Abstract
Two peptide transporter (PTR) homologs have been isolated from developing seeds of faba bean (Vicia faba). VfPTR1 was shown to be a functional peptide transporter through complementation of a yeast mutant. Expression patterns of VfPTR1 and VfPTR2 as well as of the amino acid permease VfAAP1 (Miranda et al., 2001) were compared throughout seed development and germination. In developing seeds, the highest levels of VfPTR1 transcripts were reached during midcotyledon development, whereas VfAAP1 transcripts were most abundant during early cotyledon development, before the appearance of storage protein gene transcripts, and were detectable until late cotyledon development. During early germination, VfPTR1 mRNA appeared first in cotyledons and later, during seedling growth, also in axes and roots. Expression of VfPTR2 and VfAAP1 was delayed compared with VfPTR1, and was restricted to the nascent organs of the seedlings. Localization of VfPTR1 transcripts showed that this PTR is temporally and spatially regulated during cotyledon development. In germinating seeds, VfPTR1 mRNA was localized in root hairs and root epidermal cells, suggesting a role in nutrient uptake from the soil. In seedling roots, VfPTR1 was repressed by a dipeptide and by an amino acid, whereas nitrate was without influence.
Compared with single-cell organisms, higher plants have evolved a larger proportion of genes involved in energy-dependent transport (The Arabidopsis Genome Initiative, 2000). The necessity of long distance transport in plants requires additional transport systems thus adding more complexity to our understanding on how these mechanisms function and how they are regulated. Amino acid transport is thought to be of primary importance in organic nitrogen acquisition, and might be involved in processes such as direct uptake of amino acids from the soil, phloem and xylem loading, phloem to xylem exchange, and retrieval of amino acids that “leak” from the cells (for review, see Williams and Miller, 2001). In addition to amino acid transport, peptide transport seems to play an important role in periods of rapid protein mobilization, such as redistribution of N during leaf senescence, protein deposition during seed development, and storage protein hydrolysis during germination, when the efficiency of N transport may be increased by direct uptake of peptides (Sopanen et al., 1977; Higgins and Payne, 1978; for review, see Stacey et al., 2002).
During the past decade, a number of proton-coupled amino acid and peptide transporters have been cloned from different plant species. The amino acid permease (AAP) and peptide transporter (PTR) multigene families are currently the best characterized groups of plant transporters for organic N. It has been shown, for instance, that six AAPs and one PTR isoform of Arabidopsis mediate the transport of a broad range of N compounds in heterologous systems; i.e. AtAAP1 through 6 transport a wide spectrum of amino acids including neutral, acidic, and basic amino acids, and AtPTR2(3) transports a broad range of di- and tripeptides, with little specificity toward the side chain (Frommer et al., 1994a; Fischer et al., 1995, 2002; Rentsch et al., 1995, 1996; Song et al., 1996). Moreover, the PTR family also comprises the NRT1 group of low-affinity nitrate transporters, which are structurally very similar to the peptide transporters. Thus, PTRs may have a dual function as peptide and NO3– transporters (for review, see Stacey et al., 2002). Sequence homologies indicate the existence of many other potential AAPs and PTRs in the Arabidopsis genome whose functionality remains to be tested (The Arabidopsis Genome Initiative, 2000). This apparent redundancy could be explained by tissue- and development-specific roles based on the observation that AAP isoforms have distinct expression patterns throughout the plant (for review, see Fischer et al., 1998; Rentsch et al., 1998).
Despite numerous studies on the kinetics of N uptake in sink organs, such as developing seeds, seedlings, and roots, little is known about the developmental regulation of the AAP and PTR genes in these organs (see Hirner et al., 1998). Biochemical studies revealed that during early development of pea (Pisum sativum) seeds, a nonsaturable amino acid transport system is of primary importance, and later, when approximately two-thirds of the storage proteins have been deposited, a saturable system emerges (Lanfermeijer et al., 1990). In soybeans (Glycine max), 50% of the Gln transport into developing cotyledons was shown to be energy dependent (VerNooy et al., 1986). During the early phases of germination of barley (Hordeum vulgare) grains, different peptide transport systems are present and, later on, amino acid transport activity appears (Higgins and Payne, 1978; Waterworth et al., 2000). In support of these studies, the molecular characterization of AAP and PTR genes showed that some isoforms are expressed in seeds. Briefly, PsAAP1 transcripts are localized mainly in the epidermal cells of pea cotyledons (Tegeder et al., 2000), whereas VfAAP1 mRNA is expressed throughout the storage parenchyma but not in the epidermal cell layer of developing faba bean (Vicia faba) cotyledons (Miranda et al., 2001). During germination, an AAP is expressed in the roots of castor bean (Ricinus communis) seedlings, where it may play a role in amino acid uptake from the soil and in phloem loading (Bick et al., 1998). Similarly, the peptide transporter AtPTR2 of Arabidopsis is expressed during embryo development, where it might play a role in nutrient supply (Rentsch et al., 1995; Song et al., 1996). Its antisense repression in transgenic plants leads to delayed flowering and arrested seed development (Song et al., 1997). In barley, HvPTR1 seems to be responsible for the rapid mobilization and translocation of peptides originating from the hydrolysis of endosperm storage proteins during germination (West et al., 1998). These studies highlight an important physiological role that amino acid and peptide transporters play during seed development and germination. However, temporal and spatial regulation of AAP and PTR genes is still poorly investigated.
Protein accumulation in developing seeds relies on the availability of nitrogenous compounds that are delivered by the phloem and made available to the embryo (Thorne, 1985; Barneix et al., 1992). In legumes, phloem unloading and postphloem transport through the seed coat occur symplasmically, whereas the embryo is isolated from maternal tissues. Thus, assimilates need to cross an apoplastic space before being taken up by the embryo, in a process that may be mediated by active transport systems (Offler and Patrick, 1993; Patrick and Offler, 1995). Developing faba bean seeds are typical sink organs that accumulate high amounts of storage proteins as an energy reserve. The amount of protein that is accumulated is regulated at different levels, including the availability of assimilates and genetic background (for review, see Wobus et al., 1995; Weber et al., 1997b; Golombek et al., 2001).
During germination of legume seeds, storage products are rapidly degraded and relocated to support growth of the seedling (Schlereth et al., 2000). The lack of symplasmic connections between the axis and the cotyledons during the first phases of seed germination in vetch (Vicia sativa) led to the assumption that the embryonic axis relies on the breakdown of locally stored proteins as the only N source during the first stages of germination (Tiedemann et al., 2000). On the other hand, in barley scutella, which is a tissue comparable with a cotyledon, peptides are actively transported already shortly after imbibition, most likely to provide the embryo with nutrients during the first stages of germination (Sopanen et al., 1977). During this period, the levels of peptide transport activity decrease in parallel to loss of seed viability, suggesting that peptide transport takes part of a vital process during early germination and serves as an early indicator of viability in barley seeds (Waterworth et al., 2000). To date, no reports on peptide transport in developing or germinating legume seeds have been published.
