Abstract
The genome of the budding yeast (Saccharomyces cerevisiae) provides an important paradigm for transgenomic comparisons with other eukaryotic species. Here, we report a systematic comparison of the protein kinases of yeast (119 kinases) and a reference plant Arabidopsis (1,019 kinases). Using a whole-protein-based, hierarchical clustering approach, the complete set of protein kinases from both species were clustered. We validated our clustering by three observations: (a) clustering pattern of functional orthologs proven in genetic complementation experiments, (b) consistency with reported classifications of yeast kinases, and (c) consistency with the biochemical properties of those Arabidopsis kinases already experimentally characterized. The clustering pattern identified no overlap between yeast kinases and the receptor-like kinases (RLKs) of Arabidopsis. Ten more kinase families were found to be specific for one of the two species. Among them, the calcium-dependent protein kinase and phosphoenolpyruvate carboxylase kinase families are specific for plants, whereas the Ca2+/calmodulin-dependent protein kinase and provirus insertion in mouse-like kinase families were found only in yeast and animals. Three yeast kinase families, nitrogen permease reactivator/halotolerance-5), polyamine transport kinase, and negative regulator of sexual conjugation and meiosis, are absent in both plants and animals. The majority of yeast kinase families (21 of 26) display Arabidopsis counterparts, and all are mapped into Arabidopsis families of intracellular kinases that are not related to RLKs. Representatives from 11 of the common families (54 kinases from Arabidopsis and 17 from yeast) share an extremely high degree of similarity (blast E value < 10-80), suggesting the likelihood of orthologous functions. Selective expansion of yeast kinase families was observed in Arabidopsis. This is most evident for yeast genes CBK1, HRR25, and SNF1 and the kinase family S6K. Reduction of kinase families was also observed, as in the case of the NEK-like family. The distinguishing features between the two sets of kinases are the selective expansion of yeast families and the generation of a limited number of new kinase families for new functionality in Arabidopsis, most notably, the Arabidopsis RLKs that constitute important components of plant intercellular communication apparatus.
Comparative genomics allow one to make functional projections from well-studied model organisms to species about which we know much less at the molecular and cellular level. At the same time, these studies can identify groups of genes that are unique to a species. Protein kinases are good targets for such study because they constitute a well-conserved group of proteins.
Protein kinases are important components of cellular regulatory systems. They are organized into signaling cascades, which form the backbone of the signaling network. Specific signals are restricted to specific pathways by the substrate specificity of the involved kinases. Systematic comparison of protein kinases between species can shed light on how the signaling network has been conserved and has differentiated during evolution. In this case, a comparison of yeast (Saccharomyces cerevisiae) and Arabidopsis allows us to identify groups of protein kinases that have been specifically elaborated in plants and, in some cases, to infer probable function of Arabidopsis protein kinases.
The budding yeast and Arabidopsis provide ideal candidate species for systematic trans-genomic study. Such a study requires a comprehensive list of protein kinases for each species being studied. The completeness and accuracy of the sequence information directly determines the likelihood of success of such integrative approaches. The budding yeast has long been a prototype for eukaryotic biological research. It is a much simpler system than Arabidopsis but shares cellular architecture and regulatory mechanisms with higher eukaryotic organisms. Sequencing of its genome was completed in 1996 (Goffeau et al., 1996), and the sequence is well annotated. Comprehensive functional genomic databases, such as the Yeast Protein Database (YPD; Costanzo et al., 2001) and Saccharomyces Genome Database (SGD; Weng et al., 2003), are available for this species. Arabidopsis is a model for plant molecular and cellular study. As a consequence, its genome was the first plant genome to be sequenced (Arabidopsis Genome Initiative, 2000; Martienssen and McCombie, 2001). A comprehensive classification of the Arabidopsis protein kinases has been completed and is viewable at PlantsP, a plant phosphorylation functional genomics database (Gribskov et al., 2001).
Analysis of the relationships between lineages of proteins is commonly done using programs such as ClustalW (Thompson et al., 1994) or other distance-based tree-building methods that build hierarchical clusters based on distances derived from sequence alignments. These methods are difficult to use with large and diverse protein families due to both the large number of proteins and the difficulty of determining correct multiple sequence alignments. What one needs is a method that determines pair-wise distances between sequences without requiring a multiple sequence alignment. BLAST (Altschul et al., 1997) is such a method, and we use it as the basis of our large-scale clustering.
Clustering of families of diverse multifunctional proteins such as protein kinases suffers from a second technical problem, transitivity. Commonly used methods such as unweighted pair-group method using arithmetic average (Sokal and Michener, 1958) or neighbor joining (Saitou and Nei, 1987) will often add a sequence to a group when the new sequence is similar to only one or two of the members of the group. This occurs in cases where two multidomain proteins share no common domain with each other, but each shares a domain with a third multidomain protein. This results in clusters that contain many distinctly different or only distantly related proteins. Maximum linkage clustering (Lance and Williams, 1967) is a more conservative method that adds sequences to groups only when they are close to all existing members in the group. Although not widely used for phylogenetic trees, we find it works very well for hierarchical clustering of protein families (Gribskov, 2002). On the other hand, because of its conservative nature and the presence of artifacts such as fragmentary sequences in the data, maximum linkage clustering is unable to completely join the sequences of a superfamily into a single cluster. Our approach is to combine multiple rounds of maximum linkage clustering with recalculation of average distances between generated clusters after each round. Sequentially less stringent thresholds are used for each subsequent round. This is, in effect, a hybrid maximum linkage/average linkage method that exploits the advantages of both approaches. Using this progressive, hierarchical clustering algorithm, kinases of S. cerevisiae and Arabidopsis were clustered to generate a hierarchical tree, in which cross-species orthologs and species-specific kinases can be identified. These kinases are distributed into biochemical families, as illustrated by the clustering pattern of yeast kinases.
