Summary
Recent studies have identified stem cells in brain cancer. However, their relationship to normal CNS progenitors, including dependence on common lineage-restricted pathways, is unclear. We observe expression of the CNS-restricted transcription factor, OLIG2, in human glioma stem and progenitor cells reminiscent of Type C transit amplifying cells in germinal zones of the adult brain. Olig2 function is required for proliferation of neural progenitors and for glioma formation in a genetically relevant murine model. Moreover, we show p21WAF1/CIP1, a tumor suppressor and inhibitor of stem cell proliferation, is directly repressed by OLIG2 in neural progenitors and gliomas. Our findings identify an Olig2-regulated lineage-restricted pathway critical for proliferation of normal and tumorigenic CNS stem cells.
Introduction
Investigation of highly tumorigenic cellular subpopulations in adult primary tumors has revealed fundamental similarities to normal stem and progenitor cells within the same lineage (Garraway and Sellers, 2006). Such findings support the proposition that proliferation and survival of tumor cells may be dependent on the same “lineage-restricted” pathways that govern early phases of organogenesis. However, this concept remains to be functionally tested in a broad range of solid cancers, and potential interactions between such lineage-restricted regulatory factors and classical tumor suppressors are poorly understood.
From a developmental perspective, the regulatory networks controlling stem cell behavior in the vertebrate brain are relatively well characterized. Primary cancers of the brain, therefore, provide a favorable context in which to consider the conserved features of normal and tumorigenic stem cells. The most common form of primary brain cancer is the malignant glioma, “primary glioblastoma” (Astrocytoma, WHO Grade IV) (Louis et al., 2002). These uniformly fatal cancers are characterized by stereotypical combinations of genetic lesions, commonly involving amplification/activation of the epidermal growth factor receptor (EGFR) and loss-of-function mutations in the retinoblastoma (Rb) signaling axis for cell cycle control (Louis et al., 2002; Maher et al., 2001). At a histopathological level, primary glioblastomas consist of a heterogeneous mixture of cell types with some evidence of multi-lineage differentiation, recent work has identified cells with stem-like features in glioblastoma (Bao et al., 2006; Galli et al., 2004; Singh et al., 2004; Vescovi et al., 2006). The varied appearance of these tumors has led to proposals that the “cell-of-origin” is either a developmentally arrested neural stem cell or else a committed progenitor cell that has de-differentiated to a more stem-like state (Kesari and Stiles, 2006).
Two cell populations that promote glioma growth have been identified. Rapidly cycling glioma progenitor cells, which express the proliferation marker Ki67, and generally comprise less than 30 percent of tumor cells. Even less frequent within the tumor is a highly tumorigenic subpopulation of glioma stem cells identifiable by the stem/progenitor cell marker CD133 (Bao et al., 2006; Singh et al., 2004). These two replication competent cell populations invite comparisons to progenitors in the subventricular zone (SVZ) of the normal adult brain (Sanai et al., 2005). In this stem cell niche, relatively quiescent “type B” neural stem cells give rise to rapidly dividing “type C” progenitor cells (Doetsch et al., 1999).
During central nervous system (CNS) development, the bHLH transcriptional repressor proteins Olig1 and Olig2 play essential roles in the lineage specification of progenitor cells into neuronal sub-types (somatic motor neurons and forebrain cholinergic neurons) and oligodendrocytes (Furusho et al., 2006; Hack et al., 2005a; Lu et al., 2002; Mizuguchi et al., 2001; Novitch et al., 2001; Takebayashi et al., 2002; Zhou and Anderson, 2002). At early embryonic stages, one key role of Olig2 is to maintain progenitor cells in a replication-competent state (Lee et al., 2005). The structurally related Olig1 transcription factor is required for maturation of oligodendrocyte progenitors (Arnett et al., 2004; Xin et al., 2005). Although it is co-expressed with Olig2 at early stages, Olig1 function is dispensable for specification of neurons or oligodendrocytes from replication competent progenitor cells (Lu et al., 2002; Zhou and Anderson, 2002).
Several lines of evidence suggest that the activity of Olig2 might provide a mechanistic link between growth of malignant glioma and adult neural stem cells. First, a subpopulation of type B and type C cells in the adult rodent brain express Olig2 (Bachoo et al., 2002; Hack et al., 2005a; Menn et al., 2006). Second, exposure to glioma relevant mitogens such as EGF or PDGF (Jackson et al., 2006) stimulates proliferation of Olig2+ rapidly dividing “type C” transit amplifying cells and glioma-like growths. Lastly, we and others have shown that all malignant gliomas, irrespective of grade, express Olig2 in at least some fraction of the malignant cell population (Azzarelli et al., 2004; Bouvier et al., 2003; Ligon et al., 2004; Lu et al., 2001b; Marie et al., 2001; Ohnishi et al., 2003). Here we show that Olig2 function is required for glioma formation and that a common Olig2-dependent mechanism for cell cycle control regulates growth of normal and malignant neural progenitor cells.
Results
OLIG2 is expressed in human glioma stem/progenitor cells
To determine whether OLIG2 might have a functional role in tumorigenesis we first examined whether OLIG2 is expressed in the specific cell types capable of contributing to tumor growth. We quantitated glioma progenitor cells by their expression of the proliferation marker Ki67 or CD133, which has been proposed as a marker for glioma stem cells (Bao et al., 2006; Singh et al., 2004). Immunohistochemical (IHC) co-localization of OLIG2 and Ki67 within human glioblastoma sections shows expression of OLIG2 within at least 85% of Ki67+ glioma progenitor cells by manual counting (n=3 tumors) (Figure 1A). Quantitative flow cytometry of fresh surgical glioblastoma specimens (n=11) shows that while Ki67+ cells represent a minority of the total OLIG2+ population (~37%), the vast majority (~85%) of Ki67+ glioma progenitor cells are OLIG2+ (Figure. 1A–C). In contrast, analysis of OLIG1 expression showed less correlation with cycling glioma progenitor cells, as only 57% of Ki67+ cells expressed OLIG1 (Figure 1C).
Figure 1. Human glioma progenitor cells and glioma stem cells express OLIG2.
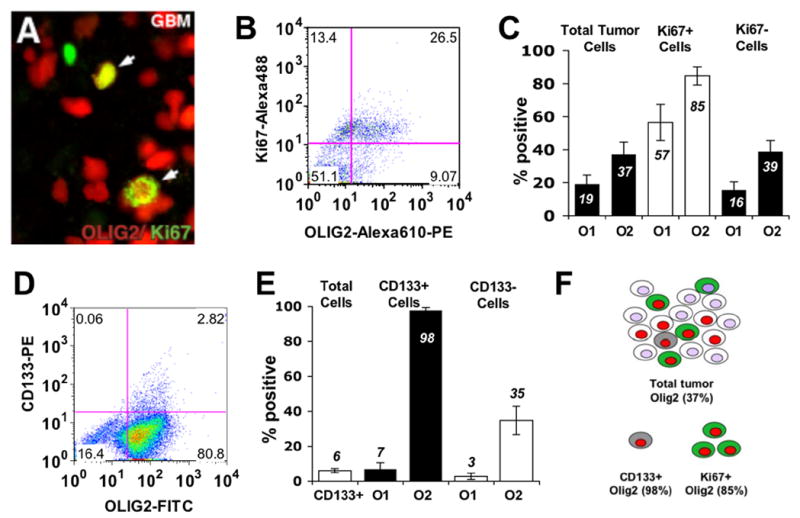
(A) Immunohistochemistry for OLIG2 (red) and the proliferation marker, Ki67 (green) in a human glioblastoma. Arrows denote co-localization of proteins (yellow) in glioma progenitor cells. (B) Scatter plot showing flow cytometric analysis of Ki67 and OLIG2 in a human glioblastoma sample. (C) Summary of flow cytometry results characterizing glioma progenitor cell populations in human glioblastomas (n=11). (D) Scatter plot showing flow cytometric analysis of CD133 and Olig2 in human glioblastoma sample. (E) Summary of flow cytometry results characterizing CD133+ putative glioma stem cell populations in fresh human GBM with respect to OLIG2 (n=11) and OLIG1 (n=4) expression. All p values < 0.001. O1=OLIG1, O2=OLIG2. Error bars indicate s.e.m. (F) Summary of the data shown in panels C and E.
Flow cytometry analysis of fresh human glioblastomas (n=11) shows that the CD133+ cells comprise only 6% of the tumor; however, almost all (98%) of these are OLIG2-positive (Figure 1D, E). In contrast, only 7% of the CD133 positive cells express Olig1 (Figure 1E). Taken together, these findings establish OLIG2 as a marker expressed in two cell types that contribute to tumor growth. However, as indicated in Figure 1C, E and summarized in Figure 1F, OLIG2 expression is not confined to glioma progenitors.