The present paper reports on the isolation of two PTR isoforms from developing seeds of faba bean, and compares their expression and regulation with VfAAP1, a preferentially cotyledon-expressed AAP, during seed development and germination. Localization of VfPTR1 transcripts showed that this PTR is temporally and spatially regulated during cotyledon development. During germination, VfPTR1 mRNA was localized in the root epidermis and hairs of seedlings, suggesting that it may take part in nutrient uptake from the soil.
RESULTS
Cloning of Two Members of the PTR Family from Faba Bean
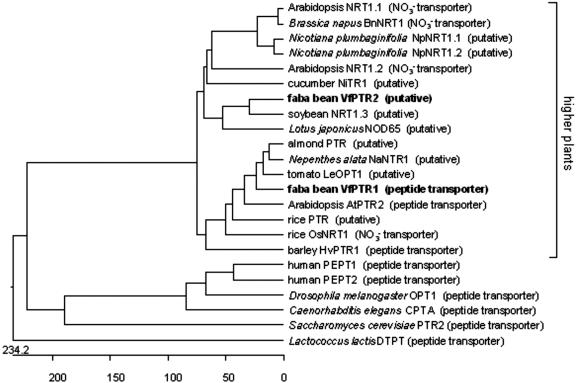
Two DNA fragments were obtained by reverse transcriptase (RT)-PCR with PTR primers using total RNA from developing cotyledons. After sequencing, they were identified as homologs of the PTR family of peptide transporters. These clones were used as probes to screen a cDNA library prepared from developing cotyledons of faba bean (Heim et al., 1993). Screening with one of the probes yielded 10 clones from an identical cDNA. Among these, a 2,028-bp long clone, denominated VfPTR1 (accession no. AY289622), was chosen for further experiments. VfPTR1 was predicted to be a full-length cDNA encoding a protein composed of 584 amino acid residues, sharing high similarity with AtPTR2 (73.8% identity) and with the putative peptide transporters of almond (Prunus dulcis; 80.3% identity; accession no. AAF20002) and tomato (Lycopersicon esculentum; 79.8% identity; accession no. AAD01600). In silico analyses according to Kyte and Doolittle (1982) and Krogh et al. (2001) suggested that the predicted VfPTR1 protein is highly hydrophobic and contains 12 transmembrane domains, which superimpose those of AtPTR2 (data not shown). A signal peptide was predicted (Nielsen et al., 1997) with the most likely cleavage site between positions 10 and 11 (SSR-LE) of the deduced VfPTR1 amino acid sequence. Attempts to clone the full-length cDNA corresponding to the second PTR isoform failed. Thus, the original RT-PCR fragment, called VfPTR2 (accession no. AY289623), was used for further analyses. VfPTR2 comprised an incomplete open reading frame encoding 316 amino acid residues, which showed highest similarity to a putative peptide/nitrate transporter of soybean GmNRT1.3 (56.3% identity; Yokoyama et al., 2001). Because the PTR gene family comprises the NRT1 subgroup of NO3– transporters, a phylogeny tree was constructed to compare the sequence distances of VfPTR1 and VfPTR2 in relation to other plant PTR/NRT1 isoforms, as well as to the peptide transporters of animals, yeast, and bacteria. A dendogram shows that VfPTR1 clusters together with AtPTR2 and the putative PTRs of almond, tomato, and Nepenthes alata (Schulze et al., 1999). VfPTR2 clusters closer to the dicotyledonous NRT1 transporters. Moreover, the plant transporters cluster apart from the peptide transporters of other organisms, whereas the plant NRT1 isoforms could not be clearly separated from the peptide transporters (Fig. 1).
Figure 1.
Dendogram based on sequence similarities among members of the PTR family. The predicted protein sequences of peptide and nitrate transporters of the PTR family from plants, animals, yeast, and bacteria were aligned by ClustalX method (MegAlign; DNAstar, Madison, WI). In parenthesis, it is indicated whether the protein function is based on experimental evidence (peptide or NO3– transporter) or if it is based on sequence similarity (putative). The units at the bottom of the tree indicate the number of substitution events.
VfPTR1 Restores Growth of a Yeast Mutant Deficient in Peptide Transport
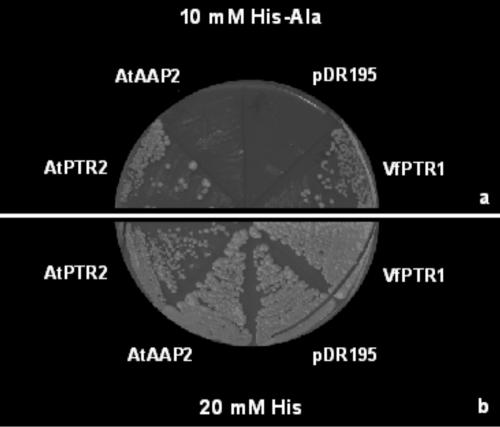
To confirm that VfPTR1 is a peptide transporter, its complete cDNA was inserted into the pDR195 vector and was expressed in the LR2 yeast mutant, which is His auxotroph and carries a mutation in the PTR2 gene and thus is unable to grow on media containing histidyl peptides as the only source of His (Rentsch et al., 1995). LR2 cells transformed with VfPTR1 exhibited efficient growth on medium containing 10 mm His-Ala, similar to cells carrying the AtPTR2 cDNA from Arabidopsis, whereas LR2 cells transformed with an amino acid permease (AtAAP2) or with an “empty” pDR195 vector, were not able to grow under similar conditions (Fig. 2A). LR2 cells grew efficiently on a nonselective medium containing 20 mm His, regardless of the construct they were carrying (Fig. 2B).
Figure 2.
Functional complementation of the LR2 yeast mutant by VfPTR1. Selective growth of mutant strains carrying the cDNAs of VfPTR1, AtPTR2 (as positive control), AtAAP2 (negative control), or pDR195 “empty” vector (negative control) on synthetic complete medium supplemented with 10 mm His-Ala (a) and nonselective growth on synthetic complete medium supplemented with 20 mm His (b).