RESULTS
Protein kinases of S. cerevisiae (119) and protein kinases of Arabidopsis (1,019) were clustered based on their amino acid sequences. Determination of inter-protein distances expressed as BLAST E values, selection of clustering thresholds, and the clustering procedure were performed as described in “Materials and Methods.” It is important to note that this clustering is based on comparison of the entire sequence. BLAST detects all conserved regions shared by two sequences (Altschul et al., 1997) so that domains outside the kinase catalytic region critically affect the result. Because the non-catalytic domains are key indicators of function, this is an important positive feature of this approach. Because the terms family, group, and class have often been used to identify various sets of kinases, throughout this manuscript, we use the neutral term “cluster” to refer to groups of kinases operationally defined by specific distance thresholds expressed as BLAST E values.
The degree to which functional inferences can be made depends on the evolutionary distance separating the sequences. In maximum linkage clustering, the members of a cluster are all guaranteed to be within a certain threshold distance of each other, that is, they constitute a clique. Our experience in this project, and in the clustering of a set of plant protein kinases that includes all Arabidopsis kinases and 117 kinases from other plant species (Gribskov, 2002), is that there are several important thresholds that can be used for inferring function. First, clusters formed at a threshold E value of 10-80 typically represent proteins with highly similar functions, often interacting with identical or closely related substrates. An example would be the two closely related (MAP2K) proteins, MKK1 and MKK2, which share the MAP kinase SLT2/MPK1 as their substrate (Waskiewicz and Cooper, 1995). Such proteins would typically be said to lie in the same subfamily. Second, clusters formed at a threshold E value of 10-35 are similar to conventional ideas of families. For instance, the calcium-dependent protein kinases (CPKs), the phosphoenolpyruvate carboxylase kinases (PPCKs), the SNF1-related kinases (SnRKs), and mitogen-activated protein kinases (MAPKs) all form clusters at this level, which we term the family level. At this level, members of a cluster have a generic similarity of domain structure but generally bind to a spectrum of related substrates.
Table I shows a synopsis of the 15 clusters produced at the threshold E value of 1. The 12 clusters of conventional protein kinases (not His kinase like and not PI kinase like, see “Materials and Methods”) are, at a threshold E value of 11.0, merged into one tree (Fig. 1). Each cluster was examined to investigate the distribution of yeast and Arabidopsis kinases at the family and subfamily level (Table I). Two types of clusters of conventional protein kinases were observed. One type of cluster (clusters 1–4) contains all of the RLKs and related cytoplasmic kinases. The other type of cluster (clusters 5–12) corresponds to intracellular protein kinases. For these families, we also list the corresponding PlantsP classification number (Table I), which is a number assigned to each protein kinase family based on a complete classification of plant protein kinases (Gribskov, 2002).
Table I.
Summary of protein kinase clusters produced at E < 1.0 threshold
| Type | Cluster | Representative Family | PlantsP Classification No. | No. of Arabidopsis Genes | No. of Yeast Genes |
|---|---|---|---|---|---|
| Receptor-like kinases (RLKs) and related kinase | 1 | N/Sa | N/S | 497 | 0 |
| 2 | N/S | N/S | 67 | 0 | |
| 3 | N/S | N/S | 54 | 0 | |
| 4 | N/S | N/S | 4 | 0 | |
| Intracellular kinase | 5 | Raf-like MAP3K | 2.1.3-4 | 22 | 0 |
| AtMRK1/ATN1 like | 2.1.5-6 | 26 | 0 | ||
| Ankyrin repeat kinase | 2.1.2 | 6 | 0 | ||
| 6 | MEKK/NPK1 like | 4.4.1-5 | 26 | 0 | |
| 7 | APG1 like | 4.2.8 | 4 | 1 | |
| ELM | 4.2.7 | 2 | 3 | ||
| SnRK (SNF1 like) | 4.2.4 | 38 | 9 | ||
| CDPK | 4.2.1, 4.2.3 | 40 | 0 | ||
| PPCK | 4.2.2 | 4 | 0 | ||
| CaMK | N/Ab | 0 | 6 | ||
| MAP4K/STE20 like | 4.1.2 | 9 | 5 | ||
| MAP3K | 4.1.1 | 12 | 5 | ||
| MAP2K | 4.1.3-4 | 10 | 4 | ||
| MAPK | 4.5.1 | 19 | 6 | ||
| Casein kinase II | 4.5.3 | 4 | 2 | ||
| CDK | 4.5.2 | 27 | 5 | ||
| GSK3/shaggy like | 4.5.4 | 10 | 4 | ||
| NIMA/NEK family | 4.1.6 | 7 | 1 | ||
| CLK | 4.5.1 | 3 | 1 (of 4) | ||
| PIM like | N/A | 0 | 2 | ||
| RAN | N/A | 0 | 2 | ||
| PTK | N/A | 0 | 2 | ||
| NPR/HAL5 | N/A | 0 | 7 | ||
| 8 | S6K | 4.2.6 | 26 | 2 | |
| DBF2 | 12 | 3 | |||
| PKA | 0 | 3 | |||
| PKC | 0 | 1 | |||
| AGC | 2 | 3 | |||
| PKA related | 2 | 3 | |||
| 9 | Casein kinase I | 3.1.1 | 17 | 4 | |
| 10 | CLK | 3.1.1 | 16 | 3 (of 4) | |
| 11 | IRE1 | 4.3.2 | 3 | 1 | |
| 12 | 5.2.1 | 1 (4g29380) | 2 | ||
| Histidine kinase | 13 | SLN1 | 0 | 1 | |
| Cytokinin receptor | 6 | 0 | |||
| Ethylene receptor | 5 | 0 | |||
| Phytochrome | 5 | 0 | |||
| 14 | PDH kinase | 1 | 1 | ||
| PI kinase like | 15 | PI kinase like | N/A | 3 | 2: |
| AtATM | MEC1 | ||||
| AtRad3 | TEL1 | ||||
| At1g50030 | |||||
| Unclustered kinase | N/A | N/A | N/A | 3: | 2: |
| At1g22870 | BUD32 | ||||
| At3g63330 | TWF1 | ||||
| At5g53450 |
N/S, Not shown
N/A, Not applicable
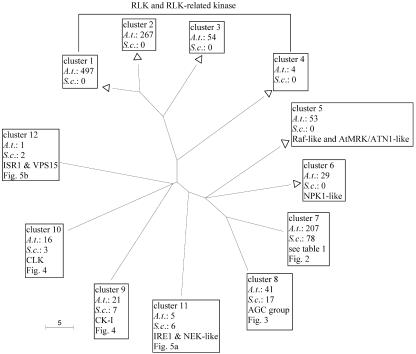
Figure 1.
