Abstract
Glucocorticoids (GCs) are used to combat inflammatory diseases. Their beneficial effect relies mainly on the inhibition of NF-κB- and/or AP-1-driven proinflammatory gene expression. Previously, we have shown that GCs repress tumor necrosis factor-induced IL-6 gene expression by an NF-κB-dependent nuclear mechanism without changing the DNA-binding capacity of NF-κB or the expression levels of the cytoplasmic inhibitor of NF-κB (IκB-α). In the present work, we investigate the effect of GC repression on different natural and/or recombinant NF-κB-driven reporter gene constructs in the presence of increasing amounts of various coactivator molecules, such as CREB-binding protein (CBP), p300, and SRC-1. We found that GCs maintain their repressive capacities, irrespective of the amount of cofactor present in the cell. Similar results were obtained for the reciprocal transrepression of a GC receptor (GR) element-driven reporter gene by p65. We demonstrate that neither the expression levels of p65 and CBP nor their physical association are affected by activated GR. Using Gal4 chimeras, we show that repression by GCs is specific for p65-mediated transactivation, ruling out competition for limiting nuclear factors as the major underlying mechanism of gene repression. In addition, the transactivation potential of a point-mutated Gal4-p65 variant with a decreased CBP interaction capability is still repressed by GR. Finally, we present evidence that the specificity of GC repression on p65-driven gene expression is codetermined by the TATA box context.
The glucocorticoid (GC) receptor (GR) is a ligand-dependent transcription factor belonging to the superfamily of steroid/thyroid hormone receptors. These receptors and their cognate hormones control various aspects of metabolic homeostasis, embryonic development, and physiological stress. Activated GRs modulate transcription by either directly binding to GR elements (GREs) in promoters of positively regulated genes or indirectly binding by association with other transcription factors, such as NF-κB or AP-1. The latter function is considered to be very important in the battle against inflammatory and immune diseases, in which GCs are used effectively as therapeutic agents (1, 2).
The transcription factor NF-κB plays a critical role in immune homeostasis, cell growth, and survival. A persistent activation of this factor compromises health and is associated with inflammatory and neoplastic diseases as well as with viral infection. The mammalian NF-κB/Rel family of proteins consists presently of five members, namely, Rel (c-Rel), p65 (Rel A), Rel B, p50 (NFKB1), and p52 (NFKB2). In general, the designation NF-κB refers to the most frequently occurring heterodimeric complex between the p50 and p65 subunits. NF-κB activation may be induced by different signals, such as viral infection, the proinflammatory cytokines tumor necrosis factor (TNF) and IL-1, phorbol esters, UV irradiation, and bacterial lipopolysaccharides. These signals lead to phosphorylation and degradation of the inhibitor of NF-κB (IκB-α), after which NF-κB can migrate to the nucleus (3). Depending on the activating stimulus, NF-κB itself is subject to posttranslational modifications that can enhance transcriptional activation of NF-κB-dependent genes (4). Cofactors have been described to bridge the gap between transcription factors and components of the basal transcription machinery. Recent studies showed that the coactivator CREB-binding protein (CBP) and its homologue p300, which contain histone acetyltransferase (HAT) activity, can enhance the transcriptional activity of p65. Moreover, CBP was found to associate with p65 via two interaction sites, one of them depending on a phosphorylated Ser residue at position 276 (5). Similarly, a potentiating effect on NF-κB- and AP-1-driven transactivations was reported for SRC-1, another HAT activity-containing coactivator that was discovered initially as a nuclear receptor coactivator (6, 7). These observations permitted a hypothetical explanation for GC repression of AP-1- and/or NF-κB-dependent gene expression: competition between GR and the driving transcription factors for a limited amount of coactivator CBP/p300 or SRC-1 in the cell would account for the repressive action by GCs. (8–10).
The potent acidic transactivation domain of p65 is also functionally able to recruit components of the basal transcription machinery (11, 12). Eukaryotic transcription is initiated by binding of TFIID to the TATA box. The multisubunit TFIID complex consists of TBP and at least eight additional proteins termed TAFs. The TBP-promoter complex is recognized subsequently by TFIIB, after which the stepwise association of additional basal transcription factors and RNA polymerase II build up a complete and functional transcription initiation complex (13, 14).
In the present paper, we report on our investigation to determine to what extent CBP and other coactivators might be responsible to relieve GC repression of various NF-κB-driven genes. Our findings show, in contrast with the previously proposed model, that competition between GR and NF-κB for limiting amounts of CBP, p300, or SRC-1 does not constitute a prevalent and universal mechanism for specific gene repression by GR. Instead, we propose a model in which GCs repress NF-κB-driven genes by interfering with the mechanistic interaction of p65 with the basal transcription machinery.