VfPTR1, VfPTR2, and VfAAP1 Differ in Steady-State mRNA Levels in Developing and Germinating Seeds
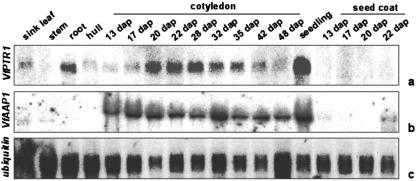
Northern hybridizations were performed to compare the mRNA profiles of VfPTR1, VfPTR2, and the amino acid permease VfAAP1 (Miranda et al., 2001). Among several tissues analyzed, VfPTR1 transcripts accumulated to highest levels in seedlings at 6 d after imbibition (dai; Fig. 3A). There was a strong accumulation of VfPTR1 transcripts in roots and in developing cotyledons as well. During cotyledon development, VfPTR1 mRNA signals were weak at 13 and 17 dap, accumulated to higher levels from 20 until 35 dap, and were weaker again at 42 and 48 dap. Sink leaves and hulls showed weak bands, and no signals were found in seed coats or stems (Fig. 3A). No signals were detected in source leaves (data not shown). The VfPTR1 mRNA steady-state levels suggest a role for this PTR in sink tissues, especially in developing cotyledons, roots, and seedlings. In contrast, when VfAAP1 was hybridized to the same blot, transcripts accumulated to high levels in the early stages of cotyledon development (13 until 20 dap), and remained at slightly lower levels later on, until the late storage phase (48 dap). A rather strong signal was also found in seedlings (Fig. 3B). The described developmental profile of VfAAP1 is in agreement with that reported previously, and suggests a role of this permease in sink tissues, primarily during early cotyledon development (Miranda et al., 2001). When compared with VfAAP1, VfPTR1 transcripts accumulated more transiently throughout cotyledon development (Fig. 3). No signals were seen when the blots were hybridized with a VfPTR2 probe. VfPTR2 transcripts could be detected by RT-PCR with VfPTR2-specific primers on total RNA from developing cotyledons at 22 and 26 dap and from roots, but not from seed coats or stems. Cloning and sequencing of the PCR products confirmed that they corresponded to VfPTR2 (data not shown).
Figure 3.
Analysis of transcript accumulation of VfPTR1 and VfAAP1 in different tissues and seed developmental stages of faba bean. Northern blots were hybridized with 32P-labeled cDNA probes of VfPTR1 (a), VfAAP1 (b), and ubiquitin (c) as loading control. Ten micrograms of total RNA isolated from different tissues and seed developmental stages was loaded per lane. dap, Days after pollination.
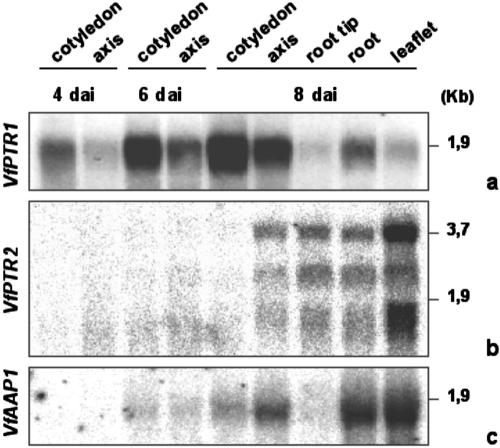
To examine the expression of VfPTR1, VfPTR2, and VfAAP1 genes during germination in more detail, northern hybridizations were performed with dissected parts of seedlings germinated on humid filter papers. RNA was extracted at three time points, which were chosen based on the exterior morphological changes: at 4 dai, before the root had ruptured the seed coat; at 6 dai, when the root was approximately 0.5 to 1.0 cm long; and at 8 dai, when roots had reached 2 to 3 cm and the first leaflets had emerged from the seed. Northern hybridizations revealed that in cotyledons, labeling with the VfPTR1 probe appeared at 4 dai, and increased steadily until 8 dai (Fig. 4A). In seedlings at 2 dai, VfPTR1 transcripts could not be detected (data not shown). In the axis, VfPTR1 mRNA levels were high at 6 dai, and further increased at 8 dai. In 8-dai seedlings, labeling with the VfPTR1 probe was also strong in the upper portion of the roots (approximately 2.5 cm), but was weak in root tips (approximately 0.5 cm) and leaflets (Fig. 4A). When VfPTR2 was used as a probe to hybridize the same blot, signals were strongest in the leaflets, moderate in the tips and upper portions of the roots, and weak in the axis of 8-dai seedlings. VfPTR2 signals in 4- and 6-dai seedlings were negligible (Fig. 4B). In addition to the bands of expected size (approximately 1.8 Kb), the VfPTR2 probe revealed two additional larger bands (Fig. 4B). The origin of these bands is unclear, but they may indicate accumulation of unspliced or alternatively spliced RNA. When hybridizations were performed with a VfAAP1 probe, mRNA levels were high in leaflets and in the upper portions of the roots, moderate in the axis, and faint in the cotyledons and root tips of 8-dai seedlings. Labeling was not seen in 4-dai seedlings, and was faint at 6 dai (Fig. 4C). Equal loading between samples was confirmed by running an ethidium bromide-stained gel (data not shown). Taken together, the mRNA steady-state levels of VfPTR1 during seedling germination and development suggest that this PTR might have a primary role at the sites of intense proteolysis, whereas VfPTR2 and VfAAP1 would play a more prominent role in the young seedling tissues.
Figure 4.
Analysis of transcript accumulation of VfPTR1, VfPTR2, and VfAAP1 in seedlings of faba bean harvested at different time points. Ten micrograms of total RNA extracted from different parts of the seedlings and at different time points was loaded per lane; i.e. before roots had ruptured the seed coats (4 dai), when roots were between 0.5 and 1.0 cm long (6 dai), and when roots were approximately 4.0 cm long (8 dai). Northern blots were hybridized with 32P-labeled cDNA probes of VfPTR1 (a), VfPTR2 (b), and VfAAP1 (c). An approximate size scale (kilobytes) is indicated. Equal loading between samples was confirmed by running an ethidium bromide-stained gel (data not shown).
