Abstract
The evolutionarily conserved Arp2/3 complex has been shown to activate actin nucleation and branching in several eukaryotes, but its biological functions are not well understood in multicellular organisms. The model plant Arabidopsis provides many advantages for genetic dissection of the function of this conserved actin-nucleating machinery, yet the existence of this complex in plants has not been determined. We have identified Arabidopsis genes encoding homologs of all of the seven Arp2/3 subunits. The function of the putative Arabidopsis Arp2/3 complex has been studied using four homozygous T-DNA insertion mutants for ARP2, ARP3, and ARPC5/p16. All four mutants display identical defects in the development of jigsaw-shaped epidermal pavement cells and branched trichomes in the leaf. These loss-of-function mutations cause mislocalization of diffuse cortical F-actin to the neck region and inhibit lobe extension in pavement cells. The mutant trichomes resemble those treated with the actin-depolymerizing drug cytochalasin D, exhibiting stunted branches but dramatically enlarged stalks due to depolarized growth suggesting defects in the formation of a fine actin network. Our data demonstrate that the putative Arabidopsis Arp2/3 complex controls cell morphogenesis through its roles in cell polarity establishment and polar cell expansion. Furthermore, our data suggest a novel function for the putative Arp2/3 complex in the modulation of the spatial distribution of cortical F-actin and provide evidence that the putative Arp2/3 complex may activate the polymerization of some types of actin filaments in specific cell types.
Actin nucleation is critical for the reorganization and dynamics of the actin cytoskeleton. Two actin nucleation factors have been identified in yeast and animals. Formins nucleate actin polymerization required for actin cable formation in yeast (Evangelista et al., 2002; Sagot et al., 2002). The Arp2/3 complex activates the polymerization of branched F-actin and promotes actin nucleation from the side of an existing actin filament, leading to the formation of a branched actin network (Mullins et al., 1998; Svitkina and Borisy, 1999; Blanchoin et al., 2000; Cooper et al., 2001; Higgs and Pollard, 2001; Robinson et al., 2001). The Arp2/3 complex consists of seven subunits (ARP2, ARP3, ARPC1/p41, ARPC2/p31, ARPC3/p21, ARPC4/p20, and ARPC5/p16; Higgs and Pollard, 2001). ARP2 and ARP3 are actin-related proteins. The structural analysis of the activated Arp2/3 complex implicates Arp2 and Arp3 as the actin nucleation sites by forming a heterodimer that resembles an actin dimer (Cooper et al., 2001; Robinson et al., 2001). The localization of the Arp2/3 complex is concentrated in the actin patches in yeast cells and at the leading edge of migrating animal cells (Welch et al., 1997a; Morrell et al., 19992; Schaerer-Brodbeck and Riezman, 2000a, 2000b; Higgs and Pollard, 2001). As a multifunctional organizer, the cellular roles of the Arp2/3 complex involve the control of actin polymerization nucleation for many cell functions such as cell movement, cell division, endocytosis, vesicle trafficking, and bacterial invasion (Welch et al., 1997b; Machesky and Gould, 1999; Schaerer-Brodbeck and Riezman, 2000a, 2000b; Higgs and Pollard, 2001). The Arp2/3 complex was discovered in Amoeba sp. cells (Machesky et al., 1994), but its function has been primarily studied genetically in the single-celled yeast and biochemically in mammalian cell culture. In complex multicellular organisms, the biological function of the Arp2/3 complex has been studied only in fruitfly (Drosophila melanogaster). A mutation of one Arp2/3 complex subunit, ARPC1, causes spindle fusion because of pseudocleavage furrow disruption in syncytical fruitfly embryos (Hudson and Cooley, 2002; Stevenson et al., 2002). Arabidopsis, with numerous advantages for genetic studies including the availability of various collections of T-DNA insertion mutants, provides a good model system to study the function of the Arp2/3 complex in higher eukaryotes.
The actin cytoskeleton is an important component of plant cell growth and development. Pharmacological and genetic studies suggest its role in a variety of cellular processes such as organelle movement, guard cell movement, cell polarity development, polarized cell growth, and cell division (Volkmann and Baluska, 1999; Belanger and Quatrano, 2000; Vantard and Blanchoin, 2002). Using live markers such as green fluorescent protein (GFP; Kost et al., 1998) fused to the actin-binding domain of the mouse talin and phalloidin staining in fixed cells, different types of actin structures have been observed in plant cells (Szymanski et al., 1999; Hepler et al., 2001; Frank and Smith, 2002). Actin cables, presumably composed of unbranched actin filaments, are detected in all living plant cells, whereas dynamic F-actin, composed of diffuse F-actin, is seen at the front of tip-growing pollen tubes and root hairs (Fu et al., 2001; Jones et al., 2002). Diffuse F-actin is also associated with the expanding parts of diffusely growing cells (Fu et al., 2002). The formation of both types of diffuse F-actin is dependent upon the Rho related GTPase from plant (ROP) subfamily of Rho GTPases, the plant relative of animal Rac/Cdc42 subfamilies of Rho GTPases known to signal the activation of the Arp2/3 complex (Fu et al., 2001, 2002; Higgs and Pollard, 2001; Jones et al., 2002). The assembly, reorganization, and dynamics of specific actin structures are regulated by intracellular and environmental signals in plants (McCurdy et al., 2001). Many of the conserved actin-binding proteins that regulate actin dynamics and organization have been identified and studied such as ADF, profilin, fimbrin, and actin motor proteins (Kovar et al., 2000; Ramachandran et al., 2000; Dong et al., 2001; McCurdy et al., 2001; McKinney et al., 2001). However, the molecular mechanism for actin nucleation in plants remains unknown.
The possible existence of the Arp2/3 complex in plants was suggested by the cloning of the AtARP2 gene encoding the ARP2 homolog (Klahre and Chua, 1999). We have now identified Arabidopsis genes encoding six additional putative subunits of the Arp2/3 complex. These genes are ubiquitously expressed in various tissues. T-DNA insertions into any of the Arp2/3 subunit genes examined induced cell shape changes in epidermal pavement cells and trichomes in the leaf. The defects were associated with mislocalization of diffuse cortical F-actin and reduction of a fine actin network in both pavement cells and trichomes. This work provides genetic evidence showing a role for the Arp2/3 complex in cell morphogenesis in higher eukaryotes.