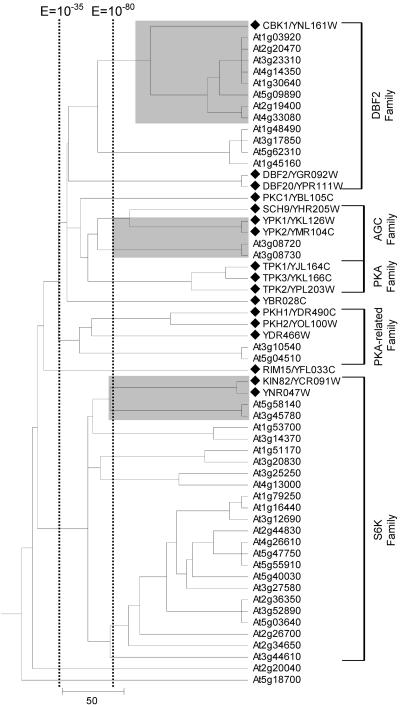
Distribution pattern of Arabidopsis and yeast kinase reveals lack of RLK and Raf-like MAP3K in yeast. The tree displays the phylogenetic relationship between the 12 conventional protein kinase clusters shown in Table II. Each branch represents a cluster described in Table II. A branch length unit corresponds to a 10-fold difference in BLAST E value. Arabidopsis-specific clusters are denoted by a white triangle. Each branch is labeled with the cluster number followed by the number of Arabidopsis (A.t.) and yeast (S.c.) kinases in the cluster. Each non-RLK cluster is briefly annotated, and the corresponding figure number is identified when a cluster is displayed later.
Six clusters (clusters 1–6) comprise only Arabidopsis protein kinases. No overlap was observed between yeast protein kinases and Arabidopsis RLK and related protein kinases (clusters 1–4). Clusters 5 and 6 also comprise only Arabidopsis kinases. These include putative Raf-related mitogen-activated protein kinase kinase kinases (MAP3Ks) such as AtCTR1, a component of the ethylene response pathway (Kieber et al., 1993). Also included is a subcluster containing protein kinases ATN1 (Tregear et al., 1996), AtMRK1 (Ichimura et al., 1997), and the Arabidopsis homolog of soybean (Glycine max) GmPK6 (Feng et al., 1993). These kinases share similarities with the catalytic domains of both Raf-like and mammalian mixed-lineage kinase (MLK) MAP3K families (Ichimura et al., 1997; Jouannic et al., 1999). S. cerevisiae lacks both raf-like and MLK MAP3K (Hunter and Plowman, 1997).
Other clusters comprise both Arabidopsis and yeast protein kinases. The trees for these clusters are shown in Figures 2 to 6. Each of Figures 7 to 11 shows the tree of a subcluster (or a kinase family) compressed in Figure 2. Within each tree, a branch representing a yeast kinase is denoted by a diamond followed by the kinase's standard name and systematic name defined in the SGD database (Weng et al., 2003). A branch representing an Arabidopsis kinase is labeled with the kinase's Arabidopsis Gene Index number. The Arabidopsis Gene Index number is followed by the name(s) given to that kinase in biological literature, if available. A bracket denotes a subcluster corresponding to a previously defined yeast kinase family or group (Hunter and Plowman, 1997). These trees are discussed below.
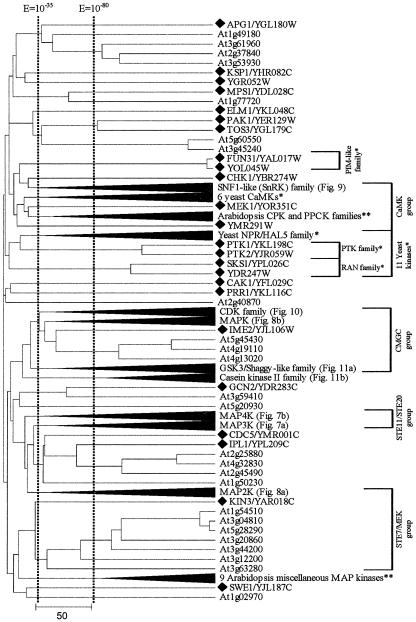
Figure 2.
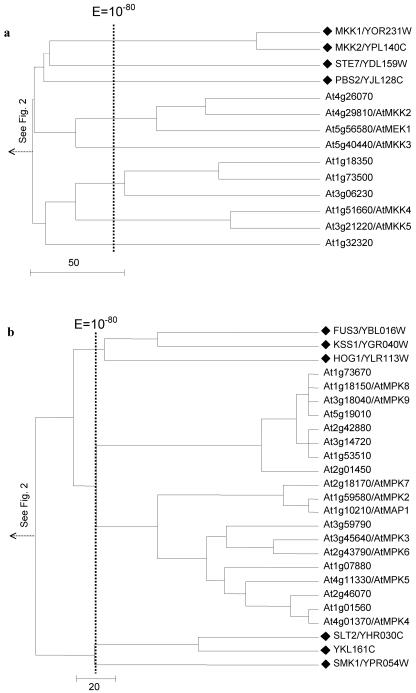
Arabidopsis lacks homologs for yeast provirus insertion in mouse (PIM)-like, Ca2+/calmodulin-dependent protein kinase (CaMK), NPR/HAL5, polyamine transport kinase (PTK), and RAN kinase families. The tree displays cluster 7 and is extensively compressed at family level due to the large number of proteins included. The black triangle branch denotes subcluster of multiple kinases. Yeast- and Arabidopsis-specific subcluster are denoted by one asterisk and two asterisks, respectively. Some of the compressed subclusters are shown later (see Figs. 7, 8, 9, 10, 11), and corresponding figure numbers are identified. A black diamond denotes branch representing a yeast kinase. Corresponding yeast kinase families and groups are labeled at the right end of the tree.