VfPTR1 mRNA Is Localized in Different Tissues during Seed Development and Germination
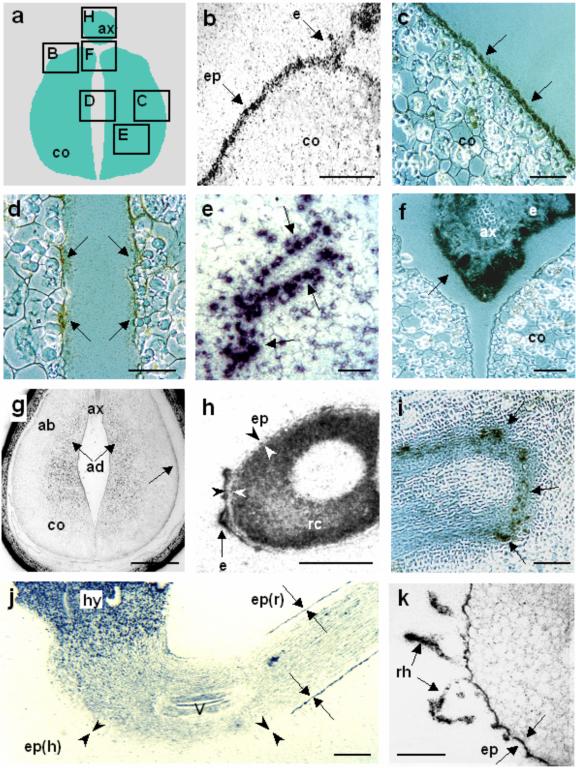
In situ hybridizations using radiolabeled probes were performed to determine the distribution of VfPTR1 transcripts within seed sections at different developing stages as defined by Borisjuk et al. (1995). During early cotyledon development, i.e. before the onset of storage protein deposition (stage IV–V), labeling by the VfPTR1 probe was found in epidermal cells of the cotyledons (Fig. 5B) and in the surrounding endosperm that retains label at all stages analyzed (Figs. 5B, f and h). At stage VI, cells within the abaxial epidermal region, differentiated into transfer cells (Weber et al., 1998), became more strongly labeled by the VfPTR1 probe than those at the adaxial epidermis (Figs. 5, C and D). Later in development, intensive labeling was also detected in cells surrounding the vascular vessels (Fig. 5E). During the main storage phase, labeling appeared in parenchyma cells underlying the transfer cell layer and in the adaxial parenchyma (Fig. 5G). Toward seed maturation (late stage VII), the cortex cells of the radicle were strongly labeled, but not the epidermal cells of this organ (Fig. 5H). Hybridization with a VfAAP1 probe revealed an even labeling of the cotyledon storage parenchyma cells at stage VI, whereas the cells in the outer epidermal region were not labeled (Miranda et al., 2001). For comparison, slides were also hybridized with the faba bean Suc transporter VfSUT1 cDNA (Weber et al., 1997a). Labeling with VfSUT1 was restricted to the transfer cells of the outer epidermal region (data not shown) in accordance with previous reports showing that the Suc and hexose transporters of faba bean are expressed in the transfer cells of the cotyledons (Weber et al., 1997a).
Figure 5.
Transcript distribution of VfPTR1 in sections of developing faba bean seeds and seedlings. a, Schematic representation of a developing faba bean embryo in a cross-section, where the boxed areas are enlarged in the following pictures; b through k, Bright-field micrographs showing in situ hybridization using a 33P-labeled VfPTR1 probe, with label seen as dark grains; b, abaxial region of a stage IV (Borisjuk et al., 1995) cotyledon with labeled epidermal cells and endosperm, but unlabeled parenchyma cells; c and d, abaxial and adaxial regions of cotyledons, respectively. The abaxial transfer cells in the outer epidermis are more strongly labeled than the cells of the inner epidermis; e, labeled cells surrounding transport vessels of a stage VI cotyledon; f, strong labeling of endosperm surrounding the axis during stage VI; g, cotyledons with labeled storage parenchyma cells and transfer cells of the outer epidermal region; h, radicle of a stage VII embryo strongly labeled within the cortex cells, but not the epidermis cells (seen as an unlabeled layer between the cortex and the endosperm). i through k, Distribution of VfPTR1 transcripts in sections of faba bean seedlings; i, section of a root showing the labeled vascular bundles; j, longitudinal section through the seedling, showing strong label in the hypocotyl and in the epidermis of the root, but not in the epidermis of the hypocotyl; k, cross-section through the root of a seedling showing labeling within the epidermis and the root hairs; ab, Abaxial region; ad, adaxial region; ax, axis; co, cotyledons; e, endosperm; ep, epidermis; ep(h), epidermis of hypocotyl; ep(r), epidermis of root; hy, hypocotyl; rc, radicle cortex; rh, root hair; v, vascular bundle. Bars in b, c, d, e, f, i, and k = 0.1 mm; g and j = 1 mm; h = 0.5 mm. Arrows point to the labeling and arrowheads point to unlabeled areas.
To examine the distribution of VfPTR1 transcripts in seedlings, in situ hybridizations were performed on sections of seedlings at the time when the roots were approximately 2.5 cm long and the first leaflets were not yet visible. VfPTR1 signals were associated with vascular bundles (Fig. 5I), and were also found in the parenchyma tissues. As shown in Figure 5J, strong labeling gradient is present toward the hypocotyl. A specific labeling pattern was observed in the epidermis across the seedling: the epidermal layer of axis and hypocotyl remained unlabeled, whereas strong labeling was found in the epidermis of roots (Fig. 5J). In cross-sections of roots, VfPTR1 transcripts were localized within epidermal cells and root hairs (Fig. 5K).
VfPTR1 mRNA Accumulation Levels in Roots Are Dependent on the N Source
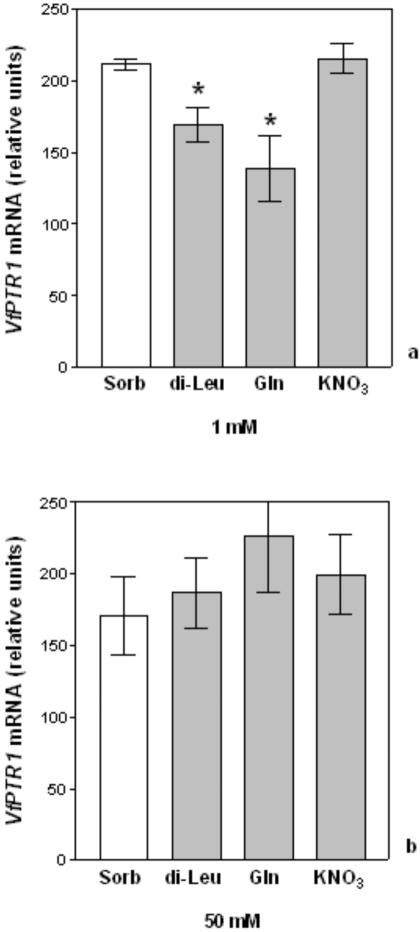
To test whether VfPTR1 mRNA steady-state levels in roots differ due to different sources of N supply, northern hybridizations were performed with RNA extracted from roots (excluding the first approximately 0.5 cm from the root tip) of 8-dai seedlings cultured in media containing sorbitol (as osmotic control), Leu-Leu, Gln, or NO3–. VfPTR1 signals were significantly lower in roots of seedlings cultured in 1 mm Leu-Leu or 1 mm Gln compared with control seedlings cultured with 1 mm sorbitol. In seedlings cultured with 1 mm KNO3, VfPTR1 mRNA accumulated to similar levels as in the control roots (Fig. 6A). Slight increases on the VfPTR1 mRNA levels in roots were observed when seeds were cultured in media containing 50 mm Leu-Leu, Gln, or KNO3 compared with control experiments, but the differences were not statistically significant (Fig. 6B). The reasons for the apparently contradictory effects of high and lower concentrations of N sources on VfPTR1 regulation are not yet known, but may be related to the substrate affinity of this transporter.