RESULTS
The Arabidopsis Genome Encodes All of the Putative Subunits of the Arp2/3 Complex
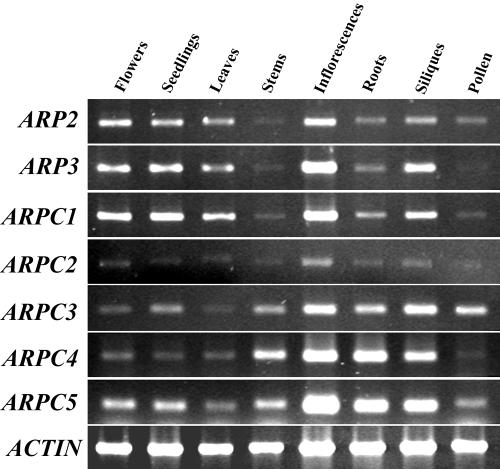
Conducting a blast search with the yeast Arp2/3 complex subunits, we identified six new putative Arp2/3 complex subunits from the Arabidopsis genome sequence. The coding sequences of Arabidopsis ARP3, ARPC1/p41, and ARPC3/p21 were predicted by The Arabidopsis Information Resource database, and the coding sequences of ARPC2/p34, ARPC4/p20, and ARPC5/p16 were predicted based on their homologous gene organizations in yeast. The coding sequences of the six subunits were confirmed through cloning and sequencing of cDNAs by reverse transcriptase (RT)-PCR using primers (supplemental data; they can be viewed at http://www.plantphysiol.org). Table I shows a comparison of the Arabidopsis homologs with those of human, Dictyostelium sp., fruitfly, budding yeast, and fission yeast (Schizosaccharomyces pombe). AtARP3, like AtARP2, shares about 50% identity with Arabidopsis actins in amino acid sequence (data not shown). All of the Arabidopsis Arp2/3 complex subunits except for ARPC1/p41 are encoded by a single gene. ARPC1/p41 is encoded by two genes arranged in reverse orientation and in close proximity. The two Arabidopsis ARPC1/p41 genes share very high nucleotide sequence identity both in the coding and promoter region. Protein motif analysis revealed seven WD-40 repeats in both ARPC1/p41 sequences. ARPC1/p41, ARPC3/p21, and ARPC4/p20 share considerable amino acid identity with their counterparts in other organisms. As in other organisms, ARPC2/p34 and ARPC5/p16 are the most diverged of the seven subunits. RT-PCR analysis shows that transcripts for all seven subunits are expressed in all tissues examined (Fig. 1). All of the subunits have low expression levels, but with relatively high expression in inflorescences. Among these seven subunits, ARPC2/p34 and ARPC4/p20 have the lowest expression.
Table I.
Comparison of Arabidopsis Arp2/3 complex subunits with their counterparts in other organisms
| Human | Fruitfly | Dictyostelium sp. | Fission Yeast | Budding Yeast | |
|---|---|---|---|---|---|
| ARP3 | 56a | 55 | 57 | 53 | 51 |
| ARPC1/p41 | 41 | 37 | 43 | 35 | 33 |
| ARPC2/p34 | 26 | 26 | 33 | 26 | 28 |
| ARPC3/p21 | 47 | 42 | 41 | 43 | 39 |
| ARPC4/p20 | 65 | 61 | 68 | 56 | 59 |
| ARPC5/p16 | 32 | 35 | 31 | 25 | 23 |
The number indicates percentage of identity of predicted amino acid sequences of Arabidopsis Arp2/3 subunits compared with those from other species
Figure 1.
RT-PCR analysis of the transcript expression of the Arabidopsis Arp2/3 subunit genes in various tissues. RNA was isolated from organs of Arabidopsis COL-0 WT plants grown in light and used for RT-PCR analysis as described in the text. A 25-cycle PCR reaction was carried out for all genes except for ARPC4/p20, for which 30 cycles were used. ACTIN2 was used as a control except for pollen RT-PCR that involves the use of the ACTIN3 gene.
Loss of Function Mutants for the Putative Arabidopsis Arp2/3 Subunits
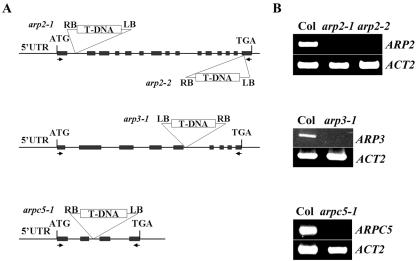
To investigate the function of Arp2/3 subunits, we obtained several mutants for Arabidopsis Arp2/3 subunits from the SALK Arabidopsis T-DNA mutant collection (Fig. 2). arp2-1 and arp2-2 were isolated with a T-DNA insertion in the first intron and the last exon, respectively. arpc5-1/p16-1 was isolated with a T-DNA insertion in the second intron, and arp3-1 contains a T-DNA insertion in the fifth intron. RT-PCR analysis revealed that none of these four mutant lines had detectable transcripts for their respective genes (Fig. 2). We consider them to be strong and possibly null alleles. Homozygous mutant plants with a single T-DNA insertion were obtained for all four mutants. By visual examination of these mutants under normal growth conditions in soil or on agar plates, we did not observe obvious changes in morphology as a whole plant in darkness or light. This is surprising, given the importance of actin nucleation in the regulation of actin organization as well as the essential role for the Arp2/3 complex in yeast.
Figure 2.
Characterization of the Arp2/3 subunit mutants. A, The location of the T-DNA insertions in arp2-1, arp2-2, arp3-1, and arpc5-1/p16-1 are shown on the maps of AtARP2, AtARP3, and AtARPC5/p16 genes. Arrows indicate the location of primers used for RT-PCR analysis of transcripts. Exons are boxed and introns and untranscribed flanking sequences are shown as lines. B, RT-PCR analysis of transcript levels for arp2-1, arp2-2, arp3-1, and arpc5/p16-1 mutants. Total RNA was extracted from inflorescences of each homozygous mutant plant and WT plants. The full-length coding sequence was amplified for each primer set.