Materials and Methods
Plasmids.
The full-size IL-6 promoter reporter gene construct p1168hu.IL6P-luc+ and the recombinant plasmid p(IL6κB)350hu.IL6P-luc+ have been described (15, 16). p(GRE)250hu.IL6P-luc+ was obtained by replacing the κB motifs in p(IL6κB)350hu.IL6P-luc+ with a linker region, containing two consensus GRE (underlined) sites flanked by a BglII and PstI restriction site. The following oligonucleotides were annealed: AGATCTCTCTGCTGTACAGGATGTTCTAGCGGATCCTGCTGTACAGGATGTTCTAGCTAC-CTGCAG and TCTAGAGAGACGACATGTCCTACAAGA-TCGCCTAGGACGACATGTCCTACAAGATCGATG-GACGTC. pNFκB-Luc was purchased from Stratagene Cloning Systems. pELAM-luc containing the E selectin promoter was a kind gift from D. Goeddel (Tularik, San Francisco). The ICAM promoter, kindly provided by K. Roebuck (Rush Presbyterian St. Luke's Medical Center, Chicago) was used to construct pICAM-luc as described (17). pCMV-CBP, pRSV and pRSVp65, pSVhGRα, and PCR3.1-SRC1a were kind gifts from R. Eckner (Institute for Molecular Biology, Zurich), G. Manfioletti (University of Trieste, Italy), W. Rombauts (University of Leuven, Belgium), and B. W. O'Malley (Baylor College of Medicine, Houston), respectively. pcDNA3, used as an empty control vector for the CBP-expressing plasmid, was purchased from Invitrogen. pGal4, pGal4-p65, and pGal4-VP16 were generously provided by M. L. Schmitz (German Cancer Research Center, Heidelberg) and, together with p(Gal)2-50hu.IL6P-luc+, have been described (15, 18).
Cytokines and Reagents.
Dexamethasone (DEX) was purchased from Sigma. The origin and activity of TNF as well as the preparation of luc reagent have been described (19). Luc assays were carried out according to the manufacturer's instructions (Promega). Normalization of luc activity, expressed as arbitrary light units, was performed by measurement of β-galactosidase (β-Gal) levels in a chemiluminescent reporter assay Galacto-Light kit (Tropix, Bedford, MA) or according to Bradford's protein determination (20). Light emission was measured in a luminescence microplate counter (Topcount, Packard).
Transfections.
HEK293T cells were transiently transfected by the calcium phosphate coprecipitation protocol (21). Briefly, 105 actively growing cells were seeded in 24-well plates 24 h before transfection, and 400, 600, or 700 ng of total DNA was transfected. At 16 h after transfection, the medium was replaced with fresh medium containing 10−6 M DEX (where appropriate) for another 24 h. Cells were lysed with lysis buffer (Tropix), and samples were assayed for their protein or β-Gal content and luc activity. L929sA cells and TC10 mouse endothelial cells were transiently transfected with diethylaminoethyl-dextrane or Lipofectamine (Life Technologies, Paisley, U.K.), respectively, as described (15).
Coimmunoprecipitation Analysis.
The conditions were essentially as described (22). Cells (n = 3 × 105) were seeded in six-well plates on day −1. After transfection of various plasmids, the cells were, where appropriate, induced with DEX for 24 h. After lysis, protein content was determined (20), and corresponding β-Gal levels were measured to determine transfection efficiencies. Luc assays were performed to analyze the regulation of promoter activities. Protein (150 μg) was used in each setup, and the lysate volume was adjusted to 800 μl with buffer as described (22). Where appropriate, 5 μg/ml anti-p65 antibody (Santa Cruz Biotechnology) or an irrelevant antibody as a control (anti-X-press; Invitrogen) was added. After immunoprecipitation, samples were loaded on a 6% denaturing polyacrylamide gel, and blotted filters were subjected to Western blot analysis with an anti-CBP antibody and, after stripping, with an anti-p65 antibody (both from Santa Cruz Biotechnology).
Western Blot Analysis.
Lysates of three or four 24-well plates from the same transfection setup were pooled; 20 μl was supplemented with Laemmli buffer, loaded onto a reducing SDS/polyacrylamide gel (6% to reveal CBP; 10% to reveal p65), and blotted onto nitrocellulose membranes. Equal protein loading was verified by staining the blots with Amido Black 10B (Sigma). The membranes were incubated with primary anti-p65 or anti-CBP antibodies; Western analysis was carried out according to the guidelines of Santa Cruz Biotechnology.