Figure 6.
Effect of different N sources on the accumulation of VfPTR1 mRNA in roots of seedlings cultured in vitro. Transcript analysis of VfPTR1 in roots after in vitro germination of seeds in Murashige and Skoog medium (Murashige and Skoog, 1962), depleted of KNO3 and NH4NO3, and supplemented with 1 mm (a) or 50 mm (b) of sorbitol (Sorb; as control), Leu-Leu, Gln, or KNO3. Northern blots were hybridized with 32P-labeled VfPTR1 probes, and signals were quantified by a phosphorimager. Loading was normalized by hybridization with an ubiquitin probe. Each bar represents the mean of three replications ± se. An asterisk denotes statistically significant results (P < 0.05).
DISCUSSION
VfPTR1 and VfPTR2, Two New Members of the PTR Family of Transporters
According to sequence analyses, VfPTR1 and VfPTR2 of faba bean are members of the PTR family of genes that include peptide and NO3– transporters (Fig. 1). To test whether VfPTR1 mediates peptide transport, functional tests were carried out with a yeast mutant that does not grow on media containing peptides as the only source of N or His in the form of a histidyl-peptide (Rentsch et al., 1995). Experiments showed that VfPTR1 restores growth of the yeast strain LR2 upon supplementation of His-Ala to the medium, implying that VfPTR1 mediates uptake of this dipeptide and complements the mutation in the PTR2 gene (Fig. 2). LR2 and a similar yeast mutant were used to characterize the substrates transported by the Arabidopsis AtPTR2 protein. AtPTR2 takes up a wide range of di- and tripeptides, with little selectivity toward their side chain (Rentsch et al., 1995; Song et al., 1996). Biochemical studies revealed that several small peptides are taken up in faba bean mesophyll cells (Jamai et al., 1996).
Phylogeny analyses show that PTRs of higher plants cluster apart from those of other organisms. However, the plant peptide and low-affinity nitrate transporters cannot be separated on basis of their sequences (Fig. 1). Despite of their high structural similarity, attempts to show that a PTR has a dual function as peptide and NO3– transporter have failed. For instance, in heterologous systems, AtPTR2 transports peptides but not NO3– (Song et al., 1996), whereas the oilseed rape (Brassica napus) and rice (Oryza sativa) NRT1 isoforms mediate transport of NO3– but not of dipeptides (Zhou et al., 1998; Lin et al., 2000). Similarly, Jamai et al. (1996) have shown that NO3– does not compete against different peptide uptake systems present in the leaves of faba bean. Nevertheless, the PTRs of Arabidopsis and oil-seed rape also have a low affinity for basic amino acids (Frommer et al., 1994a; Zhou et al., 1998). VfPTR1 is the third plant PTR shown to mediate peptide uptake, but the substrate range recognized by this transporter remains to be investigated.
In Developing Cotyledons, VfPTR1 May Function Initially in Peptide Import from the Surrounding Endospermal Apoplast and, Later, in the Transport within the Embryo
VfPTR1 was mainly expressed in cotyledons of germinating seedlings, but also to a high extent in developing cotyledons and roots. The highest accumulation of VfPTR1 transcripts during cotyledon development is delayed in comparison with the expression of VfAAP1, and occurs after the seeds have entered the storage phase, whereas the VfAAP1 expression peak precedes the storage phase (Fig. 3; Miranda et al., 2001). Activity of peptide transporters seems to be related to moments of intensive proteolysis, such as wounding, senescence, and degradation of storage proteins, when the transport of peptides is thought to be of great importance for the rapid export of organic nitrogen (Higgins and Payne, 1978; for review, see Frommer et al., 1994b). It is possible that the substrates for VfPTR1 originate mainly from the remobilization of reserves during leaf senescence, which occurs parallel to seed maturation.
In situ hybridizations revealed that VfPTR1 expression is spatially controlled during seed development. During stages V and VI, VfPTR1 mRNA is found mainly in the epidermis. Epidermal transfer cells play a key role in the active uptake of assimilates by the cotyledons (Gunning and Pate, 1974; Offler et al., 1989; Weber et al., 1998) and are the main site of expression of several transporters, such as the Suc, hexose, and amino acid transporters of faba bean and pea (Weber et al., 1997a; Tegeder et al., 1999; Tegeder et al., 2000). Later in seed development, VfPTR1 transcripts appear in cells surrounding the vascular bundles, i.e. possibly in the companion cells, of stage VI cotyledons, and in the storage parenchyma cells of stage VII cotyledons. Taken together, these observations suggest a switch in the function of VfPTR1 during development from the import of peptides from the surrounding endospermal apoplast toward the transport within the embryo. This pattern is clearly different from that described for the Suc transporter of faba bean, whose transcripts are confined to the abaxial epidermal cells throughout cotyledon development (Weber et al., 1997a). However, it resembles the expression pattern of a functionally ill-defined Suc-binding protein homolog (VfSBPL). VfSBPL mRNA is also confined to epidermal transfer cells early in development, whereas at later stages, its mRNA distribution patterns were more similar to those of storage proteins (Heim et al., 2001).
It is not clear whether and to which extent the long-distance transport of peptides and uptake by the seeds contributes to the level of storage protein accumulation in the embryos. The PTR gene of Arabidopsis is ubiquitously expressed and its antisense repression driven by a constitutive promoter led to plants with less and bigger seeds (Song et al., 1997). In legumes, a large proportion of the N used to build up the storage proteins originates from the remobilization of proteins stored transiently in maternal tissues (e.g. Wittenbach, 1982; Staswick et al., 1994), when peptide transporters are thought to play an important role. Moreover, studies with soybean embryos showed that N concentration in cotyledons increases in a direct proportion to the N concentration in the media (Hayati et al., 1996). Localization of VfPTR1 transcripts unveils a complex temporospatial regulation expression profile of this gene, pointing to an important role of this transporter during seed development of faba bean.