Olig2 function is required for glioma formation
To determine whether Olig genes are required for glioma formation we used a murine model of malignant glioma that emulates common genetic lesions underlying primary glioblastoma in humans (Bachoo et al., 2002). In this model, neural stem/progenitor cells from p16Ink4a/p19Arf null mouse embryos are transformed with a retrovirus containing a constitutively active mutation of epidermal growth factor receptor (EGFRvIII) identified in human glioblastoma multiforme (GBM) (Rasheed et al., 1999). The resulting cells can be cultured as neurospheres capable of glioma formation with 100% penetrance when transplanted into the brains of immunocompromised (SCID) mice (Figure 2A). The tumors thus formed closely model the neuropathological and immunophenotypic features of human anaplastic astrocytoma WHO Grade III (Figure 3 and Supplemental Figure 1) including prominent expression of Olig2, rapid growth, and marked single cell infiltration of normal brain structures. In this model, tumor cells can be positively identified at single cell resolution using human specific antibodies to EGFR (hEGFR).
Figure 2. Olig2 is required for glioma formation from neural stem cells.
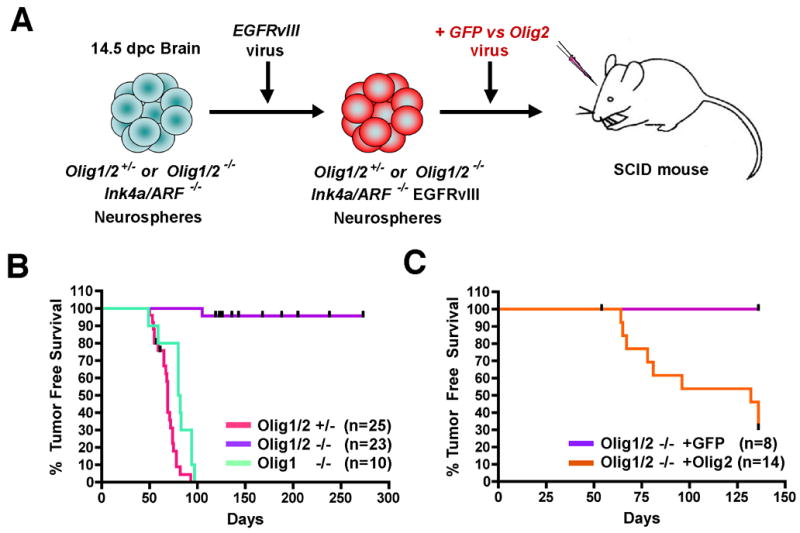
(A) Strategy for murine glioma model utilizing genetically defined neural stem cells as the cell of origin. 2 x 105 Ink4a/Arf−/−EGFRvIII neurosphere cells of the indicated genotypes were transplanted orthotopically into the striatum of SCID mice and animals monitored for symptoms of glioma formation. (B) Kaplan-Meier survival analysis. SCID mice injected with Olig1/2 +/− or Olig1−/− Ink4a/Arf−/−EGFRvIII neurospheres die from tumors with short latency (median survival=69 days and 81 days, respectively) relative to mice injected with Olig1/2-null Ink4a/Arf−/−EGFRvIII neurospheres (96% survival at 273 days). Censored animals in the Olig1/2 −/− group (up ticks) indicate non-cancer deaths. p<0.01 for Olig1/2−/− versus Olig1−/− or Olig1/2+/−. (C) Kaplan-Meier survival analysis showing restoration of tumor phenotype in Olig deficient mice by Olig2 encoding retrovirus (orange) (median TFS = 132 days), but not control GFP virus (purple) (100% survival at 136 days), p<0.01.
Figure 3. Olig2 is required for glioma growth but not neurosphere engraftment in the brain.
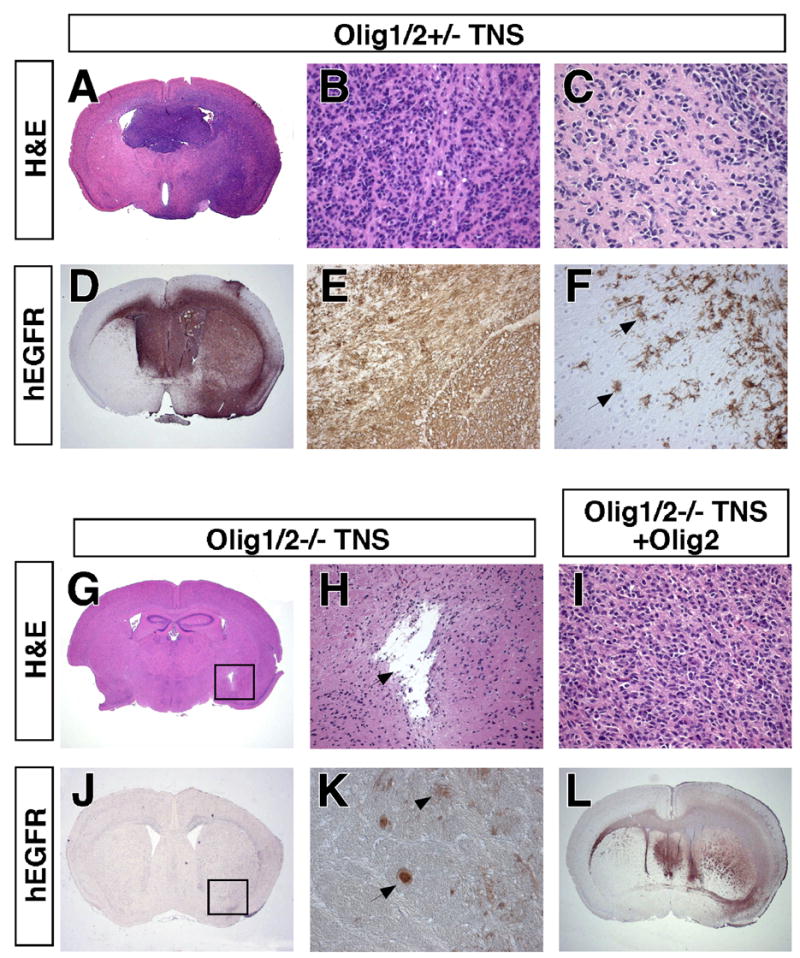
(A, D) H&E staining and hEGFR IHC on coronal section of SCID mouse brain 100 days after injection with Olig1/2+/− Ink4a/Arf−/−EGFRvIII neurospheres demonstrating growth of malignant glioma. (B, E) Tumor histologically resembles human anaplastic astrocytoma W.H.O. grade III and uniformly expresses hEGFR. (C,F) Single cell infiltration of brain parenchyma is seen by H&E and hEGFR IHC. For additional characterization see Supplemental Figure 1. (G, J) Age matched brain injected with Olig1/2−/− TNS. Box indicates injection site. (H) No tumor formation is detected by H&E staining at site of injection (arrow). (K) Rare surviving hEGFR+ Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres have morphological characteristics of astrocytes (arrowhead) and neurons (arrow). (I, L) Subsequent infection of Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres with Olig2 encoding retrovirus rescues formation of gliomas histologically identical to those from Olig1/2+/− Ink4a/Arf−/−EGFRvIII neurospheres.
To remove Olig function in this model we generated neurospheres from compound transgenic mouse embryos that were Ink4a/Arf null and either wild type, heterozygous or null with respect to combined Olig1/2 function (Zhou and Anderson, 2002) (Figure 2A). We chose to initially evaluate deletion of both Olig genes because both are expressed in malignant gliomas, and during development they have partially redundant functions (Lu et al., 2002; Zhou and Anderson, 2002). Analysis of survival cohorts showed that all animals implanted with Olig1/2 wild type (n=12) or heterozygous (n=25) Ink4a/Arf−/−EGFRvIII neurospheres developed neurological symptoms and died within 100 days post injection (dpi) (Figure 2B, Table 1). In striking contrast, 91% of the mice injected with Olig1/2-null Ink4a/Arf−/−EGFRvIII neurospheres survived without symptoms as long as 39 weeks post injection (n = 23) (Figure 2B, Table 1) and did not show evidence of tumor growth in the brain (see below).
Table 1. Survival data summary.
Total animal counts combine data from survival cohorts and timed endpoint cohorts. Median tumor free survival is calculated only using animals in survival cohorts. Survival was 96% at 273 days for animals injected with Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres and 100% at 136 days for those injected with Olig1/2−/− :GFP infected Ink4a/Arf−/−EGFRvIII neurospheres. In all experiments, the presence of tumors was scored as positive for collections of atypical cells greater than 2mm in diameter.
| Ink4a/Arf−/−EGFRvIII Genotype | Total % Developing Tumors | # Tumor Deaths/Total # Animals | Total #Independent Lines | # Animals in Survival Cohorts | Median Tumor Free Survival (days) |
|---|---|---|---|---|---|
| Olig1/2 +/+ | 100 | 12/12 | 2 | 12 | 37 |
| Olig1/2 +/− | 100 | 51/51 | 4 | 25 | 69 |
| Olig1/2 −/− | 9 | 4/45 | 4 | 23 | n/d |
| Olig1/2 −/−:GFP | 0 | 0/8 | 2 | 8 | n/d |
| Olig1/2 −/−:Olig2 | 64 | 9/14 | 2 | 14 | 132 |
| Olig1 +/− | 100 | 7/7 | 1 | 7 | 59 |
| Olig1 −/− | 94 | 16/17 | 2 | 10 | 81 |
These observations are consistent with requirements for unique or overlapping functions of Olig2 and Olig1 in glioma formation. To investigate the specific effects of Olig2, we infected Olig1/2-null Ink4a/Arf−/−EGFRvIII neurospheres with a retrovirus encoding full-length mouse Olig2 (Figure 2A). Exogenous production of Olig2 expression in Olig1/2-null Ink4a/Arf−/−EGFRvIII neurospheres effectively restores the tumorigenic phenotype (Figure 2C, Table 1) albeit with slightly increased latency and reduced penetrance compared to Ink4a/Arf−/−EGFRvIII neurospheres with intact Olig1/2 function. In addition, Ink4a/Arf−/−EGFRvIII neurospheres derived from single knockout, Olig1-null donors demonstrated robust tumor formation with 87% of animals (n=10) developing fatal tumors by 10 weeks post injection (Figure 2B). Together, these findings demonstrate that Olig2 function is specifically required for glioma formation. Further, they rule out the possibility that culturing of Ink4a/Arf−/− EGFRvIII neurospheres from Olig1/2-null animals selects for Olig-independent progenitors that are fundamentally incapable of glioma formation.