Site-Directed Mutagenesis.
Gal4-p65 was mutated to Gal4-p65S276C with a transformer site-directed mutagenesis kit (CLONTECH). The mutator oligonucleotide CGGTGCCAAGGCCTCCGCAG was used (altered nucleotides are underlined). Mutant clones were screened for the presence of a newly created StuI restriction site and verified by sequence analysis.
Results
GC Repression of the IL-6 Promoter Occurs Independently of Coactivator-Mediated Transactivation.
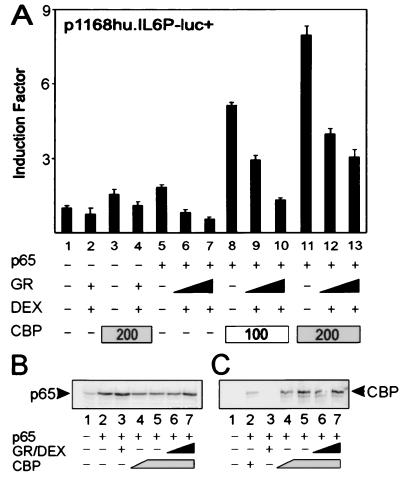
For a number of κB- and AP-1-driven gene promoters, coexpression of CBP was shown to augment p65- or c-Jun-activated gene transcription (22, 23) and to relieve repression by GR (8, 9). Correspondingly, we recently established that CBP or p300 also cooperates with p65 in a synergistic way for transcriptional induction of the IL-6 promoter (17). In the present study, we analyzed whether increased amounts of activated GR are still able to repress p65-activated reporter genes in the presence of increasing amounts of active CBP. Fig. 1A shows that, on transient overexpression of nonsaturating amounts of p65 in HEK293T cells, the cotransfected full-size IL-6 promoter is induced (lane 5 vs. lane 1), and repressed when a GR expression vector and the concomitant ligand DEX are included (lanes 6 and 7). A weak stimulation of the reporter is observed when CBP is expressed without p65 (lane 3), probably because of stimulation of the IL-6 promoter by endogenous and constitutively DNA-bound transcription factors in the cell, such as AP-1 (17). Similarly, GR is capable of down-modulating CBP-activated IL-6 promoter-dependent gene expression (lane 4), which probably results from an inhibitory effect of GR on this endogenous transcription factor activity. Coexpression of CBP and p65 stimulates the IL-6 promoter synergistically (lanes 8 and 11). However, a stepwise increase in the amount of activated GR correspondingly lowers the level of transactivation, mediated by the synergistic activity of p65 and CBP (lanes 9–10 and 12–13), and results in the same relative level of repression as that without cotransfected CBP, i.e., from 60 to 70% with 200 ng of GR expression plasmid. Similar results were obtained when p300 was coexpressed instead of CBP or when other NF-κB-driven gene promoters were tested (i.e., IL-8, ICAM, and E selectin; data not shown).
Figure 1.
(A) HEK293T cells were transiently transfected with 80 ng of p1168hu.IL6P-luc+ and with pRSV-p65 (20 ng), pSVhGRα (100 to 200 ng), pCMV-CBP (as indicated), and/or pcDNA3 or pRSV, keeping the total amount of DNA constant at 600 ng per 24-well plate. Cell lysates were assayed for luc activities and normalized for protein content. Promoter activities are expressed as “induction factor,” i.e., the ratio of expression levels recorded either under induced and noninduced conditions or under transfected and mock-transfected conditions. Assays were performed in triplicate, and results are representative of at least four independent transfection experiments. (B) Western blot analysis of lysates of transfected cells corresponding to 25 μg of protein content (the luc activities had a similar regulation on a full IL-6 promoter-dependent reporter gene as shown in A). Samples were transfected with 100 ng (lane 4) or 200 ng (lanes 5–7) of pCMV-CBP or 25 ng (lane 6) or 100 ng (lanes 3 and 7) pSVhGRα. The membrane was probed with an anti-p65 rabbit polyclonal antibody. (C) Samples were transfected as described for B (except for lane 2 where a setup with 50 ng of pCMV-CBP was included), blotted, and probed with an anti-CBP rabbit polyclonal antibody.
