonal disease under treatment with imatinib has also been described by Wolff and Ilaria (21), indicating that imatinib treatment eliminated some of the clones contributing to leukemia development. Interestingly, the integration pattern was similar for different mice with resistant ALL, suggesting that the same preexistent imatinib-resistant clone was selected. Thus, RIM emerged as the most likely cause for the reduced imatinib response, reconciling the absence of other known mechanisms of resistance, the rapid resistance development, the very aggressive phenotype seen in the resistant ALL cells, and potentially also the heterogeneous response in the earlier model by Wolff and Ilaria (21).
We were able to assign recurrent integration sites obtained by LM-PCR and FISH to recurrent integration patterns in the Southern blot analysis. Two insertions near the RUNX3 and the Kif13a locus were known CIS registered in the RTCG database (22). We focused the further functional analysis on the RUNX3 gene because the FISH analysis suggested that a duplication of the integration at the RUNX3 locus on the other allele had occurred in some cases, indicating an important functional role for this integration.
RUNX Genes Influence the Cellular Response to Imatinib.
The RUNX3 gene belongs to the core-binding factor (CBF) gene family, representing a small group of heterodimeric transcription factors comprising RUNX1/AML1, RUNX2/AML3, RUNX3/AML2, and CBF-β. Although the three RUNX genes have been shown to have nonredundant roles in murine embryonic development (23–26), defects in early hematopoiesis caused by RUNX1 deficiency could be complemented by overexpression of RUNX3 in an in vitro assay (27). In addition, the replacement of C-terminal RUNX1 sequences by RUNX3 was able to rescue early and definitive hematopoiesis in a murine knockin model, suggesting functional overlap between the two genes (28). Interestingly, RUNX1 expression peaks in early hematopoietic progenitor stem cells (HSCs), and decreased levels of RUNX1 have been shown to reduce the number of HSCs, suggesting that RUNX1 plays a role in HSC homeostasis (29, 30). Whereas RUNX1 is frequently inactivated in myeloid leukemias by translocations generating dominant-negative fusion proteins or point mutations (31, 32), there have been reports on RUNX1 amplification in pediatric B-ALL, implicating that an increased dosage of unmutated RUNX1 may also contribute to leukemogenesis (33, 34).
In our work, overexpression of RUNX3 in a Bcr-Abl-transformed murine pre-B cell line significantly protected the cells from imatinib-induced apoptosis, whereas a RUNX3 mutant unable to bind DNA did not elicit this effect. By conditionally expressing the RUNX transcription family member RUNX1, we found also that increased RUNX1 activity reduced apoptosis in this assay. Furthermore, our experiments suggested that RUNX1 expression also conveys protection from imatinib effects in primary cells in vitro and in vivo. The effect seemed to be specific for imatinib treatment because RUNX1/3 expression did not rescue Bcr-Abl-transformed Ba/F3 cells from apoptosis induced by cytarabine or etoposide. Interestingly, we found that imatinib treatment also seemed to select for cells with increased RUNX1 expression in ALL patients, further supporting a functional role for the gene in the imatinib response in humans (SI Fig. 9).
Results showing that the RUNX genes had to be active for >24 h for efficient protection and that the effect was lost in a RUNX3 mutant unable to bind DNA implicate that a transcriptional target of RUNX1/3 may be responsible for the antiapoptotic effect. The number of potential downstream targets containing RUNX-binding sites in their promoter is large (35). Defining the underlying mechanism mediating the reduced apoptotic response will therefore require further ongoing genetic analyses of the components of the RUNX1/AML1-induced transcriptional cascade important for regulating this biological process.
Our approach demonstrates the utility of RIM for the functional analysis of molecular determinants of therapeutic responses in vivo. The described method may prove to be a valuable tool for the study of disease persistence and therapy resistance in murine model systems, enabling the identification of underlying genetic as well as epigenetic aberrations. Furthermore, we have identified RUNX transcription factor genes as modulators of the cellular response toward imatinib. Our results suggest that targeting RUNX genes or its downstream effectors may help to overcome disease persistence in CML patients.
Materials and Methods
DNA Constructs and Cell Culture.
Cloning details of the constructs we used are available on request. The murine pre-B cell line Ba/F3 was transformed by retroviral infection with either MSCV-p210 or MSCV-p185 vectors. Ba/F3-MSCV-p210/p185 coexpressing MIG, MIG-RUNX3, MIG-RUNX3R193A, MIG-RUNX3ER, or MIG-RUNX1ER were established by coinfection with the respective construct and flow cytometric cell sorting.
Animal Studies and Imatinib Treatment.
Infection and transplantation of BM cells was performed as described previously (36). For the induction of CML-like disease, male mice were treated with 150 mg/kg 5-fluorouracil 4 days before BM collection. Female BALB/c recipient mice were lethally irradiated with 800 rad and transplanted with 1 × 106 infected cells by tail vein injection.