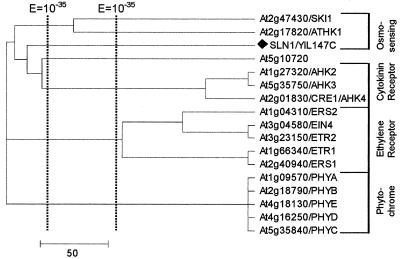
Figure 6.
Arabidopsis preserves the phospho-relay signaling mechanism. The tree of His kinases is displayed. Branch representing a yeast kinase is denoted by a black diamond and text following an Arabidopsis gene identification, when present, represents name previously assigned to the kinase. Subclusters, when corresponding to a functional group, are labeled at the right end of the tree in blue text.
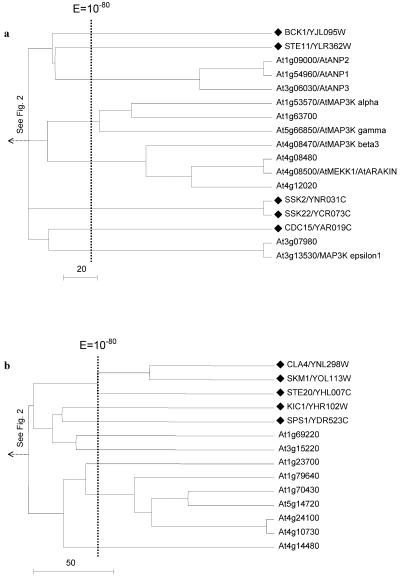
Figure 7.
Trees of MAP3K (a) and STE20-like/MAP4K kinases (b). These are subclusters from the tree shown in Figure 2. Branch representing a yeast kinase is denoted by a black diamond and name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification.
Figure 11.
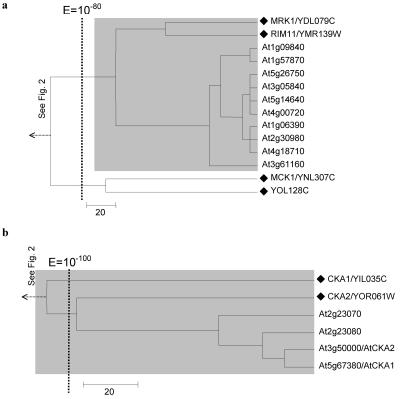
The trees of GSK3/Shaggy-like kinase family (a) and Casein kinase II family (b). It represents a subcluster from the tree shown in Figure 2. A black diamond denotes branch representing a yeast kinase. Name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification. The shaded box identified protein kinases with significant mutual E values (clustered together to the right of the vertical line indicating a BLAST E value of 10-80, see Fig. 2).
Within cluster 7 (Fig. 2), which contains 78 (of 119) yeast protein kinases, we identified five yeast-specific families (PIM like, CaMK, NPR/HAL5, PTK, and RAN) and two Arabidopsis-specific families (CPK and PPCK). Other families, such as CDK and components of the MAP kinase cascades, are common to both species. This cluster contains four yeast kinase groups, CaMK, CMGC, STE11/STE20, and STE7/MEK (Hunter and Plowman, 1997; Fig. 2).
Cluster 8 (Fig. 3) corresponds closely to the AGC kinase group, which includes the PKA (cAMP dependent), PKC (DAB activated, PL dependent), AGC, S6K (ribosomal protein S6 kinase), and DBF2 kinase families in yeast (Hunter and Plowman, 1997). No obvious plant counterparts were identified for the PKA and PKC families. However, the yeast CBK1 gene and S6K family (KIN82 and YNR047W) appear to be extensively expanded in Arabidopsis (Fig. 3).
Figure 3.
Arabidopsis lacks obvious orthologs to PKA and PKC but preserves and sometimes expands other kinase family of the yeast kinase group AGC. The tree displays cluster 8. Branch representing a yeast kinase is labeled with a black diamond, and the corresponding yeast kinase families are labeled at the right end of the tree. The shaded box identified protein kinases with significant mutual E values (clustered together to the right or near left of the vertical line indicating a BLAST E value of 10-80).
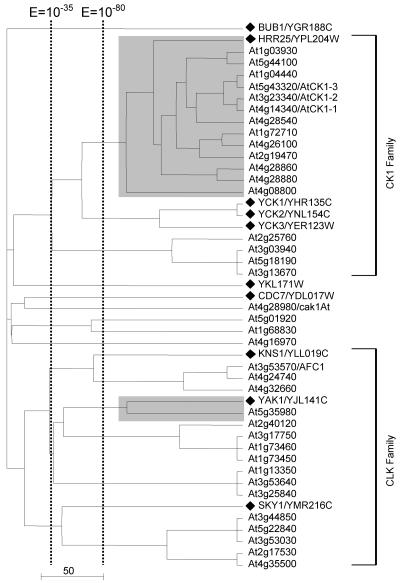
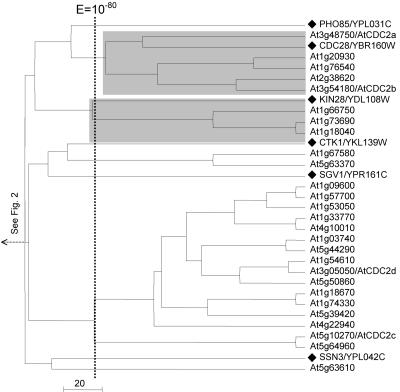
Clusters 9 and 10 are merged into one tree in Figure 4. They comprise the CK1 (casein kinase I) and the CDK-like kinase (CLK) family, respectively. Within the CK1 family, we observed the expansion of a single yeast gene, HRR25, into a subfamily of 13 closely related Arabidopsis homologs. This was the greatest gene expansion observed in our analysis.