To exclude that the stimulatory effect of CBP on p65-mediated transactivation was caused by up-regulation of p65 expression, we checked the relevant protein levels in lysates from a similar experiment. Fig. 1B shows that overexpression of CBP does not lead to higher amounts of expressed p65, whereas additional overexpression of GR does not result in lower protein levels of p65 (Fig. 1B) or of CBP (Fig. 1C).
GC Repression Acts Specifically on κB-Driven Gene Expression, Irrespective of CBP or SRC-1 Levels in the Cell.
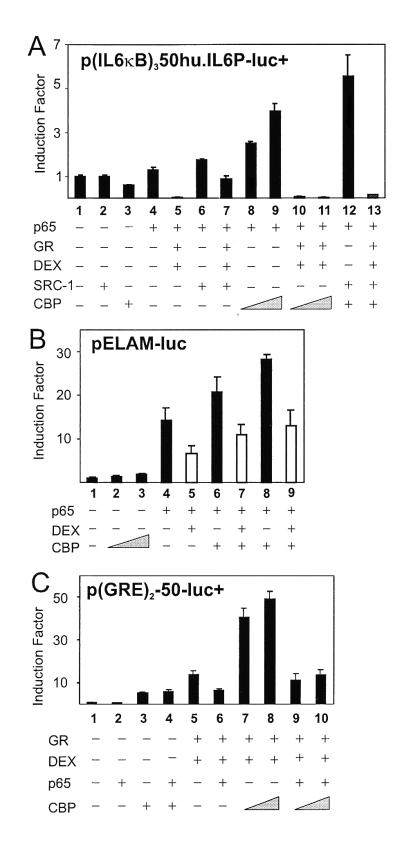
To rule out interference of transcription factors other than NF-κB in the full-size IL-6 promoter, we examined whether similar results on transrepression could be obtained with p(IL6κB)350hu.IL6P-luc+ (Fig. 2A). Transfection of nonsaturating amounts of p65 alone stimulated slightly (lane 4) but synergistically up-regulated promoter activity with increasing amounts of CBP (lanes 8 and 9). Activated GR effectively repressed p65 transactivation (lane 5) as well as the synergistic activation by CBP and p65 (lanes 10 and 11). Moreover, SRC-1 has also been reported to function as a coactivator for NF-κB-mediated gene expression (24). In this respect, Fig. 2A shows that SRC-1 enhances p65-mediated gene expression (lane 6) as well as the synergistic cooperation between CBP and p65 (lane 12), whereas this activation state is reversed totally by GCs (lane 13). Testing also other κB-dependent reporter genes, such as pELAM-luc (Fig. 2B) and pICAM-luc (data not included) in the endothelial cell line TC10 and using the endogenous GR, we confirmed that repression does not result from oversaturating and squelching concentrations of GR and, furthermore, is independent of cell type and promoter.
Figure 2.
(A) HEK293T cells were transiently transfected with 80 ng of p(IL6κB)350hu.IL6P luc+ and, where indicated, with pRSV-p65 (10 ng), pSVhGRα (50 to 100 ng), pCMV-CBP (200 to 300 ng), PCR3.1-SRC-1a (100 ng), and/or pRSV or pcDNA3. (B) TC10 cells were transiently transfected with 100 ng of pELAM-luc and, where indicated, with pRSV-p65 (50 ng), pCMV-CBP (100 to 200 ng), and/or pRSV or pcDNA3. White bars indicate the repression levels when endogenous GR was activated by DEX. (C) HEK293T cells were transiently transfected with 80 ng of p(GRE)2-50-luc+, pSVhGRα (100 ng), pRSV-p65 (100 ng), pCMV-CBP (100 to 300 ng), and/or the corresponding empty control plasmids. Total DNA content was kept at 600 ng. Cell lysates were assayed and plotted as described for Fig. 1.
Reciprocal Transrepression by p65 of GRE-Dependent Gene Expression Occurs Independently of Coactivator-Mediated Stimulation.
The influence of overexpressed CBP or p300 on transrepression exerted by p65 on GRE-mediated gene induction was investigated (Fig. 2C). Cotransfection of CBP enhances basal promoter activity of p(GRE)250hu.IL6P-luc+ (lane 3) because of cooperation of CBP with endogenous GR activated by corticosteroids in the serum of the medium. The GRE-dependent reporter is activated by GR (lane 5) and repressed by coexpressed p65 by over 50% (lane 6). With CBP, the GR-mediated transactivation is potentiated further (lanes 7 and 8), but additional cotransfection of p65 substantially transrepresses the strong synergism of GR and CBP (lanes 9 and 10). Similar results were obtained when p300 was used instead of CBP (data not shown).