In Seedlings, VfPTR1 Is Primarily Expressed at the Site of Proteolysis, whereas VfPTR2 and VfAAP1 Are Expressed in Developing Organs
VfPTR1, VfPTR2, and VfAAP1 are developmentally regulated during seed germination and early seedling growth. VfPTR1 transcripts appear first in cotyledons when the seeds show no visible signs of germination. Shortly after the root penetrates the seed coat (6 dai), VfPTR1 transcripts accumulate in the embryo axis as well. Later on, when the roots reach approximately 3.0 cm and the first leaflets are visible (8 dai), VfPTR1 is highly expressed in cotyledons, axes, and upper portions of the roots, but is low in leaflets and root tips (Fig. 4A). These data suggest that VfPTR1 plays a primary role in transporting peptides originated from proteolysis, and seems to be analogous to HvPTR1 in the germinating barley caryopsis (West et al., 1998). VfPTR2 is induced later as compared with VfPTR1, mainly in the leaflets of developing seedlings, but also in roots and axes, where it may be involved in phloem loading (Fig. 4B). The appearance of multiple bands in blots hybridized with VfPTR2 suggests the accumulation of unspliced RNA for reasons that remain unclear. A similar event has been described for a plant metal transporter, where altered substrate specificity originates from differentially spliced transcripts (Persans et al., 2001). Alternative splicing is common among human transporter genes, where it was hypothesized that although many of these isoforms may not be functional, they could be important in the functional regulation of the transporter (for review, see Gamba, 2001). Induction of VfAAP1 in growing seedling organs, mainly in axes, roots, and leaflets, suggest that this permease may function on uptake of amino acids from the soil and phloem loading, similar to the role proposed for the AAP1 of castor bean (Bick et al., 1998). The present observations are in accordance with reports showing that in germinating barley scutella, the development of peptide transport activity starts before germination and precedes that of amino acid transport, which is initiated later after germination (West et al., 1998; Waterworth et al., 2000).
In grains and legume seeds, significant storage protein mobilization happens after germination takes place. However, before the first signs of germination, there is already an increase in the amount of soluble amino compounds in cotyledons and axes, implying that proteolysis already takes place at this time point (Schlereth et al., 2000). In vetch, symplasmic connections between the embryo axis and cotyledons are absent during early germination and are established only after the switch into the seedling growth phase (Tiedemann et al., 2000). Thus, the induction of VfPTR1 transcription before germination (Fig. 4) may be important to transport protein degradation products into the storage parenchyma and possibly into the axis and epicotyl, when these tissues are still symplasmically isolated. Localization of VfPTR1 transcripts in leaflets and vascular bundles of developing seedlings (Fig. 5) suggests that this transporter also plays a role in phloem loading to provide N to the growing tissues.
Expression in Root Hairs Suggests That VfPTR1 May Function in Nutrient Uptake from the Soil
VfPTR1 is expressed in the roots of adult plants and seedlings, and in seedlings, its transcripts are localized in the root epidermis and root hairs. Similarly, a proton-coupled amino acid uptake system has been described in castor bean roots, and location of RcAAP1 transcripts in root hairs suggest that this transporter may be responsible for the uptake of amino acids from the soil (Weston et al., 1994; Bick et al., 1998). A potential role for peptide transporters on the direct uptake of small peptides from the soil has been recently hypothesized (Nishizawa and Mori, 2001; Waterworth et al., 2001). Moreover, Steiner et al. (1994) demonstrated that root growth inhibition by toxic peptides is reversed by competition with nontoxic ones, which indicates that dipeptides are taken up directly by the roots of Arabidopsis seedlings growing in vitro. There is increasing biochemical evidence indicating that some plants preferentially acquire organic N, such as amino acids or peptides, directly from the soil (Yamagata et al., 2001 and refs. therein).
VfPTR1 transcript levels in roots of seedlings growing in vitro decrease slightly when Leu-Leu or Gln is present at low concentrations in the medium, but nitrate is without influence (Fig. 6). The mode of regulation of VfPTR1 seems to differ from that of HvPTR1, where the presence of amino acids does not result in significant changes in the transcript levels (Waterworth et al., 2000). On the other hand, transcriptional down-regulation of VfAAP1 by different amino acids has been previously reported (Miranda et al., 2001). Identifying the mechanisms regulating plant amino acid and peptide transporters is essential for better understanding N uptake and relocation throughout the plant in response to N availability in the soil.
MATERIALS AND METHODS
Plant Material
Plants from faba bean (Vica faba var minor cv Fribo) were grown in pots placed in chambers supplied with artificial light (16-h light/8-h dark regime) at 20°C to 25°C. All plant material originated from the IPK GenBank (Gatersleben, Germany). Samples were collected 4 h after the beginning of the light period, and developing seeds were sorted on basis of the number of dap. For temporal analyses of germination and seedling development, dry seeds were imbibed with H2O for 1 h and were incubated on petri dishes with moist filter papers in the dark, and probes were harvested at different time points. Germination was marked by the time when the radicle of more than 50% of the seeds had ruptured the seed coat (Bewley and Black, 1994). To analyze the effect of different N sources, seeds were germinated on liquid medium with half-concentrated Murashige and Skoog medium (Murashige and Skoog, 1962), depleted of KNO3 and NH4NO3, and supplemented with 1 or 50 mm of sorbitol, Leu-Leu, Gln, or KNO3. Seeds were incubated at room temperature in the dark with gentle shaking.
Molecular Cloning and cDNA Library Screening
RT-PCR was performed with an Oligo(dT)23 primer (Sigma-Aldrich Chemie, Steinheim, Germany) and total RNA was extracted from developing cotyledons as template. RT reactions were preceded by a “hot start” (5 min at 70°C) and were carried out at 42°C for 1 h. A 3-μL aliquot was used as template for a standard PCR with degenerated primers (forward: 5′-TTYGGWGCYGAYCARTTTG-3′ and 5′-CYMTGYACDGTKACDCARGTNG; reverse: 5′-CTGGAGATTGRTCRTAGAARAAC-3′) that were designed on conserved regions among plant peptide transporters. The fragment obtained was cloned into pUC18 and sequenced. To isolate the full-length cDNA, a positively identified clone was labeled with [α-32P]dCTP (Random Primer Labeling kit; Amersham Pharmacia Biotech, Buckinghamshire, UK), and was used as probe for screening a cDNA library prepared from developing cotyledons of faba bean (Heim et al., 1993). Library screening was performed as described in Buchner et al. (1996) at a temperature of 60°C for hybridization and washing.