Figure 4.
Expansion of the yeast casein kinase I family member HRR25 and CLK kinase family in Arabidopsis. The tree displays cluster 9. A black diamond denotes a branch representing a yeast kinase, and the corresponding yeast kinase families are labeled at the right end of the tree. Name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification. The shaded box identified protein kinases with significant mutual E values (clustered together to the right of the vertical line indicating a BLAST E value of 10-80).
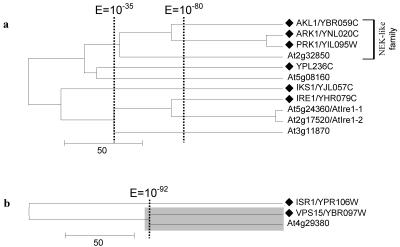
Clusters 11 and 12 comprise only five and one Arabidopsis kinases, respectively. Cluster 11 (Fig. 5A) includes the yeast NEK-like kinase family, which has three yeast members but just one Arabidopsis kinase. The yeast IRE1 kinase and three Arabidopsis homologs are also included in this cluster (Fig. 5A). Cluster 12 comprises only three genes, the yeast-specific kinase ISR1, the yeast kinase VPS15, and its Arabidopsis homolog At4g29380 (E = 2 × 10-97; Fig. 5B).
Figure 5.
Relative distribution of yeast and Arabidopsis kinases in clusters 11 and 12. The trees for the two clusters are shown in a and b, respectively, with the yeast gene denoted with a black diamond. Name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification. The shaded box identified protein kinases with significant mutual E values (10-92).
Finally, due to a lack of homology with conventional protein kinases, His kinases (clusters 13 and 14) and phosphatidylinositol kinase-like protein kinases (cluster 15) form distinct clusters (Table I). The tree for cluster 13 is shown in Figure 6.
A number of protein kinase families correspond to subclusters of cluster 7 (Table I) and are compressed in the tree shown in Figure 2. These include kinases of the MAP kinase signaling cascade, the plant CPKs, the SnRKs, the CDKs, etc. To display a map of potential cross-species orthologs, the trees for the kinases of the MAP signaling cascade (Figs. 7 and 8), the SnRKs (Fig. 9), the CDKs (Fig. 10), the shaggy/GSK-3-like kinases, and casein kinase II (CK2) kinases (Fig. 11) are shown.
Figure 8.
Trees of MAP2Ks (a) and MAPKs (b). These are subclusters from the tree shown in Figure 2. Branch representing a yeast kinase is denoted by a filled diamond and name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification.
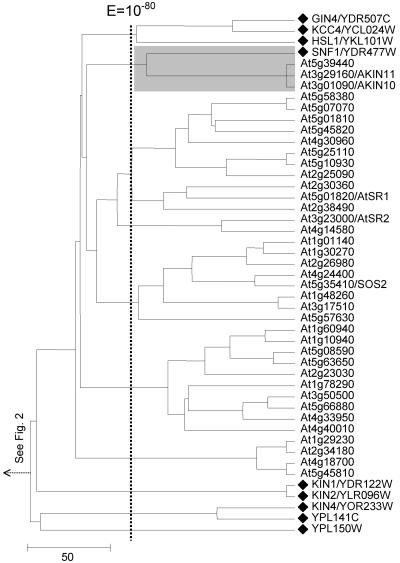
Figure 9.
Expansion of the yeast kinase SNF1 in Arabidopsis. The tree of SnRKs is displayed. It represents a subcluster from the tree shown in Figure 2. Branch representing a yeast kinase is denoted by a black diamond, and name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification. The shaded box identified protein kinases with significant mutual E values (clustered together to the right of the vertical line indicating a BLAST E value of 10-80).
Figure 10.
The tree of cyclin-dependent protein kinases (CDKs). It represents a subcluster from the tree shown in Figure 2. Branch representing a yeast kinase is denoted by a black diamond, and name(s) previously assigned to an Arabidopsis kinase through experimental characterization, when present, follow an Arabidopsis gene identification. The shaded box identified protein kinases with significant mutual E values (clustered together to the right or near left of the vertical line indicating a BLAST E value of 10-80).