GC Repression in the Gal4 One-Hybrid System Is Independent of Coexpressed CBP.
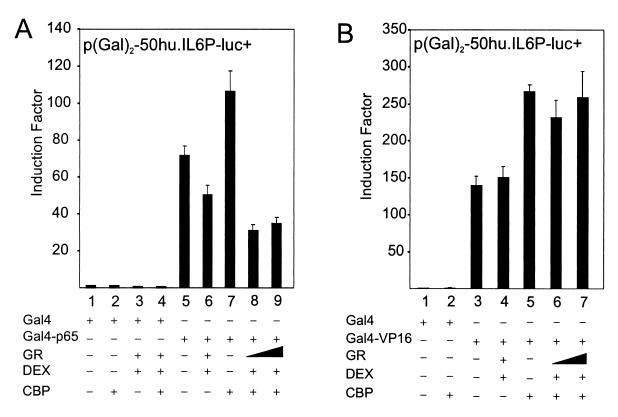
Fig. 3A shows that CBP stimulates Gal4-p65-dependent transcription on a Gal4-driven reporter in the Gal4 one-hybrid system; moreover, the level of transrepression remains largely unchanged or even increases under conditions of maximal cooperation between p65 and CBP. In this nuclear setup, interference of cytoplasmic events or other DNA-bound transcription factors, normally present in the IL-6 promoter context, are not involved. Furthermore, the Gal4-VP16 expression plasmid was also cotransfected with p(Gal)2-50hu.IL6P-luc+ and tested for GC repression (Fig. 3B). CBP also stimulates this strong viral transactivator, because its HAT activity can cooperate with the acidic transactivational domains of VP16 (25). Specific GC repression, as observed for Gal4-p65-driven transactivation, does not occur with CBP-enhanced Gal4-VP16 transcriptional activity.
Figure 3.
HEK293T cells were transiently transfected with 80 ng of p(Gal)2-50.huIL6P-luc+ and with various expression plasmids, the total amount of DNA being fixed at 400 ng, i.e., pGal4 (40 ng) or pGal4-p65 (10 ng) (A) or pGal4-VP16 (40 ng) (B), whether or not with pCMV-CBP (25 ng or 100 ng) and/or pSVhGRα (25 ng or 100 ng). In control lanes with CBP and GR, + corresponds to the highest concentration. Assays were performed in triplicate and are representative of two independent experiments.
The Physical Association Between CBP and p65 Is Not Affected Under Conditions of Gene Repression.
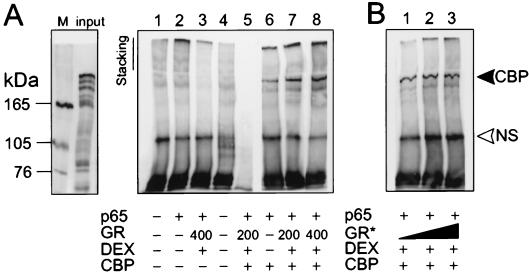
To assess whether GC repression interferes with the physical association between p65 and CBP, a coimmunoprecipitation experiment was carried out (Fig. 4). Fig. 4A shows control lanes (1–5), in which the specificity of the signal is verified. Only when CBP is overexpressed in the cell, was a concomitant association with p65 detected (lanes 6–8). Lanes 7 and 8 show that increasing amounts of activated GR do not disrupt the association between p65 and CBP. The coimmunoprecipitated band was observed reproducibly for different concentrations of transfected CBP and activated GR (data not shown). When increasing amounts of in vitro translated GR protein were added with DEX to lysates, in which only p65 and CBP are overexpressed, CBP is not squelched from immunoprecipitated p65 (Fig. 4B). Membranes were stripped subsequently and reprobed with an anti-p65 antibody to verify equal amounts of immunoprecipitated p65 (data not shown). Concomitant luc assays verified that the regulation of promoter activities was similar as in Fig. 1A (data not shown).
Figure 4.
(A) Immunoprecipitation with anti-p65 (lanes 1–4 and 6–8) or an irrelevant antibody (lane 5) of cell lysates, transfected with pRSV-p65 (200 ng), pCMV-CBP (1 μg), and/or pSVhGRα (200 ng or 400 ng), followed by Western blot analysis with anti-CBP antibody. Lane 4 is a control with lysis buffer only. The input lane represents one-third of the amount used in the assay. The M lane represents molecular mass markers. (B) Similar lysate preparation to which increasing amounts (3, 5, and 10 μl corresponding to approximately 30, 50, and 100 ng) of in vitro translated GR protein (GR*) are added, followed by immunoprecipitation and Western blot analysis as described for A. The upper arrow marks the 265-kDa band corresponding with coimmunoprecipitated CBP protein. NS, nonspecific band. The result is representative of three independent experiments.