Olig2 regulates proliferation and morphology of tumorigenic neurospheres
The intracranial tumors that form in this murine model of malignant glioma can be readily visualized using human specific EGFR antibodies. As indicated in Figure 3A–F and Supplemental Figure 1, these tumors closely recapitulate the neuropathological and immunophenotypic features of human anaplastic astrocytoma WHO Grade III. In contrast, histopathological analysis of Olig1/2 null Ink4a/Arf−/−EGFRvIII neurospheres injected animals at 70 dpi showed that these cells survive and engraft in the brain but do not form gliomas (Figure 3G, H, J, K). They were identified as scattered cells with differentiated astrocytic and neuronal morphologies immediately surrounding the injection site, but with no evidence of mass-like growth or tumor formation in nearly all (91%) cases (Table 1).
In vitro analysis to further investigate the effects of Olig loss-of-function indicates that Olig1/2-null Ink4a/Arf−/−EGFRvIII neurospheres are capable of proliferation, albeit at a significantly lower rate than Olig1/2-heterozygous cells (p <0.001) (Figure 4A). Olig1/2+/− Ink4a/Arf−/−EGFRvIII neurospheres are tri-potent (capable of expressing astrocyte, neuron and oligodendrocyte markers in differentiation conditions), whereas the Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres are bi-potent, with evidence of astrocytic and neuronal differentiation (Supplemental Figure 2), as expected based on known roles of Olig1/2 for oligodendrocyte specification in neurosphere cultures (Zhou and Anderson, 2002). Moreover, the morphology of Olig1/2-null neurospheres is dramatically altered (Figure 4C, D). Relative to their wild type counterparts, the Olig1/2-null cells show increased adhesiveness and migration on the culture dish. Cells lacking Olig function are eventually unable to sustain growth as free floating spheres and proliferate instead as a partial monolayer (Figure 4D). The aberrant adhesion of Olig-null Ink4a/Arf−/−EGFRvIII neurospheres is of interest since monolayer culture is one of the important external cues for differentiation of normal neurosphere cultures (Reynolds and Weiss, 1992).
Figure 4. Olig2 regulates the growth and morphology of Ink4a/Arf−/−EGFRvIII neurospheres.
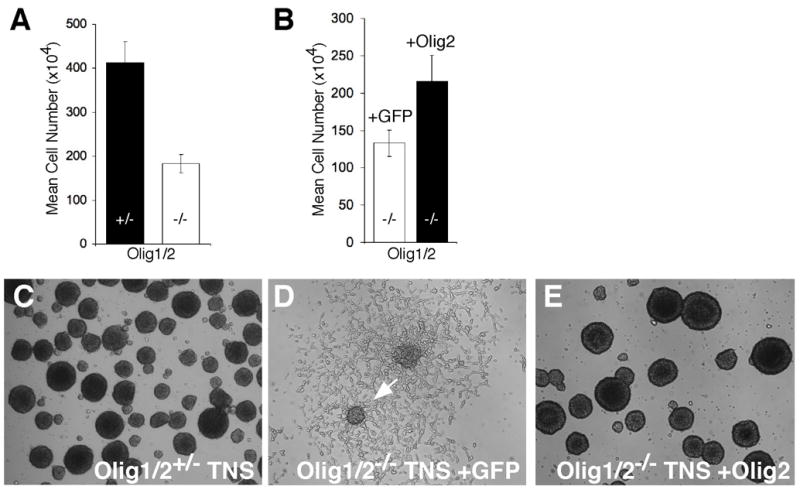
(A) The mean viable cell number of Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres is significantly reduced compared to Olig1/2+/− controls (p<0.001). (B) Partial restoration of Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurosphere growth is seen following infection with Olig2 retrovirus but not with the GFP control (p<0.05). (C) Morphology of Olig1/2+/− Ink4a/Arf−/−EGFRvIII neurospheres (Tumorigenic Neurospheres, TNS) resembles that of normal neurospheres. (D) Olig1/2−/− Ink4a/Arf−/−EGFRvIII neurospheres exhibit marked adherence and migration on underlying substrate (arrow). (E) Infection of Olig1/2 −/− Ink4a/Arf−/−EGFRvIII neurospheres with Olig2 retrovirus completely restores cells to the morphology of wild type Ink4a/Arf−/−EGFRvIII neurospheres. All neurospheres were cultured under growth conditions in the presence of EGF.
Interestingly, restoration of Olig2 activity in Olig-null Ink4a/Arf−/−EGFRvIII neurospheres by retroviral transduction restores wild type growth and morphology in vivo (Figure 3I,L) and in vitro (Figure 4B,E). Therefore, Olig2 regulates both the net proliferation and the histopathological appearance of Ink4a/Arf−/−EGFRvIII neurospheres in the brain. Loss of Olig2 function suppresses tumor formation and promotes a more mature and differentiated morphologic phenotype of the engrafted cells.
Olig2 regulates proliferation and morphology of normal neural stem cells
To better define the relationship between malignant and normal neural stem cells, we examined Olig expression in neural progenitor cells established from normal mice. Proliferating cells in S-phase were marked by a 2 hour pulse of bromodeoxyuridine (BrdU). Subsequent immunohistochemical analysis shows that Olig2 is expressed in the majority (85%) of the cycling cells (Figure 5). In contrast, relatively few of these cells express Olig1 (24%). The uncoupling of Olig1 and Olig2 expression in these normal neurospheres is reminiscent of that found in glioma stem cells of human GBM (Figure 1C).
Figure 5. Olig genes regulate the growth and morphology of normal neurospheres.
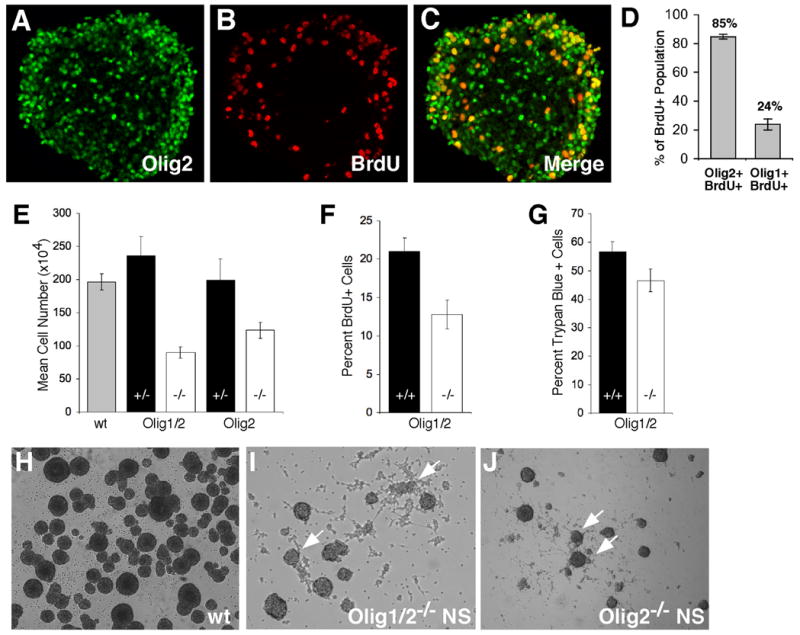
(A–D) Olig2 is highly expressed in undifferentiated and proliferative neurosphere cells. (A) IHC showing widespread Olig2 expression (green) in a cultured neurosphere. (B) IHC for BrdU. (C) Merged image showing strong co-localization of Olig2 and BrdU. (D) Manual quantitation of IHC co-localization results for Olig1 and Olig2 expression (p<0.01). (E) The mean viable cell number at various passages for Olig1/2−/− or Olig2−/− neurospheres was significantly lower than wild type or heterozygous counterparts (p<0.001). (F) The percentage of proliferating BrdU+ cells is significantly lower in the Olig1/2−/−cultures relative to wild-type (p<0.05). (G) Cell death measured by trypan blue staining is not significantly altered in the Olig1/2−/− cultures compared to wild-type. (H) Typical rounded morphology of wild type neurospheres. (I, J) Olig1/2−/− and Olig2−/− neurospheres are reduced in numbers and size. Spheres also show extensive adherence and migration on the substrate (arrows). All neurospheres were derived from ganglionic eminences of wild type or Olig mutant 14.5 dpc embryos and were cultured in the presence of EGF.