The experimentally generated knowledge in biological literature represents the best possible reference for evaluating a computational analysis. Fortunately, a substantial amount of knowledge is available in this case because yeast is an alternative genetic host for experimental studies of plant protein kinases, and the set of protein kinases of this organism is one of the best studied. We used three criteria to assess the performance of our clustering procedure. First, a search of PubMed found 11 Arabidopsis kinases, which rescued genetic deletion phenotype of the corresponding yeast kinase. In this work, seven of the 11 kinase pairs were coclustered. The other four pairs were put in close neighborhood (Table II). This is expected because some kinases will first merge with their paralogs rather than orthologs. The genetic complementation assay selects ortholog(s) only for the deleted gene. It is not informative regarding whether the selected Arabidopsis gene(s) would also complement the yeast gene's paralog(s). Further, if based on cDNA expression library screening, this assay can only select those candidates that are abundant in the cDNA expression library. However, not all isoforms of a kinase family will be abundant in a particular cDNA library if they display complementary tissue distribution patterns. For example, AtMEKK1 was selected to complement the STE11 deletion (Covic and Lew, 1996; Mizoguchi et al., 1996), whereas AtANP1 also complements the same genetic deletion (Nishihama et al., 1997; Table II). Therefore, a genetic complementation assay is a good test of functional inter-changeability but does not necessarily reveal the closest ortholog. Second, some Arabidopsis kinases have been experimentally characterized, and a number of them have been shown to possess catalytic activity, either distinctive or characteristic of a kinase family. These kinases are labeled, in the trees for the corresponding clusters, with the assigned name following their gene index number (Figs. 2, 3, 4, 5, 6, 7, 8, 9, 10, 11). A survey of these kinases found that their clustering patterns are consistent with the experimental observation. For example, the Arabidopsis kinases AtCKA1 and AtCKA2, members of the kinase family CK2 (Mizoguchi et al., 1993), are clustered together with their yeast counterparts (Fig. 11). Finally, the set of yeast kinases, most of which have been experimentally characterized, has been systematically classified before using the SAM multiple alignment program. The majority of the kinases were classified into five groups (Hunter and Plowman, 1997). We identified all of the five groups in our clustering result (Figs. 2 and 3). The consistency is also illustrated, throughout the figures of generated cluster, by the brackets denoting yeast kinase families (Figs. 2, 3, 4, 5, 6) and by the families displayed in Figures 7 to 11.
Table II.
Reported functional orthologs identified through genetic complementation analysis in yeast
The list was generated through a search of National Center for Biotechnology Information PubMed for genetic complementation that occurred between a pair of protein kinases of similar signaling pathways.
| Arabidopsis Gene | Yeast Gene | Figure | Reference(s) |
|---|---|---|---|
| At4g01370/AtMPK4 | SLT2 | 8b | Mizoguchi et al. (1998) |
| At5g56580/AtMEK1 | PBS2 | 8a | |
| At1g54960/AtANP1 | STE11 | 7a | Nishihama et al. (1997) |
| At4g08500/AtMEKK1 | Covic and Lew (1996) | ||
| Mizoguchi et al. (1996) | |||
| At3g48750/AtCDC2a | CDC28(p34cdc2) | 10 | Ferreira et al. (1991) |
| At5g24360/Atlre1-1 | IRE1 | 5a | Koizumi et al. (2001) |
| At2g17520/Atlre1-2 | |||
| At1g06390/AtGSK1 | MCK1 | 11a | Piao et al. (1999) |
| At3g01090/Akin10 | SNF1 | 9 | Alderson et al. (1991); Bhalerao et al. (1999) |
| At3g29160/Akin11 | |||
| At2g17820/ATHK1 | SLN1 | 6 | Urao et al. (1999) |
Table III lists yeast kinases for which potential Arabidopsis orthologs were identified by this hierarchical clustering approach. Overall, plant counterparts were identified for 21 of 26 yeast protein kinase families. Within these families, potential orthologs were identified and highlighted in the corresponding trees. In summary, 54 Arabidopsis protein kinases were identified as orthologous to 17 yeast protein kinases.
Table III.
Protein kinases of yeast and Arabidopsis that share significant mutual E values
They are identified by a shaded box in corresponding figures.
| Yeast Family | Yeast Gene | Function (from SGD) | Arabidopsis Orthologs | Cluster E Value |
|---|---|---|---|---|
| DBF2 | CBK1 | Pheromone response: shmooing | At1g03920 | 10-110 |
| At2g20470 | ||||
| At3g23310 | ||||
| At4g14350 | ||||
| At1g30640 | ||||
| At5g09890 | ||||
| At2g19400 | ||||
| At4g33080 | ||||
| AGC | YPK1 | Vegetative growth | At3g08720 | 10-68 |
| YPK2 | At3g08730 | |||
| S6K | KIN82 | Unknown | At5g58140 | 10-78 |
| YNR047W | At3g45780 | |||
| CK-I | HRR25 | DNA repair | At1g03930 | 10-96 |
| At5g44100 | ||||
| At1g04440 | ||||
| At5g43320 | ||||
| At3g23340 | ||||
| At4g14340 | ||||
| At4g28540 | ||||
| At1g72710 | ||||
| At4g26100 | ||||
| At2g19470 | ||||
| At4g28860 | ||||
| At4g28880 | ||||
| At4g08800 | ||||
| CLK | YAK1 | Complement PKA deletion | At5g35980 | 10-98 |
| N/A | VPS15 | Vacuolar transport | At4g29380 | 10-92 |
| SNF1 | SNF1 | Glc Metabolism | At5g59340 | 10-101 |
| At3g29160 | ||||
| At3g01090 | ||||
| CDK | CDC28 | G1/S and G2/M transition | At3g48750 | 10-86 |
| At1g20930 | ||||
| At1g76540 | ||||
| At2g38620 | ||||
| At3g54180 | ||||
| KIN28 | Transcription Regulation | At1g66750 | 10-78 | |
| At1g73690 | ||||
| At1g18040 | ||||
| GSK3 | MRK1 | Unknown | At1g09840 | 10-92 |
| At1g57870 | ||||
| RIM11 | Meiosis | At5g26750 | ||
| At3g05840 | ||||
| At5g14640 | ||||
| At4g00720 | ||||
| At1g06390 | ||||
| At2g30980 | ||||
| At4g18710 | ||||
| At3g61160 | ||||
| CK-II | CKA1 | Cell cycle, cell polarity, flocculation, ion homeostasis, transcription regulation | At2g23070 | 10-92 |
| At2g23080 | ||||
| CKA2 | At3g50000 | |||
| At5g67380 | ||||
| PI kinase like | MEC1 | Cell cycle, DNA repair | AtRAD3 | 10-103 |
| TEL1 | DNA repair, telomere maintenance | AtATM | 10-85 |
DISCUSSION
Plants are relatively less studied than other model organisms. As a consequence, the set of Arabidopsis protein kinases is not well studied at the molecular and cellular level yet. This makes it important to perform systematic comparative genomic comparison with well-studied eukaryotic species. The budding yeast is the best studied eukaryotic model organism and serves as an important paradigm for transgenomic comparisons with other eukaryotic species (Chervitz et al., 1998; Rubin et al., 2000). This is the first systematic comparative genomic study of the protein kinases of yeast and a plant multicellular species to identify the genes specifically elaborated in multicellular plant species and to provide a basis for functional inference. The budding yeast has been used as an alternative genetic host for experimental characterization of Arabidopsis kinases. The protein kinase set of this model species has been classified previously into biochemically proven families. This makes it possible to validate our results. We believe our results will be helpful for the current endeavor to learn more about these kinases and to better understand the signaling network of Arabidopsis. Our study further validates yeast as a good model system for the study of the plant intracellular kinase network. The distinguishing features of the two kinase sets are not the number of core kinase families. Instead, they are selective expansion of some families and the generation of a limited number of new kinase families in Arabidopsis, most notably the Arabidopsis RLK family.