GC Repression Still Occurs in the Absence of Optimal CBP Interaction.
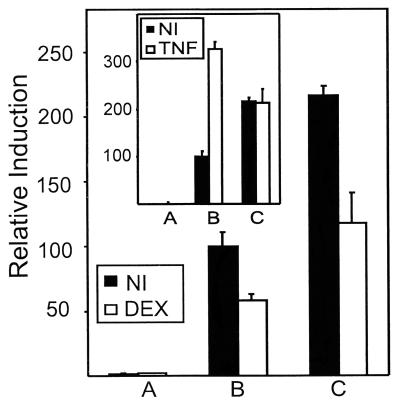
Phosphorylation of Ser-276 in p65 by protein kinase A was shown to be essential for complex formation with CBP and consecutive transactivation (5). Mutation of this residue to Cys in Gal4-p65 abolishes induction of transcriptional activity by, for example, TNF (L.V., unpublished work). Fig. 5 compares the effect of DEX and/or TNF with stably integrated wild-type and mutated Gal4-p65 in L929sA cells. Whereas the point-mutated variant is no longer inducible by TNF (Fig. 5 Inset), most likely because of a decreased interaction with CBP, the basal activity can still be repressed to the same extent as the wild-type fusion protein.
Figure 5.
L929sA cells with stably integrated pGal4 (A), pGal4-p65 (B), or pGal4-p65S276C (C) were transiently transfected with 250 ng of p(Gal)2-50-luc+ and 250 ng of β-Gal expression plasmid. Total amount of DNA was adjusted to 1,500 ng with empty vector DNA. NI, noninduced; DEX, 10−6 M at 24 h for a total of 30 h; TNF, 2,000 units/ml for a total of 6 h. The difference in activity between Gal4-p65 variants is due to different activities of the stable cell clones. The experiment is representative of three independent experiments.
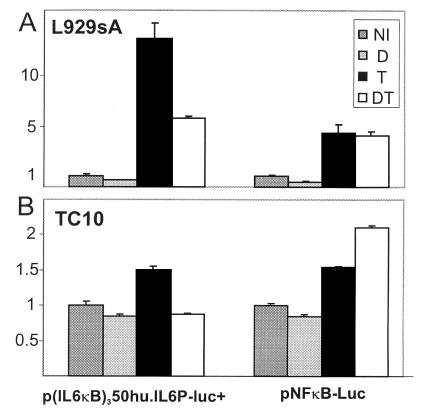
GC Repression of TNF-Induced NF-κB-Driven Genes Is Codetermined by the Identity of the TATA Box.
Fig. 6 shows that the NF-κB-driven recombinant constructs p(IL6κB)350hu.IL6P-luc and pNFκB-Luc are both inducible by TNF. However, GC repression of TNF-induced reporter gene activity is observed only with the autologous, cellular TATA box promoter construct. This result is independent of the investigated cell type, because the same result was obtained in L929sA and in TC10 cells.
Figure 6.
L929sA cells were transiently transfected with 250 ng of p(IL6κB)350hu.IL6P-luc+ or pNFκB-Luc and 250 ng of β-Gal expression plasmid, adjusting the total DNA amount to 1,500 ng with empty vector DNA. TC10 cells were transiently transfected with 100 ng of p(IL6κB)350hu.IL6P-luc+ or pNFκB-Luc and 50 ng of β-Gal expression plasmid, adjusting the total DNA amount to 400 ng with empty vector DNA. NI, noninduced; DEX, 10−6 M at 2 h for a total of 8 h; TNF, 2,000 units/ml for a total of 6 h.
Discussion
One of the great benefits of GCs is their ability to counteract the expression of several NF-κB-driven proinflammatory genes. Recently, the mechanism that underlies this efficacy has been investigated thoroughly; this investigation has led to three major hypotheses. The first hypothesis focuses on cytoplasmic events, proposing that the repressive effect of GR on NF-κB-driven gene expression is explained by GC-induced stimulation of IκB-α (26, 27). This mechanism found, however, little experimental support when several research groups, including ours, investigated various cell lines (15, 18, 28–30). Instead, our previous data showed that GC repression is an entirely nuclear phenomenon that involves a functional interference of GR with the transactivating domain of p65 (15).