The Putative Arabidopsis Arp2/3 Complex Is Required for Proper Leaf Epidermal Cell Morphogenesis
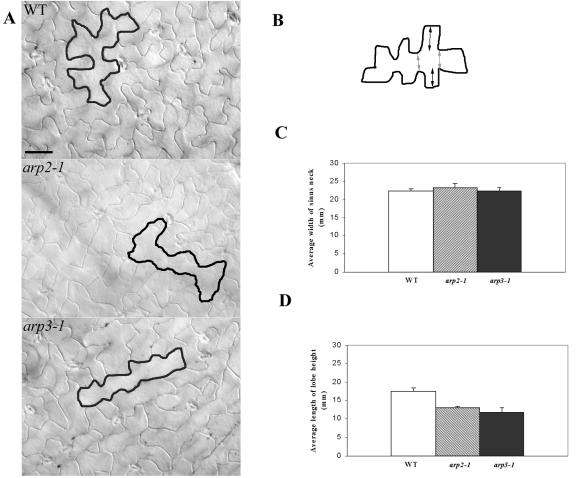
We next investigated whether the mutants are altered in cell shape formation, which has been linked to diffuse cortical F-actin (Szymanski et al., 1999; Fu et al., 2001, 2002; Frank and Smith, 2002; Jones et al., 2002; Frank et al., 2003). We first examined tip growing cells, root hairs, and pollen tubes. Surprisingly, neither the growth of pollen tubes nor root hairs was dramatically affected in all four mutants grown under normal conditions (data not shown). We then examined a possible effect of the Arp2/3 mutations on pavement cells, because their development has been linked to cortical F-actin as well. Mature pavement cells on the adaxial side of a wild-type (WT) leaf are jigsaw puzzle shaped with a striking outgrowth of lobes, giving a wavy cell outline. In all four mutants, including arp2-1, arp2-2, arp3-1, and arpc5-1, the outline of pavement cells was smoother and much less wavy (Fig. 3A). Quantitative analysis showed that the neck width was not affected, whereas the lobe height was dramatically reduced (Fig. 3, B–D).
Figure 3.
Characterization of leaf pavement cell shapes in arp2-1 and arp3-1 plants. A, Leaf pavement cell shapes. The fourth rosette leaves from 2-week-old plants were cleared, and differential interference contrast images were taken from the middle region of the cleared leaves. Leaves from WT plants and all mutant plants are of similar sizes. Tracing was used to highlight the cell shape of a typical pavement cell. Bars = 30 μm. B, A schematic diagram of a leaf pavement cell illustrates how the lobe height and neck width were measured as shown in C and D. The distance indicated by dark arrows represents the lobe height. The distance indicated by light arrows represents neck width. C, Comparison of the average width of the neck region between WT and mutant pavement cells. Measurements in both C and D were carried out using the fourth leaf of a 2-week-old plant, and 150 epidermal cells were used for each mutant line as well as WT. Statistical test (t test) shows no significant difference in neck width between WT and mutant pavement cells. D, Comparison of the average length of lobe heights between WT and mutant pavement cells. Statistical test (t test) shows that the length of lobe height is significantly reduced in the mutant pavement cell compared with the WT pavement cell.
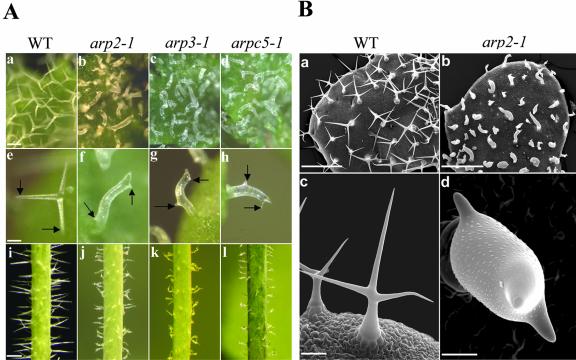
We next examined the effects of mutation in the Arp2/3 complex on trichome morphogenesis. We did this because F-actin has been shown to play an important role in morphogenesis of this epidermal cell type (Mathur et al., 1999; Szymanski et al., 1999; Schwab et al., 2003). As shown in Figure 4, we observed a very striking defect in trichome morphogenesis in the mutant plants (Fig. 4). WT Arabidopsis trichomes typically have three branches. Mutants including arp2-1, arp2-2, arp3-1, and arpc5-1 have very severe and identical trichome defects. Trichomes in these mutants had stunted branch outgrowth and unrestricted stalk expansion. Despite the striking defects in the trichome shape, trichome branch initiation was normal in all four mutants. The WT trichomes on the stem are usually without branches or only with two branches. The trichomes on the mutant stems clearly showed cell shape defects featured as a dramatic increase in radial expansion and irregular trichome shapes. The mutant trichome phenotype is very similar to that caused by treatment with actin-disrupting drugs such as cytochalasin D or by mutations in the DISTORTED genes (Mathur et al., 1999; Schwab et al., 2003). This phenotype can be attributed to the combined defects in cell polarity development in stalks and polar growth in branches. Because branches are formed, the polarity establishment for branch formation is not affected.
Figure 4.
Trichome phenotypes of arp2-1, arp3-1, and arpc5-1 mutants. A, Phenotype analyses of arp2-1, arp3-1, and arpc5-1 mutant trichomes using light microscopy. a to d, Trichomes on a young leaf from WT and mutant plants. e to h, Single trichomes with branches from WT and mutant plants. Arrows point to the branches. i to l, Trichomes on stems from WT and mutant plants. a, e, and i are from WT plants. b, f, and j are from arp2-1 plants. c, g, and k are from arp3-1 plants. d, h, and l are from arpc5-1 plants. Arrows in e to h point to the branches. Bar in a (100 μm) is the same for b through d. Bar in e (50 μm) is the same for f through h. Bar in i (500 μm) is the same for j through l. B, Scanning electron microscopic analyses of arp2-1 trichomes. a and b show mature trichomes on a young leaf from WT and arp2-1 plants, respectively. c and d show single trichomes with the typical three branches from WT and arp2-1 plants, respectively. Bar in a and b = 500 μm. Bar in c = 100 μm. Bar in d = 20 μm.