Mice were killed after CML development, and 2 × 106 leukemic cells were serially transplanted to sublethally irradiated recipients by tail vein injection. Imatinib (Novartis Pharma AG, Basel, Switzerland) was given at 50 mg/kg twice daily (bid) by gavage after WBC rose above 50,000 cells per microliter. The imatinib dose was raised to 100 mg/kg bid if mice showed signs of resistance. In some experiments, mice were given up to 200 mg/kg bid. Response was defined as decrease or stabilization of WBC over two measurements, resistance was defined as an increase in WBC and/or clinical deterioration (e.g., poor grooming, ruffled appearance, reduced movements) with evidence of leukemic disease despite imatinib treatment.
ALL disease in control mice was induced by omitting 5-fluorouracil treatment of BM donor mice according to published protocols (37). The control ALL cells were also serially transplanted to secondary mice before in vitro analysis to reproduce the settings of the imatinib-resistant mice.
For the analysis of RUNX1 expression in primary cells, BM cells were harvested from 5-fluorouracil-treated mice and coinfected with MSCV-p210 Bcr-Abl and MIG control or MIG-RUNX1 retroviral supernatant. The infected cells were subsequently plated in methylcellulose or transplanted into lethally irradiated mice.
Flow Cytometry and Western Blot Analysis.
Flow cytometry and Western blot analysis was performed essentially as described before (36). The apoptotic response of imatinib-treated cells was determined by PI staining and subsequent flow cytometric analysis for PI-positive cells.
In Vitro Colony Assays on MS-5 Stromal Feeder Cells and in Methylcellulose.
For in vitro colony assays, leukemic cells derived from spleens of imatinib-resistant or nontreated mice were cocultivated on confluent monolayers of MS-5 feeder cells (38). For in vitro resistance analysis, 2 × 104 cells were plated in 6-well plates in the presence of imatinib concentrations ranging between 0 and 10 μM. The cells were cultured for 3 weeks, dried, stained with May–Giemsa, and colony numbers were counted.
Methylcellulose assays of primary BM cells was performed as described previously (39). Cells (2 × 104, 8–10% EGFP+) were plated without growth factors in Methocult 4230 (StemCell Technologies, Vancouver, BC, Canada).
Southern Blot and PCR Analyses.
Southern blotting was performed according to standard laboratory procedures. Southern blots were hybridized with a 32P-labeled 0.7-kb EGFP or a 0.7-kb Abl cDNA fragment. For mutation analysis of the Abl kinase region, cDNA from leukemic cells derived from resistant mice was amplified with the primers 5′-catctcgctgcggtatgaagggagg-3′ and 5′-ccacctcatctgagatactggattcctgg-3′. All quantitative real-time PCRs were performed in triplicate on a 7700 sequence detector (Applied Biosystems, Weiterstadt, Germany) with SYBR green master mix according to the manufacturer's protocol.
Analysis of Provirus Integration by Extension Primer Tag Selection/Solid-Phase LM PCR.
The identification of proviral flanking sequences was performed according to published methods (12). Briefly, genomic DNA extracted from leukemic cells was digested with TaqI or Sse9I (Roche, Mannheim, Germany). After primer extension, specific fragments were purified by magnetic beads (Dynal, Hamburg, Germany), followed by adapter ligation. The flanking sequence was amplified and analyzed by automatic sequencing. Adapter and primer sequences are available on request. Sequences were deposited in the retrovirus-tagged cancer gene (RTCG) database at http://rtcgd.ncifcrf.gov (22). Sequences derived from endogenous retroviral sequences were excluded. Five cases where sequences did not contain the LTR sequence from the integrated provirus were included in SI Table 2 but not in the RTCG database.
FISH Analysis.
The M-FISH and FISH analyses were performed as described previously (40). For the detection of proviral integrations, a 1.5-kb sequence from the 5′ region of the human Bcr cDNA was used as a probe.
Supplementary Material
Acknowledgments
We thank Nancy Speck, Warren Pear, and Gary Nolan for generously providing reagents. This work was supported by a grant from the Nationales Genomforschungsnetz (funded by Bundesministerium für Bildung und Forschung Grant NGFN2 01GS0447), Dr. Mildred Scheel Stiftung Grant Verbundprojekt MSS 106291, a grant through the Sonderforschungsbereich 684 funded by Deutsche Forschungsgemeinschaft Grant SFB684: 522270 (to J.D.), and a Deutsche Jose Carreras Leukämie Stiftung Fellowship DJCLS2001/NAT-2 (to C. Miething).
Abbreviations
- ALL
acute lymphocytic leukemia
- BM
bone marrow
- CML
chronic myeloid leukemia
- HSC
hematopoietic stem cell
- LM
linker-mediated
- M-FISH
multicolor FISH
- Ph+
Philadelphia chromosome-positive
- PI
propidium iodide
- RIM
retroviral insertional mutagenesis
- RIS
retroviral integration sites
- TAM
4-OH-tamoxifen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: Sequences were deposited in the Retrovirus Tagged Cancer Gene Database, http://rtcgd.ncifcrf.gov (accession codes STIV13_1–STIV13_30).