A second hypothesis proposes that competition between nuclear factors for limited amounts of coactivator molecules might impair gene expression and, hence, that increasing coactivator concentration would counteract repression (8–10). We now present conclusive evidence that GC repression of p65-mediated gene expression is not relieved by overexpression of the coactivator molecules CBP, SRC-1, and p300 and that sustained repression by overexpressing GR does not result from down-modulation of p65 or CBP levels in the cell. Similar results were obtained in a physiological cell context by using the endogenous GR of endothelial cells. An analogous conclusion can be drawn from the reciprocal experiment, in which p65-mediated repression of a GRE-driven reporter was independent of increasing amounts of CBP/p300. As a matter of fact, repression of the GC response by STAT-5 has also been shown not to result from competition for limiting amounts of p300/CBP (31).
Our observations contradict previous conclusions that GR-mediated repression of NF-κB is relieved by overexpression of CBP/p300 or SRC-1 (9). A control experiment showing the combined activity of p65 and CBP together in the absence of activated GR is, however, not shown in the latter paper. Actually, pure competition for cofactors to explain GC repression lacks specificity, because various transcription factors converge at the level of CBP/p300 for their transcriptional activity. A limitation in the amount of cofactors, resulting in internal competition, would not appear and thus lie at the basis of the synergism observed between GR and p65 on a GRE-NFκB-combined response element-driven promoter (9). Alternatively, one can imagine that GR adopts a different conformation when working as a monomer in trans to inhibit NF-κB activity or when it is bound to DNA as a homodimer to transactivate (32). The existence of dissociating ligands and of various receptor point mutants of GR (18, 33–35), which separate transactivation and transrepression, supports this notion and also disfavors the competition model. This perception is corroborated further by the generation of GR dimerization-defective mice, in which GR can still work in transrepression but no longer transactivate (36). Another argument in favor of our conclusions is the fact that Gal4-VP16 activity, although enhanced by overexpressed CBP, is, unlike Gal4-p65, not repressed by GR. This fact rules out that the mechanism of gene repression relies on a general and aspecific squelching of and competition for common cofactors. In support, under conditions of GC repression, the physical association between p65 and CBP is not disrupted by repressing amounts of activated GR both in vivo and in vitro. The Gal4p65S276C variant, which is assumed not to interact optimally and functionally with CBP (5) and which shows no responsivity to TNF induction, was as efficiently repressed by the endogenous GR as wild-type Gal4-p65; this result excludes a preponderant role for CBP in the repression phenomenon. Because simple competition for common cofactors is not the main mechanism of GC repression, the question arises as to what the effective mechanism might be.
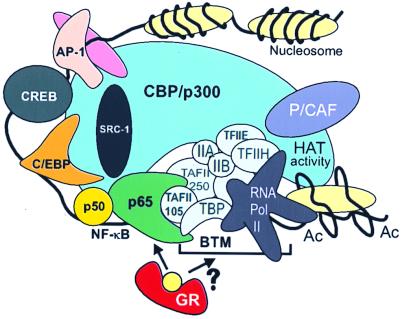
A third hypothesis involves direct protein–protein interaction (reviewed in refs. 1 and 37) and relies on the in vitro association of GR with p65 as a possible way to explain their mutual inhibition (38). Furthermore, the transactivating C terminus of p65 was already shown to bind directly to the general TFIIB and TBP in vitro. This binding could help to stabilize TFIID interactions with promoter DNA and to build up a productive transcription initiation complex (11). In addition, TAFII105 and a specific activator-recruited cofactor complex were found to interact directly with p65 and to enhance transcription (39, 40), whereas TAF mutations have been described that can disrupt the activation of Dorsal, the p65 homologue in Drosophila (41). In this respect, it is extremely interesting that, in different cell types and under the physiological conditions of induction, we found a highly selective specificity of gene repression by GR depending on the TATA box environment. In agreement with our data, we propose a model (Fig. 7) in which gene repression results from direct interference of GR with p65 and its associating targets of the basal transcription machinery, irrespective of the cell type used and the set of coactivator molecules present. Identification of the actual component(s) involved awaits further elucidation.
Figure 7.
A model for GC repression of NF-κB-driven genes. The cointegrator molecule CBP/p300 contacts various transcription factors along the IL-6 promoter DNA and shows HAT activity when NF-κB is activated (17). The latter factor complex also has direct contacts with various components of the basal transcription machinery (see Discussion). Activated GR targets p65 and may thus disturb the necessary conformation or interactions necessary for transcriptional enhancement. BTM, basal transcription machinery; Ac, acetylated nucleosome.