The fact that arp2-1, arp2-2, arp3-1, and arpc5-1 show almost identical phenotypes in leaf epidermal cell morphogenesis strongly suggests that the observed phenotype is due to a defect in the activity of the putative Arp2/3 complex. In fact, we have just obtained a T-DNA insertion mutant for ARPC4/p20 from the SALK collection that shows an identical phenotype to the four mutants (S. Li, unpublished data). Furthermore, a second T-DNA insertion mutant for ARP3 obtained from a different T-DNA insertion population showed the same phenotype as arp3-1. Therefore, we conclude that the putative ARP2/3 complex plays a critical role in the control of cell morphogenesis in Arabidopsis leaf epidermal pavement cells and trichomes. In the root epidermis, root hairs are slightly wavy, but other epidermal cells in the root appear to be normal in shape and size (data not shown).
The Spatial Distribution and Formation of Cortical F-Actin Is Altered in the Arp2/3 Subunit Mutants
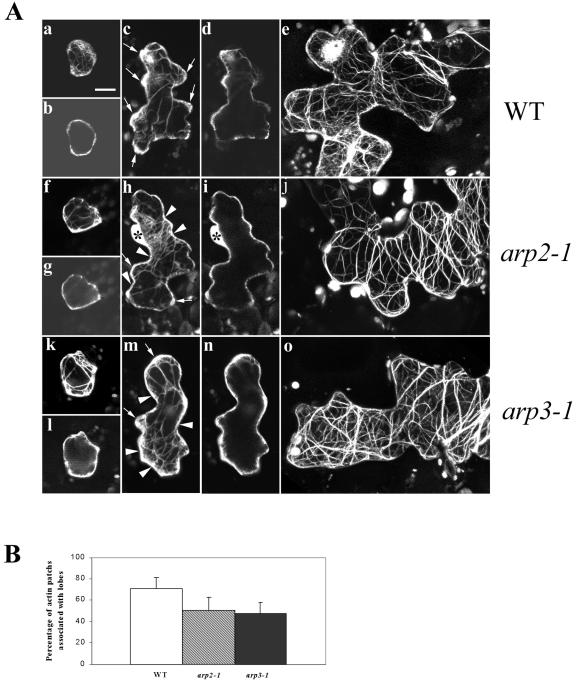
To understand the mechanism by which the putative Arabidopsis Arp2/3 complex modulates cell morphogenesis, we investigated the actin cytoskeleton structure in these Arp2/3 subunit mutants. To visualize F-actin, an actin-binding domain of the mouse talin tagged with GFP (GFP-mTalin) was transiently expressed in leaf epidermal cells of mutant arp2-1 and arp3-1 as well as WT plants (Fu et al., 2002). In WT plants, the young pavement cells capable of expanding in all directions characteristically contain strong diffuse F-actin throughout the entire cell periphery, and a few actin cables localize to the cell cortex and the cytoplasm. At this stage, no obvious F-actin structural change is detected in the pavement cells of either arp2-1 or arp3-1 mutants (Fig. 5A). When the WT pavement cells develop expanding lobes, patches of strong diffuse F-actin are found at the tips of the extending lobes. These actin patches are rarely observed in the nonexpanding neck region of WT pavement cells. In both arp2-1 and arp3-1 mutants, the lobes are initiated, but their growth is inhibited. Interestingly, the patches of strong cortical diffuse F-actin typically associated with lobes were reduced. Instead, cortical diffuse F-actin was more evenly distributed throughout the cell periphery. A quantitative analysis was performed to measure the percentage of association of strong diffuse F-actin with lobes in stage II cells from WT and mutant plants. In the mutant plants, the percentage of association of strong diffuse F-actin with lobes is significantly reduced compared with that of WT plants. Consequently, in the mutant cells with extending lobes, cortical diffuse F-actin was frequently found in the neck region. At this stage, actin cables are not extensive and seem to be randomly distributed both in WT and mutant pavement cells. In mature WT leaf pavement cells, F-actin exists mainly as extensive long actin cables that appear to be oriented randomly. In the mature pavement cells of arp2-1 and arp3-1 mutants, the actin cables were also quite extensive but tended to be oriented perpendicularly to the long axis of the cell. These observations suggest that mutations in the Arp2/3 subunit genes affect the spatial distribution, but not the assembly, of either cortical diffuse F-actin or actin bundles in Arabidopsis leaf pavement cells.
Figure 5.
Analysis of actin organization in leaf pavement cells of arp2-1 and arp3-1 plants. A, F-actin organization in leaf pavement cells from arp2-1 and arp3-1 plants visualized by GFP-mTalin at different developmental stages. Leaves from 10-d-old plants were bombarded with GFP-mTalin construct, and cells expressing GFP-mTalin were imaged using confocal microscopy as described in the text. Pavement cells at three developmental stages are shown: b, g, and k, Stage I cells before lobe initiation; c, h, and m, stage II cells forming lobes; and e, j, and o, stage III cells with fully extended lobes. More than 60 cells were imaged for each stage of cell in every genotype. Shown are cells with representative actin organization. a to e, Leaf pavement cells of WT plants. f to j, Leaf epidermal cells of arp3-1 plants. k to o, Leaf epidermal cells of arp2-1 plants. a, c, e, f, h, j, k, m, and o show three-dimensional images projected from laser scanning. b, d, g, i, l, and n are single medial sections of a, c, f, h, k, and m, respectively. Arrows in c, h, and m point to the diffuse F-actin patches associated with growing lobes. Arrowheads in h and m point to diffuse F-actin associated with neck regions. Asterisks in h and i indicate the nucleus of the cell. Bar = 15 μm for all images. B, Quantitative analysis of association of diffuse cortical F-actin with the tip of expanding lobes in pavement cells. Measurements were done on 30 stage II cells by visual examination. The percentage of strong diffuse cortical F-actin associated with lobes for WT, arp2-1, and arp3-1 pavement cells are 70.83 ± 10.57, 50.22 ± 12.08, and 47.08 ± 10.91, respectively. Statistical analyses (t test) showed that the percentage of strong diffuse F-actin associated with lobes of both mutant pavement cells is significantly reduced (n = 30, P ≤ 0.05).