This study uses a hybrid maximum linkage/average linkage clustering of full-length protein sequences. The use of stringent thresholds in initial steps, combined with the maximum linkage methodology, ensures that the clustering procedure discriminates between two unrelated multidomain proteins when each of them shares a domain with a third multidomain protein. Proteins that are closely related in yeast and Arabidopsis will share extensive sequence similarity both within and outside the kinase catalytic domain. Therefore, we expect that molecules whose function is truly conserved will be merged in the maximum linkage clustering procedure because the initial proliferation of the protein kinase family preceded the divergence of the plant and fungal lineages. This is supported by the clustering patterns of Arabidopsis and yeast proteins in reported cases, where the Arabidopsis protein can complement the corresponding yeast mutant and, thus, is a functional homolog (Table II). In this work, additional potential Arabidopsis orthologs were identified. They might have been missed by the genetic complementation assays due to their low abundance in the screened cDNA libraries or due to the assay's tendency to favor constitutive (or more active) isoforms. Our result is further validated by its consistency with the biochemical activities of a number of Arabidopsis kinases and its consistency with reported systematic classification of yeast kinases, most of which have been experimentally proven. This clustering effort identifies potential orthologs whose biochemical function can be directly tested in future genetic complementation assays.
Lack of Yeast Homolog for Arabidopsis RLK
This is the first conclusive observation, to our knowledge, that yeast lacks homologs to plant RLK (Fig. 1). In animals, there are numerous receptor Tyr kinases and relatively few receptor Ser/Thr kinases. Multicellular species, such as Arabidopsis, require elaborate intercellular signaling mechanism to regulate growth, development, and response to the environment. Transmembrane receptor protein kinases play key roles in making the proximal response to intercellular signals. The absence of transmembrane receptor-like protein kinases in yeast reflects a reduced need to respond to extracellular signals due to unicellular habit.
Cross-Species Comparison of Intracellular Kinase
The MAP Kinase Cascade
The MAP kinase cascades correspond to a set of protein kinases, namely the MAPKs, the MAP2Ks, MAP3Ks, and in some cases, MAP4Ks. It has become apparent over the past few years that plant MAP kinase cascades are involved in many vital cellular signal propagation and integration processes (Tena et al., 2001). Many of the components of these signaling cascades seem to be shared by yeast and Arabidopsis (Figs. 7 and 8), especially the MAPKs.
Nevertheless, there are Arabidopsis MAP3Ks that have no yeast counterpart. Arabidopsis MAP3Ks can be divided into three subfamilies, namely the Raf like, the STE11/MEKK like, and the CDC15-like MAP3K. The Arabidopsis-specific Raf-related family (Table II) includes AtMAP3Kδ1, AtEDR1, and AtCTR1 in a single subcluster of 12 kinases. There are two additional subclusters of seven and three Raflike kinases, respectively (data not shown). These kinases currently have not been characterized, and functional annotation awaits genetic and/or biochemical data. The STE11/MEKK-like family includes a number of previously described proteins, AtANP 1 to 3; AtMAP3K-α, -β, and -γ; and AtMEKK1. The CDC15-like family contains the yeast CDC15 gene and two Arabidopsis kinases, the AtMAP3Kε1 (Jouannic et al., 2001) and a close homolog (Fig. 7A).
In addition, Arabidopsis possesses two clusters of short MAP3K-related proteins, with the sizes of most of them ranging from 300 to 450 amino acid residues. These putative kinases seem to display a plant-specific feature in that they contain MAP3K-like catalytic domains but lack the usually long stretch of regulatory domains flanking the catalytic region. The first cluster, the GmPK6/AtMRK1/ATN-1 like, shares similarity with both Raf-like MAP3K and mammalian MLK in the catalytic domain but lacks obvious regulatory domains, except that a subset of them contains ankyrin repeats in the N-terminal one-half (Table II, cluster 5). For example, the Arabidopsis homolog of GmPK6 shares 39% identity (56% positive) with the catalytic region of human MLK-2 but lacks the Leu zipper and the large C-terminal domains typical of MLKs (Dorow et al., 1995). The proteins in the second cluster contain MEKK-like catalytic domains but lack the catalytic domain-flanking region of MEKKs (Table II, cluster 6). They are currently annotated as putative NPK1/MEKK-like kinases in public biological databases. Whether and how the two groups of short putative kinases are involved in plant MAP kinase signaling cascades remain to be determined by genetic and biochemical approaches.