Acknowledgments
We thank K. Van Wesemael for excellent technical assistance. K.D.B. and L.V. hold a fellowship from the Vlaams Instituut voor de Bevordering van het Wetenschappelijk-technologisch Onderzoek in de Industrie. G.H. is a Research Director with the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. Research was supported by the Interuniversitaire Attractiepolen.
Abbreviations
- β-Gal
β-galactosidase
- DEX
dexamethasone
- GC
glucocorticoid
- GR
glucocorticoid receptor
- GRE
GR element
- TNF
tumor necrosis factor
- HAT
histone acetyltransferase
References
- 1.Göttlicher M, Heck S, Herrlich P. J Mol Med. 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- 2.Katzenellenbogen J A, Katzenellenbogen B S. Chem Biol. 1996;3:529–536. doi: 10.1016/s1074-5521(96)90143-x. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 4.Mercurio F, Manning A M. Curr Opin Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhong H, Voll R E, Ghosh S. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 6.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O'Malley B W. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 7.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 8.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 9.Sheppard K A, Phelp K M, Williams A J, Thanos D, Glass C K, Rosenfeld M G, Gerritsen M E, Collins T. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 10.Lee S K, Kim H J, Na S Y, Kim T S, Choi H S, Im S Y, Lee J W. J Biol Chem. 1998;273:16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz M L, Stelzer G, Altmann H, Meisterernst M, Baeuerle P A. J Biol Chem. 1995;270:7219–7226. doi: 10.1074/jbc.270.13.7219. [DOI] [PubMed] [Google Scholar]
- 12.Hahn S. Nature (London) 1993;363:744–747. [Google Scholar]
- 13.Hoffmann A, Oelgeschlager T, Roeder R G. Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roeder R G. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 15.De Bosscher K, Schmitz M L, Vanden Berghe W, Plaisance S, Fiers W, Haegeman G. Proc Natl Acad Sci USA. 1997;94:13504–13509. doi: 10.1073/pnas.94.25.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanden Berghe W, De Bosscher K, Plaisance S, Boone E, Haegeman G. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 18.Vanden Berghe W, Francesconi E, De Bosscher K, Rèsche-Rigon M, Haegeman G. Mol Pharmacol. 1999;56:797–806. [PubMed] [Google Scholar]
- 19.VandenBerghe W, Plaisance S, Boone E, De Bosscher K, Schmitz M L, Fiers W, Haegeman G. J Biol Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Graham F L, van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arias J, Alberts A S, Brindle P, Claret F, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 24.Na S, Lee S, Han S, Choi H, Im S, Lee J. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 25.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 26.Auphan N, DiDonato J A, Rosette C, Helmberg A, Karin M. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 27.Scheinman R, Cogswell P C, Lofquist A, Baldwin A S., Jr Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 28.Brostjan C, Anrather J, Csizmadia V, Stroka D, Soares M, Bach F H, Winkler H. J Biol Chem. 1996;271:19612–19616. doi: 10.1074/jbc.271.32.19612. [DOI] [PubMed] [Google Scholar]
- 29.Heck S, Bender K, Kullmann M, Gottlicher M, Herrlich P, Cato A C B. EMBO J. 1997;16:4698–4707. doi: 10.1093/emboj/16.15.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wissink S, van Heerde E C, van der Burg B, van der Saag P T. Mol Endocrinol. 1998;12:355–363. doi: 10.1210/mend.12.3.0081. [DOI] [PubMed] [Google Scholar]
- 31.Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. Mol Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 32.Lefstin J A, Yamamoto K R. Nature (London) 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 33.Vayssière B M, Dupont S, Choquart A, Petit F, Garcia T, Marchandeau C, Gronemeyer H, Resche Rigon M. Mol Endocrinol. 1997;11:1245–1255. doi: 10.1210/mend.11.9.9979. [DOI] [PubMed] [Google Scholar]
- 34.Liden J, Delaunay F, Rafter I, Gustafsson J, Okret S. J Biol Chem. 1997;272:21467–21472. doi: 10.1074/jbc.272.34.21467. [DOI] [PubMed] [Google Scholar]
- 35.Iniguez Lluhi J A, Lou D Y, Yamamoto K R. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 36.Reichardt H M, Kaestner K H, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 37.McEwan I J, Wright A P, Gustafsson J A. BioEssays. 1997;19:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 38.Ray A, Prefontaine K E. Proc Natl Acad Sci USA. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamit-Hezi A, Dikstein R. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]