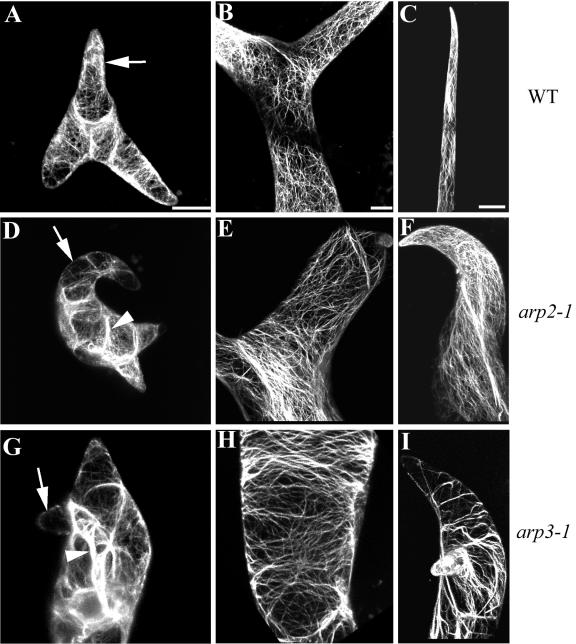
Actin organization in the trichomes of distorted mutants has been extensively characterized (Mathur et al., 1999; Szymanski et al., 1999; Schwab et al., 2003). To assess whether the Arabidopsis Arp2/3 complex affects the same pathway as the DISTORTED genes, we examined F-actin in the arp2-1 and arp3-1 mutant trichomes. Young WT trichomes contain network-like fine actin filaments that are oriented randomly and diffuse cortical F-actin in growing branches as well as the stalk. Thick actin cables are rarely detected in these cells. A network of fine actin bundles is found in the stalk of WT mature trichomes, whereas the mature branches usually contain longitudinal actin cables (Fig. 6). In both arp2-1 and arp3-1 mutants, actin structural defects were detected in the young as well as in the mature trichomes. Thick actin cables occurred in young mutant trichomes, whereas both fine actin network and diffuse F-actin were greatly reduced in their swollen stalks and stunted branches. In the mature mutant trichome, the severity of the F-actin structure alteration was also correlated with the deformation of the trichome. When the trichome shape was severely deformed with very limited growth of branches, transverse actin bundles were observed in both the stalk and the branches, analogous to the actin cytoskeleton in the mature pavement cells of the mutants. All of these actin cytoskeleton structure defects observed in Arp2/3 subunit mutants are similar to the description of DISTORTED mutants (Mathur et al., 1999; Schwab et al., 2003). Previous work has shown that disruption of the actin cytoskeleton by cytochalasin D induces the same cell shape changes in trichomes, suggesting that it is the loss of a certain actin population such as the fine actin network and diffuse actin that are responsible for the trichome phenotypes.
Figure 6.
Actin organization in trichomes of arp3-1 and arp2-1 plants. F-actin was visualized as described in Figure 5. All images shown are projections. Bar in A is for A, D, and G. Bar in B is for B, E, and H. Bar in C is for C, F, and I. Bar = 20 μm. A, D, and G, Young trichomes from a WT, arp2-1, and arp3-1 plants, respectively. Arrow in A points to diffuse cortical F-actin signal in a rapid growing branch. Arrows in D and G indicate the loss of fine F-actin in the mutant branches. Arrowheads in D and G point to thick actin cables. B, E, and H, Stalks of mature trichomes from WT, arp2-1, and arp3-1 plants, respectively. C, F, and I, Branches of mature trichomes from WT, arp2-1, and arp3-1 plant, respectively.
DISCUSSION
In this work, we have identified all subunits of the putative Arabidopsis Arp2/3 complex and investigated the function of three subunit genes using T-DNA insertion mutants. This work represents an important step of the description and functional analysis of a conserved actin-nucleating machinery in the plant kingdom. Importantly, we have demonstrated a critical role for the putative Arabidopsis Arp2/3 complex in cell polarity development and cell morphogenesis. Similar functional roles for this complex have been reported in yeast but not in any multicellular organisms. Interestingly, our results suggest that the putative Arabidopsis Arp2/3 complex modulates polar cell expansion probably through localizing diffuse cortical actin to the site of growth, a role previously unknown for the Arp2/3 complex in other organisms. However, this complex appears to control cell polarity development in trichomes through activating the formation of a network of cortical fine actin bundles. Similarly, this complex is known to activate the formation of cortical actin patches in yeast and a dense actin network in the leading edge of migrating Dictyostelium sp. and animal cells.
Genetic studies in yeast have shown that the Arp2/3 complex is an essential actin nucleation machinery (Winter et al., 1999). Deletion of any subunit gene causes defects in either actin patch formation or actin patch movement, leading to lethality. However, none of the homozygous T-DNA mutants for the Arabidopsis Arp2/3 subunits affect the viability of the plants or cause dramatic defects in organ development. This situation is somewhat similar to the observations in multicellular animals. The Arp2/3 complex has been identified in human and various animal model systems (fruitfly, Caenorhabditis elegans, and Dictyostelium sp.). The loss-of-function mutations in fruitfly ARPC1 and ARP3 genes only affect a subset of actin rearrangements and cause defects in ring canal expansion during oogenesis (Hudson and Cooley, 2002), whereas RNAi suppression of any of the Arp2/3 subunit genes in C. elegans only affect ventral closure (Sawa et al., 2003). Interestingly, none of our arp2/3 subunit mutants show dramatic developmental defects. It is possible that loss of Arp2, Arp3, or ARPC5 gene alone does not completely disrupt the activity of the complex. It has been shown that the human Arp2/3 complex subunits p34 and p16 are not essential for the activity of the complex in a reconstitution assay system (Gournier et al., 2001). It will be interesting to see whether a phenotype related to actin cytoskeleton dynamics is more severe in a mutant in which more than one Arabidopsis Arp2/3 subunit units are knocked out.