Analysis of the growth rates of Olig1/2- or Olig2-null neural progenitors demonstrates a significant reduction (2.2- and 1.6-fold, respectively) in growth for both genotypes relative to heterozygous or wild type controls (Figure 5E). In addition, Olig1/2 null neural stem cells show a marked reduction in the number of rapidly cycling cells in S phase as determined by BrdU labeling (1.6 fold reduction) (Figure 5F). Cell death (as measured by Trypan blue exclusion) is not affected by Olig status (Figure 5G). We observed that single cell suspensions of Olig1/2-null neurospheres are capable of self-renewal (data not shown). However, the morphologic appearance of neurospheres is clearly regulated by Olig status in a fashion similar to that noted for Ink4a/Arf−/−EGFRvIII neurospheres (Figure 5H–J).
A computational screen for direct genetic targets of Olig2
An oppositional relationship between growth and differentiation is controlled by pRB, p53 and their downstream effector proteins (Adams and Kaelin, 1998; Cheng et al., 2000; Kippin et al., 2005; Meletis et al., 2006). Olig2 is known to function as a transcriptional repressor (Mizuguchi et al., 2001; Novitch et al., 2001). Using a strategy summarized in Figure 6, we conducted a genome-wide computational screen for Olig2 repressible genes that are enriched for cis-acting E-box regulatory elements and might function as components of the pRB or p53 signaling networks.
Figure 6. A computational screen for direct genetic targets of Olig2.
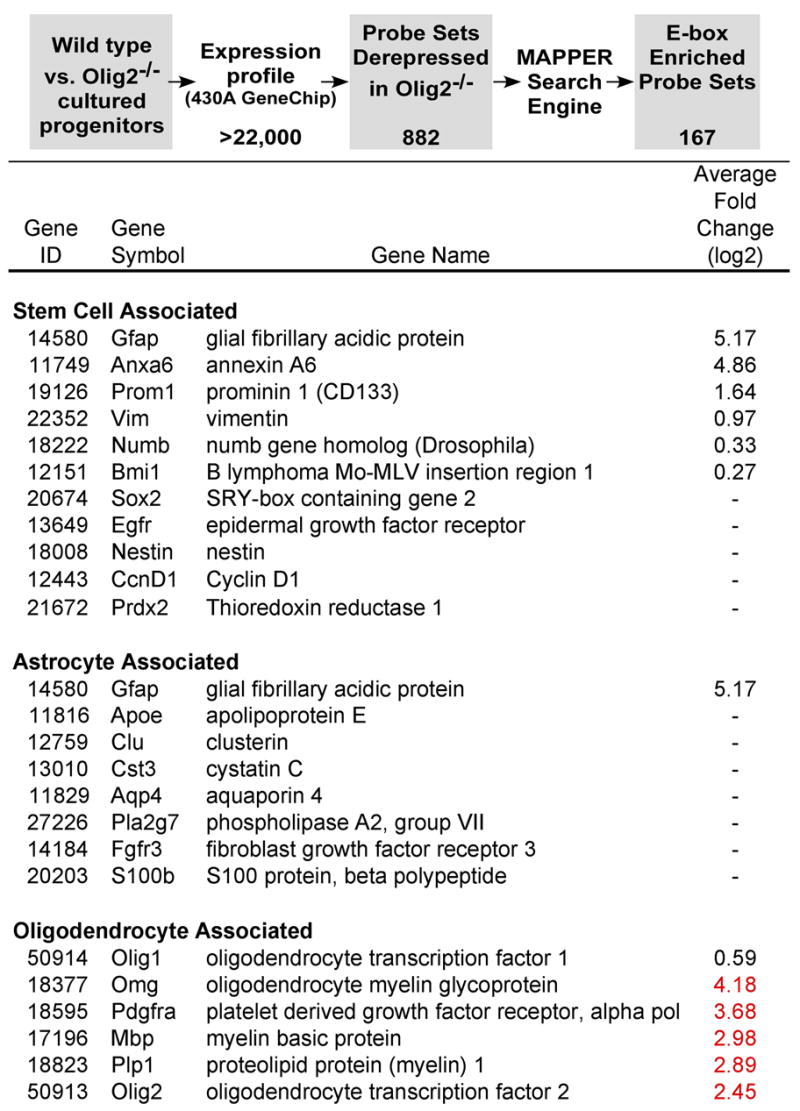
The screen is predicated on the functions of Olig2 protein as a transcriptional repressor (Mizuguchi et al., 2001; Novitch et al., 2001) that binds to canonical E-box regulatory elements of its target genes (Lee et al., 2005). Microarray expression profiling identified 882 probe sets that were derepressed (up-regulated) in Olig2-null neural stem cells relative to their wild type counterparts (see Supplementary Table 1). An additional 1021 probe sets corresponded to mRNAs that were down-regulated in Olig2-null cells relative to wild type (Supplementary Table 2). The list of derepressed probe sets was filtered through the MAPPER tool for binding site analysis to identified 167 genes with promoters enriched for E-box control elements. Analysis of this list of potential Olig2 targets revealed the presence of p21 and three additional p53-inducible genes (see Supplementary Table 3). A sampling of marker genes for stem cells, oligodendrocytes, astrocytes and neurons that are either derepressed (black font), repressed (red font) or unchanged (−) in Olig2-null cells relative to wild type is shown.
Entry-level expression profiling data (Figure 6 and Supplementary Tables 1 and 2) showed, as expected, that Olig2-null neurospheres lacked oligodendroglial lineage gene expression signatures (e.g. PDGFRα, PLP, MBP). Some genes indicative of stem cell character (Ivanova et al., 2002; Livesey et al., 2004; Parker et al., 2005; Ramalho-Santos et al., 2002) were not significantly changed in Olig2-null neurospheres relative to wild type whereas others, including prominin 1/CD133, were actually more prevalent in the Olig2-null cells. Astrocyte specific markers were generally unaffected by Olig2 expression with the exception of Gfap which can mark both fibrous astrocytes in the brain (Bignami et al., 1972) and neural progenitor cells (Doetsch et al., 1999). Canonical neuronal marker genes showed no apparent trend, with some genes being up-regulated and others down-regulated in the absence of Olig2 (Figure 6 and Supplementary Tables 1 and 2).
Filtering the expression profiling data for Olig2-repressible, E-box enriched probe sets (Figure 6) identified four p53-inducible genes (Figure 6 and Supplemental Table 3. Three of these p53-responsive genes are poorly characterized from a functional perspective. However the fourth is p21WAF1/CIP1 (hereafter termed “p21”), which encodes a p53-inducible inhibitor of cell cycle progression. The p21 gene product antagonizes phosphorylation of pRB by inhibiting the activity of cyclin-dependent kinase 2 (Adams and Kaelin, 1998). We focused our subsequent studies on the relationship between Olig2 and p21 because targeted disruption of p21 has been shown to enhance the proliferation of adult neural progenitor cells (Kippin et al., 2005).
The tumor suppressor p21 is a direct target of Olig2 repression in normal stem cells and malignant glioma
The activity of p21 is regulated primarily at the transcriptional level through canonical E-box DNA-binding elements in the upstream promoter/enhancer region of the gene (Gartel and Radhakrishnan, 2005) leading to the hypothesis that Olig2 might directly regulate p21 through such sites (Figure 7A).
Figure 7. p21 is a direct target of Olig2 repression in neural stem/progenitor cells and human glioblastoma.
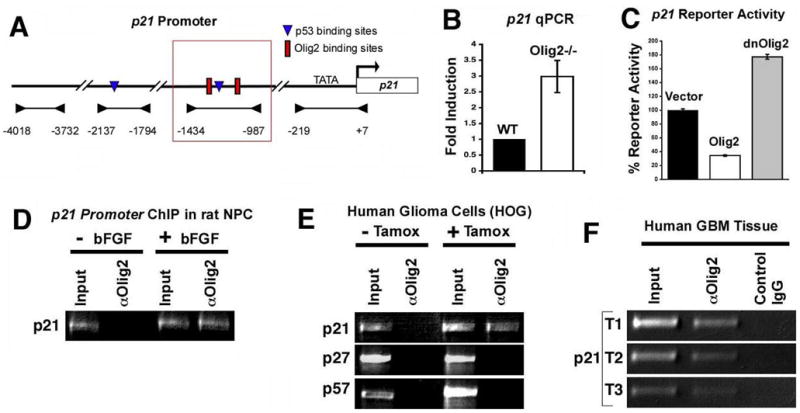
(A) Diagram of P21 promoter region. Arrowheads indicate promoter regions analyzed by PCR in ChIP assays. E-boxes (red) and a p53 binding site (blue) are shown within the region that is bound by Olig2 (red box). (B) Quantitative PCR validates increased expression of p21 mRNA in Olig2-null (n=6) versus wild type (n=3) neurospheres (p<0.036). C) p21 luciferase reporter activity is repressed by Olig2 expression in a human GBM cell line (U87) while a dominant negative Olig2 (dnOlig2) defective in binding DNA yields increased expression (p values< 0.0001). (D) ChIP analysis using anti-Olig2 antibody to detect Olig2 binding to the p21 promoter in E14 rat cortical progenitor cells. Olig2 binding was only detected upon induction of Olig2 expression by FGF. Input is total DNA. (E) ChIP analysis of p21 promoter in a human glioma cell line transfected with a construct encoding a tamoxifen-activated Olig2 fusion protein. No binding to the related cell cycle inhibitors p27 or p57 is detected. Olig2 is detected using anti-ER antibody. (F) ChIP analysis demonstrating OLIG2 is bound to the p21 locus in vivo within freshly isolated human GBM specimens (n=3, T1–T3). Mock ChIP was conducted with normal rabbit IgG as control.