This article contains supporting information online at www.pnas.org/cgi/content/full/0604716104/DC1.
References
- 1.Goldman JM, Melo JV. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Relling MV, Downing JR. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 3.Deininger M, Buchdunger E, Druker BJ. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 4.Cortes J, O'Brien S, Kantarjian H. Blood. 2004;104:2204–2205. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 5.Holtz MS, Slovak ML, Zhang F, Sawyers CL, Forman SJ, Bhatia R. Blood. 2002;99:3792–3800. doi: 10.1182/blood.v99.10.3792. [DOI] [PubMed] [Google Scholar]
- 6.Graham SM, Jorgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, Holyoake TL. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 7.Nowicki MO, Pawlowski P, Fischer T, Hess G, Pawlowski T, Skorski T. Oncogene. 2003;22:3952–3963. doi: 10.1038/sj.onc.1206620. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson CM, Largaespada DA. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 11.Lu SJ, Man S, Bani MR, Adachi D, Hawley RG, Kerbel RS, Ben-David Y. Cancer Res. 1995;55:1139–1145. [PubMed] [Google Scholar]
- 12.Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H, Williams DA, Ragg S, Hesemann CU, Von Kalle C. Hum Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- 13.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A, Romeyn L. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 14.Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, Van Lohuizen M. Nat Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 15.Vijaya S, Steffen DL, Kozak C, Robinson HL. J Virol. 1987;61:1164–1170. doi: 10.1128/jvi.61.4.1164-1170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart M, Mackay N, Cameron ER, Neil JC. J Virol. 2002;76:4364–4369. doi: 10.1128/JVI.76.9.4364-4369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Yan J, Matheny CJ, Corpora T, Bravo J, Warren AJ, Bushweller JH, Speck NA. J Biol Chem. 2003;278:33088–33096. doi: 10.1074/jbc.M303973200. [DOI] [PubMed] [Google Scholar]
- 18.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 20.Hochhaus A, Kreil S, Corbin AS, La Rosee P, Muller MC, Lahaye T, Hanfstein B, Schoch C, Cross NC, Berger U, et al. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 21.Wolff NC, Ilaria RL. Blood. 2001;98:2808–2816. doi: 10.1182/blood.v98.9.2808. [DOI] [PubMed] [Google Scholar]
- 22.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 24.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 25.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, et al. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 27.Goyama S, Yamaguchi Y, Imai Y, Kawazu M, Nakagawa M, Asai T, Kumano K, Mitani K, Ogawa S, Chiba S, et al. Blood. 2004;104:3558–3564. doi: 10.1182/blood-2004-04-1535. [DOI] [PubMed] [Google Scholar]
- 28.Fukushima-Nakase Y, Naoe Y, Taniuchi I, Hosoi H, Sugimoto T, Okuda T. Blood. 2005;105:4298–4307. doi: 10.1182/blood-2004-08-3372. [DOI] [PubMed] [Google Scholar]
- 29.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Blood. 2004;103:2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 30.Sun W, Downing JR. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, et al. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 33.Mikhail FM, Serry KA, Hatem N, Mourad ZI, Farawela HM, El Kaffash DM, Coignet L, Nucifora G. Leukemia. 2002;16:658–668. doi: 10.1038/sj.leu.2402399. [DOI] [PubMed] [Google Scholar]
- 34.Soulier J, Trakhtenbrot L, Najfeld V, Lipton JM, Mathew S, Avet-Loiseau H, De Braekeleer M, Salem S, Baruchel A, Raimondi SC, et al. Leukemia. 2003;17:1679–1682. doi: 10.1038/sj.leu.2403000. [DOI] [PubMed] [Google Scholar]
- 35.Otto F, Lubbert M, Stock M. J Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- 36.Miething C, Grundler R, Fend F, Hoepfl J, Mugler C, von Schilling C, Morris SW, Peschel C, Duyster J. Oncogene. 2003;22:4642–4647. doi: 10.1038/sj.onc.1206575. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawaguchi Y, Jinnai I, Nagai K, Yagasaki F, Yakata Y, Matsuo T, Kuriyama K, Tomonaga M. Leukemia. 2001;15:590–594. doi: 10.1038/sj.leu.2402068. [DOI] [PubMed] [Google Scholar]
- 39.Miething C, Mugler C, Grundler R, Hoepfl J, Bai RY, Peschel C, Duyster J. Leukemia. 2003;17:1695–1699. doi: 10.1038/sj.leu.2403040. [DOI] [PubMed] [Google Scholar]
- 40.Jentsch I, Geigl J, Klein CA, Speicher MR. Cytogenet Genome Res. 2003;103:84–88. doi: 10.1159/000076294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.