In animal cell systems, it has been proposed that a predominant role of this actin-nucleating complex is to promote the polymerization of branched actin filaments and the formation of a highly dense branched actin network in the leading edge that allows membrane protrusion and cell migration. Indeed, genetic studies in fly and worm have shown that the Arp2/3 complex is important for cell migration (Hudson and Cooley, 2002; Sawa et al., 2003). Therefore, our findings about the function of the Arp2/3 complex in cell morphogenesis in pavement cells and trichomes are interesting. Shape formation in these cells is dependent upon the spatial pattern of deposition and remodeling of cell walls, which bear the turgor pressure that drives cell expansion. This raises an interesting question: whether the plant Arp2/3 complex has a distinctly different cellular and biochemical function from the animal counterpart, or whether shape formation in leaf epidermal cells such as pavement cells and trichomes necessitates a more complex model than previously posed, one in which cells can accommodate targeted expansion requiring the Arp2/3 complex-mediated actin polymerization or remodeling.
The defects in the expansion of pavement cell lobes and trichome branches observed in all mutants examined define a role for the putative Arp2/3 complex in the modulation of polar cell expansion, which is crucial for cell morphogenesis in plants. How might the Arp2/3 complex control polar cell expansion? In leaf pavement cells, lobe expansion is thought to require a population of ROP GTPase-dependent diffuse F-actin localized to the tip of lobes, which might function to deliver cell wall and cell membrane materials to the active growing site via post-Golgi vesicles. In the Arp2/3 subunit mutant plants, lobes are formed, but their extension is severely inhibited. This is consistent with our observation that the diffuse cortical F-actin, although decreased in amount due to a more random distribution, is found in the lobes of the mutant pavement cells. The ectopic distribution of the diffuse cortical F-actin in the neck region in the mutant did not lead to increased expansion in that region, most likely due to the presence of a mechanism to suppress cell expansion here, such as transverse cortical microtubules (Mathur and Hulskamp, 2002). We propose that the random distribution of the diffuse cortical F-actin is the direct result of the defect in the Arp2/3 complex activity and that it is not due to cell shape changes per se. Evidence to support this is the fact that DN-rop2 expression also inhibits lobe extension but does not cause redistribution of the cortical F-actin (Fu et al., 2002). From these observations, we conclude that at least one function of the Arp2/3 complex is to modulate the localization of diffuse cortical F-actin in plant cells.
During trichome branch formation, a diffuse form of F-actin is also apparent near the tip of the branch in WT plants and is reduced in the stunted mutant branches. This reduction could be due to its random distribution, as occurs in pavement cells, or to its reduced assembly. The dramatic isotropic growth in the mutant trichome stalks makes it difficult to distinguish between these possibilities or to determine whether the change in the actin cytoskeleton is the result or the cause of cell shape changes. In maize (Zea mays) leaf pavement cells, mutations in BRK1, which encodes a homolog of HSPC300 that regulates the Arp2/3 complex in mammalian cells, eliminate lobes and associated diffuse cortical actin (Frank et al., 2003). This raises the possibility that the Arp2/3 complex may be required for the assembly of a fine actin network in some plant cells. Additionally or alternatively, the Arabidopsis Arp2/3 complex could regulate polar cell expansion through other mechanisms, e.g. targeting of post-Golgi vesicles or movement of other endomembrane structures or organelles (which ultimately affect polar secretion). Arp2/3 complex-mediated actin polymerization has been shown to regulate the trafficking of vesicles (Rozelle et al., 2000; Fucini et al., 2002; Luna et al., 2002; Stamnes, 2002) and pathogen movement in animal cells (Welch et al., 1997b). In budding yeast, arc35-1, a mutation in the ARPC2 subunit, affects calmodulin localization to the active growing sites where rapid endocytosis and exocytosis occur. Overexpression of calmodulin in the arc35-1 cells suppresses the defects both in the actin and microtubule cytoskeleton. Given that so many proteins need to be specifically targeted to their destination, the Arp2/3 complex may receive various signals to activate actin polymerization during multiple steps of vesicle trafficking to confer the specificity of vesicle targeting.
The isotropic growth in the trichome stalk of the Arp2/3 subunit mutants is very interesting because it suggests a role for the Arp2/3 complex in the control of cell polarity development. This role is similar to one of the roles of the yeast Arp2/3 complex (Morrell et al., 1999). Mutations in several Arp2/3 subunits cause defects in the establishment of cell polarity in the mother cell during cell division, leading to the unrestricted isotropic growth of the mother cell. This defect is thought to be due to the abolishment of actin patch-mediated endocytosis, which is essential for cell polarity establishment. Interestingly, we found that the swollen mutant stalk lacks the extensive cortical network of fine actin bundles normally found in WT trichome stalks, but contains some thick actin bundles. Complete disruption of F-actin by cytochalasin D phenocopies the Arp2/3 subunit mutants (Mathur et al., 1999), suggesting that it is the loss of some actin population(s) that is responsible for the depolarization of the stalk. The formation of thick actin bundles is probably an indirect effect due to the increased G-actin pool from the loss of a fine actin network. Thus, these observations imply that the Arabidopsis Arp2/3 complex controls the establishment of cell polarity by regulating the assembly of a fine actin network.
An interesting question would be: How can this actin network control cell polarity, through endocytosis as in yeast or by other mechanisms? It was proposed that the phenotype of distorted mutants is a result of disrupted directional vesicular trafficking (Mathur et al., 1999). Directed vesicle targeting could be controlled by the fine actin network or directly by the Arp2/3 complex-mediated actin polymerization. Interestingly, a recent study on the distorted mutants combined with cytochalasin treatments suggest that F-actin may regulate the orientation of cortical microtubules, which could directly control cell polarity (Mathur et al., 1999). Given the conserved functional role for the Arp2/3 complex in cell polarity (Mellman and Warren, 2000), it is important and interesting to determine the mechanism by which the Arabidopsis Arp2/3 complex regulates this fundamental process.