Quantitative PCR was used to validate the original expression profiling data implicating p21 as an Olig2 repressible gene. As indicated in Figure 7B, p21 expression is upregulated nearly three-fold in Olig2-null neurosphere cells relative to their wild type counterparts. Moreover, in a human glioma cell line (U87), ectopic Olig2 suppresses the expression of a p21/luciferase reporter construct (Figure 7B) while a dominant-negative Olig2 defective in binding DNA causes increased expression (p< 0.0001) (Figure 7C).
Chromatin immunoprecipitation (ChIP) assays show that p21 is a direct genetic target of Olig2. For these assays a variety of controls were used to exclude adventitious interactions between Olig2 antibodies and the p21 gene. First, we took advantage of the fact that cortical progenitor cells from E14 rat embryo do not express Olig2 unless they are exposed to FGF or Sonic hedgehog proteins (Gabay et al., 2003; Kessaris et al., 2004). As shown (Figure 7D), ChIP assays detect an FGF-dependent Olig2/p21 interaction in these cortical progenitor cells. Second, we transfected a tamoxifen-activated Olig2 fusion protein into a human glioma cell line (HOG) that does not express endogenous Olig2. Using ChIP assays, we found that this ectopic Olig2 binds to human p21 regulatory elements in the presence of tamoxifen but not in its absence (Figure 7 E). Moreover, the ectopic Olig2 does not interact with upstream regulatory elements of two other p21-like CDK2 inhibitors, p27 and p57, that are regulated primarily by post transcriptional mechanisms (Agrawal et al., 1996; Gartel and Radhakrishnan, 2005; Hengst and Reed, 1996; Pagano et al., 1995) (Figure 7F). Finally, we used ChIP assays to show endogenous Olig2 interacting with p21 regulatory elements in fresh surgical isolates of human malignant gliomas (glioblastoma) but not with control IgG (Figure 7F). Collectively these data show that Olig2 interacts directly with upstream regulatory elements of p21 and functionally can repress its expression.
Discussion
Genetic lesions that underlie malignant glioma in humans include activating mutations in the EGFR and PI-3 kinase pathways and loss-of-function mutations in tumor suppressors such as p53 and p16INK4A/p19ARF (Barber et al., 2004; Barker et al., 1997; Duerr et al., 1998; Rasheed et al., 1997; Rasheed et al., 1999; Samuels et al., 2004; Steck et al., 1988). However, these mutations are not unique to cancers of the CNS. Lineage-restricted pathways that regulate brain tumor behavior may represent more specific therapeutic targets with little potential to affect off target cell types. We show here that a stereotypic combination of mutant EGFR and Ink4a/Arf loss, two genetic lesions commonly found in primary gliomas in humans, can evidently only cause brain tumors in the context of Olig2 function. In both normal and malignant neural stem cells, Olig2 functions as a direct repressor of the cell cycle inhibitor gene, p21. Together, our findings demonstrate an Olig2-regulated lineage-restricted pathway that is critical for proliferation of normal and tumorigenic CNS stem cells.
Olig2 expression is a unifying feature of normal stem cells and malignant glioma
Analogies have been drawn between formation of blood and development of the brain, a common underlying theme being the replication competent stem cell, which occasionally divides asymmetrically to give rise to committed progenitor cells (Anderson, 2001). Recent evidence indicates that the cell-of-origin for some leukemias resembles in many respects normal hematopoietic progenitors (Krivtsov et al., 2006). This observation and other data support the emerging view that critical lineage specific determinates of cancer progenitor cell growth may be found in their normal developmental counterparts (Garraway and Sellers, 2006; Kho et al., 2004).
Although the cell of origin for malignant glioma remains obscure (Kesari and Stiles, 2006; Sanai et al., 2005), recent studies have highlighted similarities with mutipotent neural stem and progenitor cells found in the developing and adult mammalian brain (Jackson et al., 2006). Highly tumorigenic subpopulations of cells in malignant gliomas have been identified that express the hematopoietic stem cell marker CD133. As is true of normal progenitor cells from the germinal zones of the embryonic and adult CNS, these tumor stem cells can be cultured as self-renewing “spheres” with multipotent characteristics (Bao et al., 2006; Galli et al., 2004; Singh et al., 2003; Yuan et al., 2004). In analogy to neural progenitor cells of the adult brain (Sanai et al., 2005), malignant glial tumors might be viewed as populations of (1) developmentally uncommitted stem cells marked by CD133, (2) rapidly cycling, transit amplifying cells marked by Ki67 that contribute significantly to tumor growth and (3) CD133-negative, non cycling progeny cells that make up the bulk of the tumor mass. We show here that all three of these tumor cell populations contain cells that are positive for OLIG2.
Analogies can be drawn between the OLIG2 expression pattern in glioma and in normal adult brains where expression is observed in (1) a subset of “type B” stem cells, (2) in transit amplifying (“type C”) progenitors of the subventricular zone (Hack et al., 2005b; Menn et al., 2006) and (3) in terminally differentiated oligodendrocytes throughout the brain. In preliminary studies (data not shown) we have noted that not all of the CD133-positive cells in our clinical specimens are positive also for Ki67. The mixture of Ki67-positive and negative cancer stem cells might suggest that the OLIG2-positive stem cells represent a transition state between the B and the transit amplifying C cell of the normal brain.
Because it is expressed in three biologically distinct cell cohorts, OLIG2 alone cannot serve as a specific marker for any functional subset of glioma cells. However, the critical role of Olig2 in murine gliomagenesis provides incentive to look for additional markers that could be used in combinatorial fashion to resolve these functional subsets in human cancers. A better resolution of the putative stem cell population marked by CD133 would be of special interest. Additional markers might also be used to better define the relationship between glioma stem cells and the type B and type C cells within the germinal zone of the adult brain.
An Olig2-dependent pathway underlies glioma formation and regulates growth of normal neural stem cells
Both normal and tumorigenic progenitor cells share an Olig2 requirement for growth. At what developmental time point might such effects be relevant? Loss of Olig2 function in the embryonic forebrain ganglionic eminence does not result in global changes to ventricular zone proliferation or cell death. Rather, one observes a failure to develop oligodendrocytes (Lu et al., 2002; Zhou and Anderson, 2002) and subsets of neurons (Furusho et al., 2006), suggesting primarily an early role in cell fate specification. However, sustained expression of Olig2 at adult stages, in a subset of “type B” stem cells and transit amplifying (“type C”) progenitors (Menn et al., 2006) suggests additional roles for this transcription factor in late-stage proliferating progenitor cells found in the adult SVZ. We further speculate that Olig2 could also serve cell cycle regulatory roles in slowly-replicating NG2 progenitor cells that are abundant in the adult cerebral cortex (Ligon et al., 2006).
To better understand the mechanisms critical for growth of glioma we focused on the proliferating subset of cells in human GBM. Approximately 10–15% of cells in these tumors are actively dividing and of these we found that 85% expressed OLIG2. Both OLIG1 and OLIG2 are expressed in human glioma (Lu et al., 2001b; Marie et al., 2001). However, our findings indicate that Olig2 function in particular is necessary for tumorigenesis in a robust murine model of primary glioma that incorporates activation of EGFR and mutation of the Ink4a/Arf locus. Interestingly, similar findings are obtained when Olig2 is conditionally deleted from an independent glioma model based on mutations of the Nf1 and p53 loci (R. Lu and L. Parada, personal communication). Thus, the requirement for Olig2 function may apply to glioma formation in general.
Olig2 functions as a “gateway” gene for brain tumor development
Olig2 fulfills criteria of a lineage-restricted competence factor for brain cancer and in this respect it bears striking analogy to the bHLH-LZ transcription factor, MITF, in melanoma (Du et al., 2004; Garraway and Sellers, 2006). First, Olig2 function is crucial for the development of neural progenitors and progeny cells specifically in the CNS. Second, its expression is deregulated in brain cancer. Finally, its function is required for tumor formation.