Functional Complementation of a Yeast Mutant
For functional characterization, the complete VfPTR1 cDNA was cloned into pDR195 for transformation of the yeast mutant LR2 (MATα, hip1-614, his4-401, can1, ino1, ura3–52, and PTR2Δ::hisG; Rentsch et al., 1995). Tests for complementation of this mutant by VfPTR1 were performed by streaking the transformed cells on synthetic complete (SC) medium supplemented with 10 mm His-Ala. For nonselective growth, cells were plated on SC medium supplemented with 20 mm His. The positive control consisted of cells carrying the peptide transporter AtPTR2 cDNA of Arabidopsis (Rentsch et al., 1995). Cells transformed with AtAAP2 (Kwart et al., 1993) or with an “empty” pDR195 vector were used as negative controls.
RNA Isolation and Northern Analyses
Total RNA isolation was performed as described by Heim et al. (1993) or with the Purescript RNA Isolation kit (Gentra Systems, Minneapolis) and were separated in 1% (w/v) agarose gels containing 15% (w/v) formaldehyde, and blotted overnight onto Hybond-N+ nylon membranes (Amersham Pharmacia Biotech). DNA fragments that excluded the poly(A) tail were randomly labeled with [α-32P]dCTP and were used as probes. Hybridizations were performed at 65°C and were washed at high stringency according to Church and Gilbert (1984). For quantitative northern analyses, signals on the membranes were quantified using a phosphorimager (Fuji-BAS; Fuji Photo Film, Tokyo), and loading was normalized by hybridizing the filters with a cDNA probe corresponding to the ubiquitin gene.
In Situ Hybridization
Sample fixation and slide preparation followed the protocol described in Weber et al. (1995) with the only exception being that poly(A)-coated slides (Sigma-Aldrich Chemie) were used. VfAAP1, VfPTR1, and VfSUT1 were enzymatically digested to exclude the poly(A) tails. The resultant cDNA fragments were randomly labeled with [α-33P]dCTP and were used as probes. Slides were hybridized overnight at 42°C and were washed with 50% (w/v) formamide in 0.1× SSC. After drying, slides were coated with photoemulsion (LM-1; Amersham Pharmacia Biotech), exposed for 3 to 10 d at 4°C, and developed with D-19 developer (Eastman-Kodak, Rochester, NY). If necessary, slides were counterstained with 0.05% (w/v) toluidine blue.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.024422.
This work was supported by the Deutsche Forschungsgemein-schaft (project no. WE1614/5–1)
Footnotes
In the present report, AtPTR2-A (Steiner et al., 1994) will not be regarded as a plant PTR following the author's correction (Steiner et al., 2000). The denomination AtPTR2 will be used as a synonym for AtPTR2-B (Song et al., 1996), AtNTR1 (Frommer et al., 1994a), and AtOPT1 as in Rentsch et al. (1998), as they all correspond to the same PTR isoform.
References
- Barneix AJ, Arnozis PA, Guitman MR (1992) The regulation of nitrogen accumulation in the grain of wheat plants (Triticum aestivum). Physiol Plant 86: 609–615 [Google Scholar]
- Bewley JD, Black M (1994) Seeds. Physiology of Development and Germination. Plenum Press, New York
- Bick JA, Neelam A, Hall JL, Williams LE (1998) Amino acid carriers of Ricinus communis expressed during seedling development: molecular cloning and expression analysis of two putative amino acid transporters, RcAAP1 and RcAAP2. Plant Mol Biol 36: 377–385 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Weber H, Panitz R, Manteuffel R, Wobus U (1995) Embryogenesis in Vicia faba L.: histodifferentiation in relation to starch and storage protein synthesis. J Plant Physiol 147: 203–218 [Google Scholar]
- Buchner P, Borisjuk L, Wobus U (1996) Glucan phosphorylases in Vicia faba L.: cloning, structural analyses and expression patterns of cytosolic and plastidic forms in relation to starch. Planta 199: 64–73 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W-N, André B, Rentsch D, Krolklewicz S, Tegeder M, Breitkreuz K, Frommer WB (1998) Amino acid transport in plants. Trends Plant Sci 3: 188–195 [Google Scholar]
- Fischer W-N, Kwart M, Hummel S, Frommer WB (1995) Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Fischer W-N, Loo DDF, Koch W, Ludewig U, Boorer KJ, Tegeder M, Rentsch D, Wright EM, Frommer WB (2002) Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J 29: 717–731 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D (1994a) Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett 347: 185–189 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Kwart M, Hirner B, Fischer WN, Hummel S, Ninnemann O (1994b) Transport for nitrogenous compounds in plants. Plant Mol Biol 26: 1651–1670 [DOI] [PubMed] [Google Scholar]
- Gamba G (2001) Alternative splicing and diversity of renal transporters. Am J Physiol Renal Physiol 281: F781–F794 [DOI] [PubMed] [Google Scholar]
- Golombek S, Rolletschek H, Wobus U, Weber H (2001) Control of storage protein accumulation during legume seed development. J Plant Physiol 158: 457–464 [Google Scholar]
- Gunning BES, Pate JS (1974) Transfer cells. In AW Robards, ed, Dynamic Aspects of Plant Ultrastructure. McGraw-Hill, London, pp 441–480
- Hayati R, Egli DB, Crafts-Brandner SJ (1996) Independence of nitrogen supply and seed growth in soybean: studies using an in vitro culture system. J Exp Bot 47: 33–40 [Google Scholar]
- Heim U, Wang Q, Kurz T, Borisjuk L, Golombek S, Neubohn B, Adler K, Gahrtz M, Sauer N, Weber H et al. (2001) Expression patterns and subcellular localization of a 52 kDa sucrose-binding protein homologue of Vicia faba (VfSBPL) suggest different functions during development. Plant Mol Biol 47: 461–474 [DOI] [PubMed] [Google Scholar]
- Heim U, Weber H, Bäumlein H, Wobus U (1993) A sucrose-synthase gene of Vicia faba L.: expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta 191: 394–401 [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW (1978) Peptide transport by germinating barley embryos: evidence for a single common carrier for di- and oligopeptides. Planta 138: 217–221 [DOI] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14: 535–544 [DOI] [PubMed] [Google Scholar]
- Jamai A, Laloi M, Bourbouloux A, Valantin M, Delrot S (1996) Characterization of leucine-leucine transport in leaf tissues. J Exp Bot 47: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer W (1993) Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J 4: 993–1002 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Koerselman-Kooij JW, Borslap AC (1990) Changing kinetics of l-valine uptake by immature pea cotyledons during development: An unsaturable pathway is supplemented by a saturable system. Planta 181: 576–582 [DOI] [PubMed] [Google Scholar]