Another interesting question that needs future attention is how the Arabidopsis Arp2/3 complex is regulated. The Wiskott-Aldrich Syndrome domains family of proteins, the effectors of Cdc42 and the activators of the Arp2/3 complex, have not been found in the Arabidopsis genome. Furthermore, ROP GTPases do not seem to activate the formation of diffuse cortical F-actin through the Arp2/3 complex in pavement cells, and the DN-rop2 expression does not produce trichome defects, although it eliminates diffuse cortical F-actin in pavement cells (Fu et al., 2002). Additionally, arp2-1 and arp3-1 mutant plants have minor defects in root hair and pollen tube growth (S. Li, unpublished data), processes that show tip growth dependence on actin and ROP GTPases (Fu et al., 2001; Jones et al., 2002). These observations suggest that additional actin polymerization mechanism(s) exist in plants. In yeast and animals, various Arp2/3 activators that are structurally divergent have been identified besides the Cdc42/Wiskott-Aldrich Syndrome domains (Higgs and Pollard, 2001). Therefore, the plant Arp2/3 complex is likely to be regulated by novel mechanisms. The identification of the plant activators of the Arp2/3 complex will be of tremendous interest in the future. Development of an in vitro assay for the plant Arp2/3 complex will be essential for the identification of its activators.
MATERIALS AND METHODS
Database Search and Bioinformatics
To identify additional putative Arabidopsis Arp2/3 subunits, the yeast Arp2/3 subunit cDNAs (Winter et al., 1997) were used as queries for a BLAST search of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) and The Arabidopsis Information Resource database (http://www.arabidopsis.org). From this search, we identified putatively annotated coding sequences for genes of ARP3, ARPC1, and ARPC3. The coding sequences for ARPC2, ARPC4, and ARPC5 were predicted based on the gene organizations of their counterparts in yeast. The accession numbers for ARP3 ARPC1, ARPC2, ARPC3, ARPC4, and ARPC5 are: At1g13180, At2g30910/At2g31300, At1g30825, At1g60430, At4g14140, and At4g01710, respectively. The amino acid alignment was conducted by ClustalW program (http://www.ebi.ac.uk/clustalw).
DNA Manipulation and Plasmid Construction
To confirm that these six annotated Arp2/3 subunit sequences are indeed expressed, we amplified cDNAs for each subunit from a flower cDNA library (a gift from Hong Ma [Pennsylvania State University, University Park, PA]) using PCR primers covering the predicted Arp2/3 subunit-coding sequences (see supplemental data). PCR fragments of each were then subcloned into an AT cloning vector pGEM-T (Invitrogen, Carlsbad, CA). The resulting six clones were then sequenced.
T-DNA Mutant Identification and Reverse Transcription and PCR Analysis
Putative T-DNA lines were ordered from the Arabidopsis Biological Resource Center. The accession numbers for arp2-1, arp2-2, arp3-1, and arpc5-1/p16-1 are SALK_03448, SALK_077920, SALK_010045, and SALK_123936, respectively. For arp2-1, arp2-2, and arpc5-1/p16-1, T-DNA insertions were verified by PCR reactions using T-DNA LB primers and gene-specific primers. Homozygotes of each line were identified by RT-PCR using RNA extracted from inflorescences.
For the analysis of the transcription of each subunit, total RNA was isolated from different Arabidopsis tissues using the TRIZOL Reagent (Invitrogen). For silique and stem RNAs, two additional chloroform extractions were performed. Reverse transcription and PCR amplification were carried out as described previously. For the ARPC4 gene, 30 cycles of PCR amplification were used. For other Arp2/3 subunit genes, 25 cycles of PCR amplification (94°C for 45s, at 54°C for 45s, and at 72°C for 1 min 30s) were carried out using primers shown in Table I. As PCR amplification and loading controls, the same template cDNA was amplified using primers for the constitutive actin-2 gene. The actin-3 gene was used as a control in pollen RT-PCR. Twenty microliters of each PCR product was loaded on an 0.8% (w/v) agarose gel to visualize the amplified subunit cDNAs. Five microliters of actin PCR product was loaded as a control.
Light Microscopy Analysis of Leaf Epidermal Cell Shapes and Trichome Phenotypes
All experiments described in this study involve Arabidopsis ecotype Col-0. Arabidopsis plants were grown in growth chambers at 25°C under a light regime of 8 h of darkness and 16 h of light. The morphology of leaf pavement cells from WT and mutant plants was analyzed using cleared intact leaves as described previously (Fu et al., 2002). The fourth rosette leaf from 2-week-old plants was immersed in 5% (w/v) NaOH, boiled for 1 min, washed with distilled water, and incubated in bleach until it became clear. Cleared leaves were observed with an inverted microscope (Eclipse TE300, Nikon, Tokyo) equipped with a cooled CCD camera (C4742–95, Hamamatsu, Hamamatsu City, Japan). The measurement of neck width and lobe height of pavement cells was conducted using Metamorph 4.5 software (Universal Imaging, West Chester, PA). Trichome phenotypes were analyzed using a M2 Bio Epi-fluorescence microscope (Zeiss, Thornwood, NY) with a digital camera.
Scanning Electron Microscopy
Ten-day-old seedlings were fixed in 37% (w/v) formaldehyde, glacial acetic acid, 70% (v/v) ethanol (5:5:90, v/v) solution for 48 h and dehydrated in an ethanol series (70%, 75%, 80%, 85%, 90%, 95%, and 100%, v/v). After critical point drying and coating, samples were observed with a Philips XL30-FEG scanning electron microscope (FEI Company, Hillsboro, OR). Images were taken and processed using Adobe Photoshop 5.5 (Adobe Systems, Mountain View, CA)
Particle Bombardment-Mediated Transient Expression in Arabidopsis Leaves
Rosette leaves collected from 10-d-old plants were used for transient expression using a particle bombardment procedure described previously (Fu et al., 2002). All plasmid DNAs were amplified in the Escherichia coli strain Top 10 and purified using Qiagen plasmid Midi or Mini Kits according to the manufacturer's instructions (Qiagen USA, Valencia, CA). Routinely, 0.5-mg gold particles were coated with 0.5 μg of pBI221:GFP-mTalin DNA. Bombarded leaves were kept on damp filter paper before observation under the confocal microscope as described below.