However, the notion of Olig2 as a critical lineage-restricted competence factor for brain cancer does not depend on functions as an oncogene per se. Although OLIG2 ectopic expression in the T-cell lineage is associated with T-cell leukemia (Ferrando et al., 2004; Wang et al., 2000), gain-of-function studies in mice suggest it is unlikely to be sufficient for brain cancer formation (Lu et al., 2001a, KL, EH and DHR, unpublished observations). Further, our data argue against critical functions for Olig2 in specification of stem cells. We and others (Zhou and Anderson, 2002) have noted that Olig1/2-null neurospheres are capable of self-renewal and differentiation into two of the three principal neural cell types. Expression of stem cell marker genes is either unaffected in Olig2-null neurospheres compared to controls or actually up-regulated in the Olig2-null cells (Figure 6 and Supplemental Table 1). However, we cannot rule out the possibility that Olig2 co-localization in type B cells of the SVZ (Menn et al., 2006) and with nearly all CD133-positive cells in human glioma (this study) might regulate the transition from quiescent stem cell to transit amplifying progenitor cell in the context of normal development and tumorigenesis, respectively.
Our genetic data establish that Olig function is critical for the glioma promoting effects of mutation of the p16INK4A/p19ARF tumor suppressor locus and resonate with recent reports that age-related competence of such neural progenitors is regulated by p16Ink4a function (Molofsky et al., 2006). Together, these data suggest that possibility that Olig function is critical for several cell cycle regulatory mechanisms underlying growth and survival of both normal and tumorigenic CNS progenitors.
Molecular mechanisms of Olig2 gateway function
The p21 cell cycle inhibitor gene is notable for its important role in maintaining the relatively quiescent state of stem cells for both blood (Cheng et al., 2000) and brain (Kippin et al., 2005). In addition, p53 regulation of neural stem cell growth is also mediated in part through its effects on p21 (Meletis et al., 2006). Targeted disruption of p21 enhances the proliferation rate of neural stem cells in the adult mammalian forebrain (Kippin et al., 2005). Our data show that the p21 locus is subject to direct Olig2 transcriptional repression thus providing a direct link between Olig2 and the cell cycle regulatory apparatus in normal and gliomagenic progenitors. Collectively, these observations raise the interesting possibility that OLIG2 function might be dispensable for tumor formation in a p21 null context. However an alternative, more complex scenario is suggested by our computational screen, which reveals three additional p53-inducible genes that may serve as direct genetic targets for Olig2 (Supplemental Table 3). The p21 gene product does function as a tumor suppressor in knockout mice but its activity as such is rather weak. Moreover, p21 loss-of-function mutations are infrequent in human cancers (Adams and Kaelin, 1998; Gartel and Radhakrishnan, 2005). The laboratory and clinical data suggest that loss of p53 gene function upstream of p21 is a far more effective route to cancer. Thus OLIG2 might suppress multiple p53 targets and/or other non-p53 target genes in its role as a “gateway” gene for brain cancer.
Interestingly, primary glioblastomas are highly resistant to radiation and cytotoxic drugs despite the fact that p53 is generally intact within these tumors. Although there are multiple routes to suppression of p53 function (Besson and Yong, 2001), our data suggest a novel, OLIG2-dependent avenue to p53 pathway antagonism which is potentially active even at early stages of gliomagenesis. One might imagine that increased OLIG2 and decreased p21 expression would predict more aggressive tumor and shorter survival times. However, the literature on p21 and glioma survival is ambivalent. Some studies show that p21 expression is higher in low-grade than in high-grade astrocytomas (Kamiya and Nakazato, 2002) and that the presence of p21 is associated with prolonged survival (Korshunov et al., 2002). Other studies indicate that higher p21 staining is associated strongly with higher histological grade (p < 0.001), a higher rate of proliferation (p = 0.021), and worse survival (Miettinen et al., 2001). Still other studies report that p21 levels have no impact at all on survival in high grade gliomas (Korshunov et al., 2002; Kraus et al., 2001). There are many pitfalls to interpreting the impact of p21 (or any gene, for that matter) on survival in gliomas. However one obvious problem is that previous studies have monitored p21 expression in the whole tumor. It is conceivable that important prognostic insights have been hitherto obscured for this reason. It is possible that useful prognostic insights could be derived from an analysis of p21 expression in CD133/OLIG2-positive glioma cells.
We should also note that, in apparent contradistinction to our findings, it has recently been reported that ectopic OLIG2 expression retards growth of an immortalized glioma cell line, binds to promoter elements of p27 and stimulates expression of p27 mRNA and protein (Tabu et al., 2006). These observations of a linkage between OLIG2 and p27 gene transcription in a glioma cell line stand apart from a wide range of data showing that p27 is regulated primarily by post transcriptional mechanisms in other cell types (Agrawal et al., 1996; Gartel and Radhakrishnan, 2005; Hengst and Reed, 1996; Pagano et al., 1995). The p27 primers used for our ChIP assays amplify in the region identified by Tabu et al and yet we did not detect any direct OLIG2:p27 interactions in primary human tumor samples. Further, p27 expression in our OLIG2 null neurospheres was not altered. Some of the discrepancies between the data reported here and that of Tabu et al may reflect the differential properties of serum-cultured glioma cell lines versus primary neurospheres and fresh human tumors that have been noted in recent studies (Lee et al., 2006).
The Olig2-regulated lineage-restricted pathway: a potential therapeutic target in brain cancer
Brain tumors remain a major cause of cancer-related death despite advances in surgery, imaging and conventional treatment modalities (Louis et al., 2002; Maher et al., 2001). This emphasizes the need to develop novel medical strategies based on a comprehensive understanding of the biological mechanisms underlying gliomagenesis (Rich and Bigner, 2004). Recent findings defining progression from normal progenitor to cancer stem cell in leukemias have suggested that therapeutic targeting of a self-renewal program expressed in an abnormal context may be possible (Krivtsov et al., 2006). Identification of conserved transcriptional regulatory mechanisms within normal and neoplastic hematopoietic progenitor cells has facilitated diagnosis of acute myelogenous leukemia and suggested novel therapeutic avenues (Tenen, 2003). Indeed, recent studies have identified OLIG2 expression as a negative prognostic indicator in human glioblastoma (Liang et al., 2005), and our findings identify this core transcriptional regulator as an important candidate for anti-tumor therapeutics.
In preliminary experiments, stem cells derived from a primary human glioma were transduced with hairpin RNAi expression vectors or dominant negative mutation constructs targeted towards OLIG2. Genetic knockdown of OLIG2 by these means inhibited the ability of these human glioma stem cells to form intracranial tumors in SCID mice (S.K. and C.D.S, unpublished observations). Further experiments with multiple human tumors are needed to comprehensively assess the requirement for OLIG2 function in the settings of the various genetic lesions associated with human glioma such as PTEN, INK4a/ARF and p53. While transcription factors are not generally considered ‘drugable’ targets in anti-tumor therapy, OLIG2 activity could be inhibited at the level of expression, post-translational modification or protein-protein interactions (Kondo and Raff, 2004; Setoguchi and Kondo, 2004; Sun et al., 2003; Walensky et al., 2004). Equally, it should be possible to define downstream factors in the Olig-regulated pathway that could prove useful in this regard.
Experimental Procedures
Mouse procedures
Animal husbandry was performed according to DFCI ACUC approved protocols for all experiments reported. Olig1-cre and Olig1/2 (Lu et al., 2002; Zhou and Anderson, 2002) mouse lines were used on a mixed Swiss and C57Bl6/J background. Ink4a/Arf mice were obtained from the NCI mouse depository on a C57Bl6/J background. Immunocompromised Icr-SCID homozygous mice were obtained from Taconic Inc. (Model #ICRSC-M). For injections, neurospheres were dissociated and resuspended in HBSS at a concentration of 100,000 viable cells/μl. Two μl were injected into the right striatum as previously described (Bachoo et al., 2002). No effects of passage number were noted for all cell lines injected. Animals were placed into survival or time point cohorts and were sacrificed at the onset of neurological symptoms or once moribund.
Neurosphere cultures
Normal neural stem cells (NSC) were isolated from the ganglionic eminences of E14.5 C57Bl6/J embryos, using techniques previously described (Laywell et al., 2002). Cells were cultured in DMEM/F12 (with L-Glutamine, Invitrogen) medium containing glucose (0.3%), penicillin/streptomycin (50μg/mL), Apo-Transferrin (0.1mg/mL), Progesterone (20nM), sodium selenite (30nM), putrescine (60μM), insulin (25μg/mL) and 20ng/mL EGF (all reagents from Sigma). NSC were cultured on plates coated with anti-adhesive (Laywell et al., 2002) and neurospheres were passaged once a week by mechanical dissociation. Live cells were counted using a hemocytometer and trypan blue exclusion and re-plated at equivalent densities (2x104 cells/well) in 6-well plates.
Ink4a/Arf−/−EGFRvIII neurospheres were generated from various genetic backgrounds using identical techniques. Ink4a/Arf−/−EGFRvIII neurospheres were generated from Ink4a/Arf−/− NSCs (on Olig1/2+/− or Olig1/2−/− background). These cells were subsequently infected with EGFRvIII encoding retrovirus at first passage (P1) and treated with puromycin (2μg/mL) 48 hours after infection. For the rescue experiment, Ink4a/Arf−/−EGFRvIII neurospheres of various genetic backgrounds were transduced with Olig2 or GFP encoding retroviruses at P3 and selected with blasticidin (2μg/mL). Expression of Olig2 or GFP was confirmed by immunocytochemistry. Selection drugs were maintained at all times during in vitro growth. Viable Ink4a/Arf−/−EGFRvIII cell numbers at passage were generally higher for all genetic backgrounds than normal neurospheres and no changes in growth properties were noted up to 15 passages including Olig1/2−/− lines, which showed partial growth deficiencies. Ink4a/Arf−/−EGFRvIII neurospheres were injected at least 2 passages after last infection. No difference in growth or tumor formation was noted upon testing of frozen neurosphere lines (1 heterozygous and1 knockout line tested).