- Lin C-M, Koh S, Stacey G, Yu S-M, Lin T-Y, Tsay Y-F (2000) Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol 122: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Heim U, Sauer N, Wobus U, Weber H (2001) Amino acid permeases in developing seeds of Vicia faba L.: expression precedes storage protein genes and is regulated by amino acid supply. Plant J 28: 61–71 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineer 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Nishizawa NK, Mori S (2001) Direct uptake of macro-organic molecules. In N Ae, J Arihara, K Okada, A Srinivasan, eds, Plant Nutrient Acquisition: New Perspectives. Springer-Verlag, Tokyo, pp 421–444
- Offler CE, Nerlich SM, Patrick JW (1989) Pathway of photosynthate transfer in the developing seed of Vicia faba L.: transfer in relation to seed anatomy. J Exp Bot 40: 769–780 [Google Scholar]
- Offler CE, Patrick JW (1993) Pathway of photosynthate transfer in the developing seed of Vicia faba L.: a structural assessment of the role of transfer cells in unloading from the seed coat. J Exp Bot 44: 711–724 [Google Scholar]
- Patrick JW, Offler CE (1995) Post-sieve element transport of sucrose in developing seeds. Aust J Plant Physiol 22: 681–702 [Google Scholar]
- Persans MW, Nieman K, Salt DE (2001) Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA 98: 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Boorer KJ, Frommer WB (1998) Structure and function of plasma membrane amino acid, oligopeptide and sucrose transporters from higher plants. J Membrane Biol 162: 177–190 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB (1996) Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permease genes identified by suppression of an amino acid targeting mutant. Plant Cell 8: 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Schlereth A, Becker C, Horstmann C Tiedemann J, Müntz K (2000) Comparison of globulin and cysteine proteinases in embryonic axes and cotyledons during germination and seedling growth of vetch (Vicia sativa L.). J Exp Bot 51: 1423–1433 [PubMed] [Google Scholar]
- Schulze W, Frommer WB, Ward JM (1999) Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. Plant J 17: 637–646 [DOI] [PubMed] [Google Scholar]
- Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G (1997) Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol 114: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Steiner HY, Zhang L, Naider F, Stacey G, Becker JM (1996) Cloning of a second Arabidopsis peptide transport gene. Plant Physiol 110: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopanen T, Burston D, Matthews DM (1977) Uptake of small peptides by the scutellum of germinating barley. FEBS Lett 79: 4–7 [DOI] [PubMed] [Google Scholar]
- Stacey G, Koh S, Granger C, Becker JM (2002) Peptide transport in plants. Trends Plant Sci 7: 257–263 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Papa C, Huang JF, Rhee Y (1994) Purification of the major soybean leaf acid phosphatase that is increased by seed-pod removal. Plant Physiol 104: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner HY, Song W, Zhang L, Naider F, Becker JM, Stacey G (1994) An Arabidopsis peptide transporter is a member of a new class of membrane transport proteins. Plant Cell 6: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner HY, Song W, Zhang L, Naider F, Becker JM, Stacey G (2000) Corrections. Plant Cell 12: 2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Wang XD, Frommer WB, Offler CE, Patrick JW (1999) Sucrose transport into developing seeds of Pisum sativum L. Plant J 18: 151–161 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analyses of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Thorne JH (1985) Phloem unloading of C and N assimilates in developing seeds. Annu Rev Plant Physiol 36: 317–343 [Google Scholar]
- Tiedemann J, Neubohn B, Müntz K (2000) Different functions of vicilin and legumin are reflected in the histopattern of globulin mobilization during germination of vetch (Vicia sativa L.). Planta 211: 1–12 [DOI] [PubMed] [Google Scholar]
- VerNooy CD, Thorne JH, Lin W, Rainbird M (1986) Cessation of assimilate uptake in maturing soybean seeds. Plant Physiol 82: 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, West CE, Bray CM (2001) The physiology and molecular biology of peptide transport in seeds. Seed Sci Res 11: 275–284 [Google Scholar]
- Waterworth WM, West CE, Daws MI, Bray CM (2000) The barley scutellar peptide transporter: relationship to germination and lost of seed viability. In M Black, KJ Bradford, J Vázquez-Ramos, eds, Seed Biology: Advances and Applications. CAB International, Wallingford, UK pp 297–308
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U (1997a) A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in faba bean seeds. Plant Cell 9: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (1997b) Sugar import and metabolism during seed development. Trends Plant Sci 2: 169–174 [Google Scholar]
- Weber H, Heim U, Borisjuk L, Wobus U (1995) Cell-type specific, coordinate expression of two ADP-glucose pyrophosphorylase genes in relation to starch biosynthesis during seed development in Vicia faba L. Planta 195: 352–361 [DOI] [PubMed] [Google Scholar]
- Weber H, Heim U, Golombek S, Borisjuk L, Wobus U (1998) Assimilate uptake and the regulation of seed development. Seed Sci Res 8: 331–345 [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM (1998) Cloning and functional characterization of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. Plant J 15: 221–229 [DOI] [PubMed] [Google Scholar]
- Weston K, Hall JL, Williams LE (1994) Characterization of a glutamine/proton co-transporter from Ricinus communis roots using isolated plasma membrane vesicles. Physiol Plant 91: 623–630 [Google Scholar]
- Williams L, Miller A (2001) Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Physiol Plant Mol Biol 52: 659–688 [DOI] [PubMed] [Google Scholar]
- Wittenbach VA (1982) Effect of pod removal on leaf senescence in soybeans. Plant Physiol 70: 1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus U, Borisjuk L, Panitz R, Manteuffel R, Bäumlein H, Wohlfahrt T, Heim U, Weber H, Misera S, Weschke W (1995) Control of seed storage protein gene expression: new aspects on an old problem. J Plant Physiol 145: 592–599 [Google Scholar]
- Yamagata M, Matsumoto S, Ae N (2001) Possibility of direct acquisition of organic nitrogen by crops. In N Ae, J Arihara, K Okada, A Srinivasan, eds, Plant Nutrient Acquisition: New Perspectives. Springer-Verlag, Tokyo, pp 399–420
- Yokoyama T, Kodama N, Aoshima H, Izu H, Matsushita K, Yamada M (2001) Cloning of a cDNA for a constitutive NRT1 transporter from soybean and comparison of gene expression of soybean NRT1 transporter. Biochim Biophys Acta 1518: 79–86 [DOI] [PubMed] [Google Scholar]
- Zhou J-J, Theodoulou FL, Muldin I, Ingemarsson B, Miller AJ (1998) Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J Biol Chem 273: 12017–12023 [DOI] [PubMed] [Google Scholar]