Visualization of F-Actin and Confocal Microscopy
To visualize F-actin in the leaf pavement cells and trichomes, GFP-mTalin was expressed transiently as described above. Sixteen to 24 h after bombardment, leaf pavement cells or trichomes expressing GFP-mTalin were observed using a confocal microscope (MRC 600, Bio-Rad Laboratories, Hercules, CA) as described previously (Fu et al., 2001). Optical sections (0.5 μm) were used to collect a three-dimensional projection. Confocal images were analyzed using Metamorph 4.5 software and processed using Adobe Photoshop 5.5. For the quantification of association of strong diffuse F-actin with lobes, confocal projection images of actin cytoskeleton in stage II pavement cells from both WT and both mutant plants were randomly mixed together. The counting was based on visual examination. After sorting, the percentage of diffuse F-actin associated with lobes was assigned to individual samples.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We are grateful to Bin Shuai and Ying Gu for their assistance with the scanning electron microscopy. We thank Dr. Martha Orozco-Cardenas for her help with the microscopy.
This work was supported by the National Science Foundation (grant nos. IBN–0115078 and MCB–0111082 to Z.Y. and IBN 0077886 to E.M.L.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Belanger KD, Quatrano RS (2000) Polarity: the role of localized secretion. Curr Opin Plant Biol 3: 67-72 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD (2000) Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007-1011 [DOI] [PubMed] [Google Scholar]
- Cooper JA, Wear MA, Weaver AM (2001) Arp2/3 complex: advances on the inner workings of a molecular machine. Cell 107: 703-705 [DOI] [PubMed] [Google Scholar]
- Dong CH, Xia GX, Hong Y, Ramachandran S, Kost B, Chua NH (2001) ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A (2002) Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol 4: 260-269 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG (2003) Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130: 753-762 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Smith LG (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12: 849-853 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang Z (2001) Rop GTPase-dependent dynamics of tiplocalized F-actin controls tip growth in pollen tubes. J Cell Biol 152: 1019-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini RV, Chen JL, Sharma C, Kessels MM, Stamnes M (2002) Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol Biol Cell 13: 621-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD (2001) Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell 8: 1041-1052 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159-187 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70: 649-676 [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L (2002) A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol 156: 677-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Chua NH (1999) The Arabidopsis actin-related protein 2 (AtARP2) promoter directs expression in xylem precursor cells and pollen. Plant Mol Biol 41: 65-73 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393-401 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Staiger CJ, Weaver EA, McCurdy DW (2000) AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J 24: 625-636 [DOI] [PubMed] [Google Scholar]
- Luna A, Matas OB, Martinez-Menarguez JA, Mato E, Duran JM, Ballesta J, Way M, Egea G (2002) Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell 13: 866-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD (1994) Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilinagarose. J Cell Biol 127: 107-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Gould KL (1999) The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol 11: 117-121 [DOI] [PubMed] [Google Scholar]
- Mathur J, Hulskamp M (2002) Microtubules and microfilaments in cell morphogenesis in higher plants. Curr Biol 12: R669-R676 [DOI] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua N (1999) The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126: 5559-5568 [DOI] [PubMed] [Google Scholar]
- McCurdy DW, Kovar DR, Staiger CJ (2001) Actin and actin-binding proteins in higher plants. Protoplasma 215: 89-104 [DOI] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB (2001) Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development. Plant Cell 13: 1179-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Warren G (2000) The road taken: past and future foundations of membrane traffic. Cell 100: 99-112 [DOI] [PubMed] [Google Scholar]
- Morrell JL, Morphew M, Gould KL (1999) A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast. Mol Biol Cell 10: 4201-4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA 95: 6181-6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124: 1637-1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD (2001) Crystal structure of Arp2/3 complex. Science 294: 1679-1684 [DOI] [PubMed] [Google Scholar]
- Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL (2000) Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol 10: 311-320 [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D (2002) An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol 4: 626-631 [DOI] [PubMed] [Google Scholar]
- Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T (2003) Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci 116: 1505-1518 [DOI] [PubMed] [Google Scholar]
- Schaerer-Brodbeck C, Riezman H (2000a) Functional interactions between the p35 subunit of the Arp2/3 complex and calmodulin in yeast. Mol Biol Cell 11: 1113-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer-Brodbeck C, Riezman H (2000b) Saccharomyces cerevisiae Arc35p works through two genetically separable calmodulin functions to regulate the actin and tubulin cytoskeletons. J Cell Sci 113: 521-532 [DOI] [PubMed] [Google Scholar]
- Schwab B, Mathur J, Saedler RR, Schwarz H, Frey B, Scheidegger C, Hulskamp M (2003) Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: actin controls the spatial organization of microtubules. Mol Genet Genomics 11: (in press) [DOI] [PubMed]
- Stamnes M (2002) Regulating the actin cytoskeleton during vesicular transport. Curr Opin Cell Biol 14: 428-433 [DOI] [PubMed] [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE (2002) Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol 12: 705-711 [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11: 2331-2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard M, Blanchoin L (2002) Actin polymerization processes in plant cells. Curr Opin Plant Biol 5: 502-506 [DOI] [PubMed] [Google Scholar]
- Volkmann D, Baluska F (1999) Actin cytoskeleton in plants: from transport networks to signaling networks. Microsc Res Tech 47: 135-154 [DOI] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ (1997a) The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138: 375-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ (1997b) Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385: 265-269 [DOI] [PubMed] [Google Scholar]
- Winter D, Podtelejnikov AV, Mann M, Li R (1997) The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol 7: 519-529 [DOI] [PubMed] [Google Scholar]
- Winter DC, Choe EY, Li R (1999) Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA 96: 7288-7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.