Determination of neurosphere growth rates were performed by counting the total viable cell numbers at time of passage over several passages (from 4 to 10 passages) and averaged to calculate mean cell number at passage. To estimate the number of proliferating cells, neurospheres were pulsed with BrdU (1μM) during the last 1 hour of culturing, dissociated, and processed as previously described (Kippin et al., 2005).
For Olig2 and BrdU immunostaining of neurosphere sections, neurospheres were pulsed with 1μM BrdU for two hours prior to fixation, washed in PBS and fixed in 4% PFA. Spheres were embedded in HistoGel (Richard Allan Scientific) subsequently paraffin embedded, and sectioned at 5μm. Rabbit polyclonal anti-Olig2 (1/10,000) and mouse monoclonal anti-BrdU (BD Biosciences; 1/100) were used in a standard immunohistochemistry protocol. All numbers are expressed as mean +/− s.e.m.
Retroviral vectors and virus production
Construction of the retroviral vector encoding the constitutively active mutant of EGFR (EGFRvIII) has been described (Bachoo et al., 2002). Full-length mouse Olig2 was PCR cloned and an XbaI site added via primer sequence to allow cloning into the EcoRI/XbaI sites of the pWZL-BLAST retroviral vector (Morgenstern and Land, 1990). GFP was similarly PCR cloned with primers that added EcoRI and XbaI sites to the 5’ and 3’ ends respectively.
Chromatin immunoprecipitation
Chromatin immunoprecipitations were performed as described by Shang et. al. (Shang et al., 2002) with the following modifications: For HOG cells stably transfected with Olig2-estrogen receptor fusion construct, tamoxifen (0.5μM) was added for four hours prior to the crosslinking step. Rat cortical progenitor cells were isolated from E14 embryos in serum free media as previously described (Williams et al., 1997). Proliferation and Olig2 expression were induced by addition of fibroblast growth factor 2 (30ng/ml). After four days cells were processed for chromatin immunoprecipitation. Fresh surgical isolates of human glioblastoma were snap frozen in liquid nitrogen. Frozen tissue was homogenized and fixed with formaldehyde and processed for chromatin immunoprecipitations.
The primers used for ChIP analysis were as follows: Rat p21 F(forward), AGGTGTCTAGACTCCAGATT, R (reverse) AAAATCAAGGCTTTGCTGG (Olig2 binding region). Human p21 SM19F TCCTGCAGCACGCGAGGTT, SM19R TGTGAGCAGCTGCCGAAGTC (−219 to +7, TATA box region); SM18F GGGCAGGAGGCAAAAGTCCT, SM18R GAAGCCTGTCCTCCCCGAGG (−1434 to −987, Olig-2 binding region); SM17F GAACAGGGTATGTGATCTGC, SM17R TGGTACTGAGCTTCACAATG (−2137 to −1794, near p53 binding sites); and SM29F TCTGTGAAAACATGCCCAGC, SM29R TTGAAACAGGGGACCGTGTC (−4018 to −3732, far upstream region).
Luciferase Assay
The p21 promoter-luciferase reporter plasmid was constructed using 2.4kB fragment from the upstream region containing Olig2 binding sites in the human p21 promoter. The construct was cotransfected with pcDNA3.1/myc/HIS-A, mouse Olig2 expression plasmid or mouse DN-Olig2 expression plasmid using Lipofectamine 2000 (Invitrogen) in U87 cells. Cells were harvested and assayed for luciferase activity 2 days after transfection using a luciferase assay kit (Promega). Luciferase activity for each construct was normalized using the protein concentrations and analyzed using a Berthold Mithras LB940 platereader for luminescence.
Real-time PCR for p21
RNA was extracted from neural progenitor cells using TRIzol reagent (Invitrogen) and further purified using the RNeasy kit (Qiagen). One μg of total RNA was used to generate cDNA, which was then used for the quantitative PCR using pre-made TaqMan gene expression assays (Applied Biosystems) for p21 and rodent GAPDH.
Computational Screen for Olig2 Target Genes
The Affymetrix Mouse 430A2 microarray data of the murine Olig2-null neural stem cells (four separate lines) were analyzed in comparison with wild-type neural stem cells (three separate lines) using the non-parametric wilcoxon ranksum test. A p-value < 0.05 was considered significant. At this level of significance there were 882 probe sets corresponding to genes that were over-expressed in Olig2-null cells relative to their wild type counterparts. An additional 1021 probe sets corresponded to genes that were down-regulated in Olig2-null cells relative to wild type cells. From the long list of 882 over-expressed probe sets, we used the “MAPPER” search engine and database (Marinescu et al., 2005a; Marinescu et al., 2005b) to identify 167 probe sets that contained an E-box motif.
Flow Cytometry
Dissected tissues from 11 independent cases of glioma were placed in DMEM solution with antibiotics on ice until processing. Tissue was washed in artificial cerebrospinal fluid (CSF) and acutely dissociated as described previously (Singh et al., 2004) using a modified tissue dissociation medium containing Collagenase IV (1mg/ml; Worthington), Hyaluronidase (0.67mg/ml; Sigma), DNase I (0.4mg/ml; Worthington), kynurenic acid and N-acetylcysteine (60 ug/ml; Sigma). Cells were washed and used fresh for flow cytometry analysis or resuspended in media for culture. Trypsin was avoided due to higher proteolytic activity and potential cleavage of cell surface proteins (Anderson et al., 2002). Red blood cells were removed using lympholyte-M (Cedarlane).
Fluorescent activated cytometry was performed on a FACSCalibur machine (BD Biosciences). Antibodies used included: mouse IgG1 anti-CD133 antibody (AC133, unconjugated and PE- and APC-conjugated, Miltenyi Biotech, Auburn, CA), rabbit polyclonal Olig2 and Olig1 antibodies (Arnett et al., 2004), mouse IgG1 and anti-Ki67 (M0722, Dako, Carpinteria, CA). Cells were first stained for surface CD133 (antibody dilution 1:100) for 30 minutes at 4°C then washed (PBS with 1% human serum), fixed, and permeabilized to allow access to intracellular antigens (Cytofix/Cytoperm solution; BD Biosciences). Cells were then further incubated with antibody for intracellular antigens (dilution 1:100) for 30–90 minutes at 4°C and washed. Cells were incubated with secondary antibodies (anti-rabbit Alexa488 or Alexa610-PE, and anti-mouse IgG Cy5 or Alexa488, Molecular Probes) for 30 minutes at 4°C and washed twice before FACS analysis. Single positives and isotype stains were used as controls for compensation and gating thresholds. Data was analyzed using FLOWJO software (v6.2.1; Tree Star, Inc).
Histology Analysis, Immunohistochemistry, and in situ Hybridization
Histological screening of tumors was performed by serial analysis of H&E coronal sections taken at approximately 2mm intervals along the entire brain. Tumors were scored as present based on identification of a collection of atypical cells greater than 2mm in diameter, although generally tumors at 2 months exceeded 1.5cm in size and infiltrated the entire right hemisphere. Selected animals that lacked histologic evidence of tumors were also more stringently screened for tumor growth by IHC for hEGFR.
Immunohistochemistry was performed according to standard protocols (Ligon et al., 2004). A new monoclonal antibody to Olig2 was developed by standard methods. Polyclonal antibodies to Olig1 and Olig2 were previously described (Arnett et al., 2004). Other antibodies were Ki67 (rabbit polyclonal, Novocastra, Ki67p), p21 (mouse monoclonal, SantaCruz F5), NeuN (Chemicon), TuJ1 (Sigma), hEGFR (Dako), Nestin (BD Biosciences), GFAP (Sigma) and BrdU (Chemicon). In situ hybridization was performed on human tumors as previously described (Lu et al., 2001b) using full length cDNA encoding human CD133 (obtained from OpenBiosystems Inc.).
Collection and use of fresh and discarded human tumor tissue was approved through Brigham and Women’s Hospital (BWH) Institutional Review Board. After frozen section diagnosis of glioma by the attending neuropathologist, representative tissue samples were dissected. Portions of the tumors were collected in chilled media for the studies described here and other portions were allocated for paraffin embedding for histological diagnosis.
Supplementary Material
Acknowledgments
The authors are grateful to R. Lu and L. Parada (University of Texas Southwestern) for helpful conversations and sharing unpublished data. The authors wish to thank Dong-in Yuk, Emily Learner, and Emily Jerczyk for expert technical assistance. E.H. is the recipient of a Robert K. Olendski American Brain Tumor Association fellowship. S.K. is supported by a Sontag Foundation Distinguished Scientist Award. R.A.D. is an American Cancer Society Research Professor and an Ellison Medical Foundation Senior Scholar and is supported by the Belfer Foundation for Innovative Cancer Science. D.J.A. is a HHMI Investigator and D.H.R. is recipient of a James S. McDonnell Research Award. S.M is the recipient of an NRSA fellowship NS05563 from NINDS. This work was supported by grants from the NIH to K.L.L. (K08NS047213), R.A.D. (P01 CA95616), C.D.S. (PO1NS047572) and D.H.R. (R01NS40511), and by a grant from the Goldhirsh Foundation, Boston, MA to C.D.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Kaelin WG., Jr Negative control elements of the cell cycle in human tumors. Curr Opin Cell Biol. 1998;10:791–797. doi: 10.1016/s0955-0674(98)80123-3. [DOI] [PubMed] [Google Scholar]
- Agrawal D, Hauser P, McPherson F, Dong F, Garcia A, Pledger WJ. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol Cell Biol. 1996;16:4327–4336. doi: 10.1128/mcb.16.8.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron. 2001;30:19–35. doi: 10.1016/s0896-6273(01)00260-4. [DOI] [PubMed] [Google Scholar]
- Anderson RC, Elder JB, Brown MD, Mandigo CE, Parsa AT, Kim PD, Senatus P, Anderson DE, Bruce JN. Changes in the immunologic phenotype of human malignant glioma cells after passaging in vitro. Clin Immunol. 2002;102:84–95. doi: 10.1006/clim.2001.5152. [DOI] [PubMed] [Google Scholar]
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Azzarelli B, Miravalle L, Vidal R. Immunolocalization of the oligodendrocyte transcription factor 1 (Olig1) in brain tumors. J Neuropathol Exp Neurol. 2004;63:170–179. doi: 10.1093/jnen/63.2.170. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006 doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- Barker FG, Chen P, Furman F, Aldape KD, Edwards MS, Israel MA. P16 deletion and mutation analysis in human brain tumors. J Neurooncol. 1997;31:17–23. doi: 10.1023/a:1005768910871. [DOI] [PubMed] [Google Scholar]
- Besson A, Yong VW. Mitogenic signaling and the relationship to cell cycle regulation in astrocytomas. J Neurooncol. 2001;51:245–264. doi: 10.1023/a:1010657030494. [DOI] [PubMed] [Google Scholar]
- Bignami A, Eng LF, Dahl D, Uyeda CT. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Research. 1972;43:429–435. doi: 10.1016/0006-8993(72)90398-8. [DOI] [PubMed] [Google Scholar]
- Bouvier C, Bartoli C, Aguirre-Cruz L, Virard I, Colin C, Fernandez C, Gouvernet J, Figarella-Branger D. Shared oligodendrocyte lineage gene expression in gliomas and oligodendrocyte progenitor cells. J Neurosurg. 2003;99:344–350. doi: 10.3171/jns.2003.99.2.0344. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Duerr EM, Rollbrocker B, Hayashi Y, Peters N, Meyer-Puttlitz B, Louis DN, Schramm J, Wiestler OD, Parsons R, Eng C, von Deimling A. PTEN mutations in gliomas and glioneuronal tumors. Oncogene. 1998;16:2259–2264. doi: 10.1038/sj.onc.1201756. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Dodge RK, Paietta E, Larson RA, Wiernik PH, Rowe JM, Caligiuri MA, Bloomfield CD, Look AT. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet. 2004;363:535–536. doi: 10.1016/S0140-6736(04)15542-6. [DOI] [PubMed] [Google Scholar]
- Furusho M, Ono K, Takebayashi H, Masahira N, Kagawa T, Ikeda K, Ikenaka K. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293:348–357. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- Hack MA, Saghatelyan A, de Chevigny A, Pfeifer A, Ashery-Padan R, Lledo PM, Gotz M. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005a;8:865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- Hack MA, Saghatelyan A, de Chevigny A, Pfeifer A, Ashery-Padan R, Lledo PM, Gotz M. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci . 2005b doi: 10.1038/nn1479. In Press. [DOI] [PubMed] [Google Scholar]
- Hengst L, Reed SI. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Kamiya M, Nakazato Y. The expression of p73, p21 and MDM2 proteins in gliomas. J Neurooncol. 2002;59:143–149. doi: 10.1023/a:1019633910603. [DOI] [PubMed] [Google Scholar]
- Kesari S, Stiles CD. The bad seed: PDGF receptors link adult neural progenitors to glioma stem cells. Neuron. 2006;51:151–153. doi: 10.1016/j.neuron.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Jamen F, Rubin LL, Richardson WD. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development. 2004;131:1289–1298. doi: 10.1242/dev.01027. [DOI] [PubMed] [Google Scholar]
- Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, Rowitch DH, Kohane IS. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov A, Golanov A, Sycheva R. Immunohistochemical markers for prognosis of oligodendroglial neoplasms. J Neurooncol. 2002;58:237–253. doi: 10.1023/a:1016270101321. [DOI] [PubMed] [Google Scholar]
- Kraus JA, Wenghoefer M, Glesmann N, Mohr S, Beck M, Schmidt MC, Schroder R, Berweiler U, Roggendorf W, Diete S, et al. TP53 gene mutations, nuclear p53 accumulation, expression of Waf/p21, Bcl-2, and CD95 (APO-1/Fas) proteins are not prognostic factors in de novo glioblastoma multiforme. J Neurooncol. 2001;52:263–272. doi: 10.1023/a:1010684203704. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Laywell ED, Kukekov VG, Suslov O, Zheng T, Steindler DA. Production and analysis of neurospheres from acutely dissociated and postmortem CNS specimens. Methods Mol Biol. 2002;198:15–27. doi: 10.1385/1-59259-186-8:015. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Kesari S, Kitada M, Sun T, Arnett HA, Alberta JA, Anderson DJ, Stiles CD, Rowitch DH. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A. 2006;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Young TL, Cepko CL. An analysis of the gene expression program of mammalian neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:1374–1379. doi: 10.1073/pnas.0307014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Pomeroy SL, Cairncross JG. Focus on central nervous system neoplasia. Cancer Cell. 2002;1:125–128. doi: 10.1016/s1535-6108(02)00040-5. [DOI] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001a;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Park JK, Noll E, Chan JA, Alberta J, Yuk D, Alzamora MG, Louis DN, Stiles CD, Rowitch DH, Black PM. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001b;98:10851–10856. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Marie Y, Sanson M, Mokhtari K, Leuraud P, Kujas M, Delattre JY, Poirier J, Zalc B, Hoang-Xuan K. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358:298–300. doi: 10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005a;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005b;33:D91–97. doi: 10.1093/nar/gki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen HE, Paunu N, Rantala I, Kalimo H, Paljarvi L, Helin H, Haapasalo H. Cell cycle regulators (p21, p53, pRb) in oligodendrocytic tumors: a study by novel tumor microarray technique. J Neurooncol. 2001;55:29–37. doi: 10.1023/a:1012961918848. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ohnishi A, Sawa H, Tsuda M, Sawamura Y, Itoh T, Iwasaki Y, Nagashima K. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62:1052–1059. doi: 10.1093/jnen/62.10.1052. [DOI] [PubMed] [Google Scholar]
- Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Parker MA, Anderson JK, Corliss DA, Abraria VE, Sidman RL, Park KI, Teng YD, Cotanche DA, Snyder EY. Expression profile of an operationally-defined neural stem cell clone. Exp Neurol. 2005;194:320–332. doi: 10.1016/j.expneurol.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rasheed BK, Stenzel TT, McLendon RE, Parsons R, Friedman AH, Friedman HS, Bigner DD, Bigner SH. PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer Res. 1997;57:4187–4190. [PubMed] [Google Scholar]
- Rasheed BK, Wiltshire RN, Bigner SH, Bigner DD. Molecular pathogenesis of malignant gliomas. Curr Opin Oncol. 1999;11:162–167. doi: 10.1097/00001622-199905000-00004. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rich JN, Bigner DD. Development of novel targeted therapies in the treatment of malignant glioma. Nat Rev Drug Discov. 2004;3:430–446. doi: 10.1038/nrd1380. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–968. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Steck P, Lee P, Hung MC, Yung W. Expression of an altered epidermal growth factor receptor by human glioblastoma cells. Cancer Res. 1988;48:5433–5439. [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23:9547–9556. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabu K, Ohnishi A, Sunden Y, Suzuki T, Tsuda M, Tanaka S, Sakai T, Nagashima K, Sawa H. A novel function of OLIG2 to suppress human glial tumor cell growth via p27Kip1 transactivation. J Cell Sci. 2006;119:1433–1441. doi: 10.1242/jcs.02854. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jani-Sait SN, Escalon EA, Carroll AJ, de Jong PJ, Kirsch IR, Aplan PD. The t(14;21)(q11.2;q22) chromosomal translocation associated with T-cell acute lymphoblastic leukemia activates the BHLHB1 gene. Proc Natl Acad Sci U S A. 2000;97:3497–3502. doi: 10.1073/pnas.97.7.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Park JK, Alberta JA, Muhlebach SG, Hwang GY, Roberts TM, Stiles CD. A PDGF-regulated immediate early gene response initiates neuronal differentiation in ventricular zone progenitor cells. Neuron. 1997;18:553–562. doi: 10.1016/s0896-6273(00)80297-4. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