The SNF1-Like Kinase (SnRK) Family
Plants possess yeast SNF1 homologs that can complement the SNF1 deletion mutation in yeast (Alderson et al., 1991; Bhalerao et al., 1999). Two isoforms, Akin10 and Akin11, have been experimentally identified in Arabidopsis (Bhalerao et al., 1999). A potential third isoform, At5g39440, which is closely related to Akin11, was identified by this clustering effort (Fig. 9). A large group of proteins similar to SNF1, the SnRKs, has been identified in Arabidopsis and other plants. These proteins have pleiotropic roles in response to hormonal, nutritional, and environmental stresses. In addition to SNF1, the other five members of yeast SNF1/AMPK family and the yeast GIN4 family (Hunter and Plowman, 1997) coclustered with the plant SnRK sequences (Fig. 9). These observations suggest further functional parallels between yeast and Arabidopsis in addition to preservation of the SNF1 Suc/Glc signaling pathway. This is supported by the interactions observed between the kinase AtSOS2 and AtSOS3 (calcium sensor) and between kinase AtSR1 and AtCBL2 (calcium sensor) in Arabidopsis (Halfter et al., 2000; Nozawa et al., 2001), which are similar to the interaction between kinase HSL1 and CNB1 (calcium sensor) in yeast (Mizunuma et al., 2001).
The CDK Family
Six yeast kinases and 27 Arabidopsis homologs were identified in this family. The four Arabidopsis CDK genes described by Joubès et al. (2000) are all found in this subcluster (Fig. 10). Included in this group is the yeast CTK1, the catalytic subunit (subunit-α) of the RNA polymerase II C-terminal kinase complex. In yeast, this complex is composed of three components. However, no homolog to the γ-subunit has been identified in other species and the β-subunit (the cyclin-like subunit) shares only marginal similarity with its counterparts in other species, such as fission yeast (Schizosaccharomyces pombe; Sterner et al., 1995).
The Two-Component Phospho-Relay System (His Kinase)
This signaling machinery is utilized by prokaryotic species to sense and respond to environmental stimuli (Schaller, 2000). To our knowledge, no such mechanism has been observed in mammalian species. Yeast preserves this mechanism in the upstream portion of its osmosensing pathway, which bifurcates at the YPD1 protein to regulate two response regulators, SSK1 and SKN7. Arabidopsis, however, possesses several such signaling pathways, namely the ethylene response, osmosensing, and cytokinin response pathways (Fig. 6). At least in cytokinin signaling, the phospho-relay system, like that of bacteria, can extend from cell surface into the nucleus, where ARR1, the response regulator component of the two-component system, acts as a transcription factor (Sakai et al., 2001).
Due to a lack of homology with conventional protein kinases, these kinases form their own cluster (Fig. 6). The five Arabidopsis phytochromes are also included in this cluster due to their possession of a His kinase-like domain. However, phytochrome His kinase domains are very divergent from those of prokaryotes, and, in the case of PhyA, have been shown to exhibit Ser/Thr as opposed to His kinase activity (Fankhauser, 2001). Similarly, both yeast and Arabidopsis pyruvate dehydrogenase kinases possess the His kinase-like domain but exhibit Ser/Thr kinase activity (Table II).
Cross-Species Preservation and Divergence of Protein Kinase Family
Besides the Arabidopsis RLK, five species-specific protein kinase families were found for each species. The Arabidopsis-specific families include CPK, PPCK, Raf-like kinase, and the two clusters of short MAP3K-like kinases discussed earlier. Further, the CPK and PPCK kinase families are not found in animals either. The plant CPKs appear to have originated from the fusion of a CaMK-like ancestor with its calmodulin-like regulatory subunit (Harmon et al., 2000). This family has been extensively elaborated in plants and appears to fill roles equivalent to that of yeast CaMKs and additional plant-specific functions. The yeast PIM-like, CaMK, NPR/HAL5, PTK, and RAN kinase families are not found in Arabidopsis (Fig. 2). It is noteworthy that the CaMK and PIM-like kinase families lack Arabidopsis counterparts but have homologs in animals, whereas the NPR/HAL5, PTK, and RAN kinase families are absent in both animals and plants. Although no obvious plant counterpart was identified for the PKA and PKC families, we are reluctant to call them yeast specific because they cluster with other Arabidopsis and yeast kinases at E value < 10-35 (Fig. 3).
The majority of yeast protein kinase families (21 of 26) exhibit obvious counterparts and sometimes are extensively expanded in Arabidopsis. This is most evident in cases where one yeast kinase shares high similarity with multiple Arabidopsis kinases. Examples include the yeast kinase CBK1 (Fig. 3), HRR25 (Fig. 4), and SNF1 (Fig. 9). The two yeast CK2 isoforms, CKA1 and CKA2, coclustered with four Arabidopsis homologs at an E value of 10-92 (Fig. 11B). The yeast GSK3/shaggy-like kinase MRK1 and RIM1, as another example, coclustered with 10 Arabidopsis homologs at an E value of 10-92 (Fig. 11A). Our observation is consistent with previous reports that the distinguishing feature of the protein sets of yeast and animals is not the size of their “core proteome” but the selective expansion of some families and the generation of a limited number of new protein families in multicellular organisms (Chervitz et al., 1998; Rubin et al., 2000).
However, these observations show that gene expansion is not always evenly distributed across yeast kinases. The expansion of kinase CBK1, HRR25, and SNF1 all represent events where one member of a protein kinase family is selectively expanded. As discussed above, only two of the four yeast GSK3/shaggy-like kinases were merged with Arabidopsis homologs at an E value of 10-92. The other two, MCK1 and YOL128c, were not until an E value of 10-48 was reached (Fig. 11). The Arabidopsis SnRKs (Fig. 9) and MAPKs (Fig. 8B) coclustered with only a subset of the corresponding yeast protein kinases. The yeast NEK-like family, for example, is actually reduced in Arabidopsis (Fig. 5A). It remains to be seen whether other plant species share this selective kinase preservation and expansion pattern with Arabidopsis and whether non-plant multicellular species display complementary gene preservation and duplication patterns during evolution. Along with RLK kinases and the preservation of the His kinase signaling mechanism, these gene preservation and expansion patterns may prove to be important characteristics distinguishing the plant and animal signaling network.
MATERIALS AND METHODS
Sequence Set of Protein Kinases
Yeast (Saccharomyces cerevisiae) and Arabidopsis protein kinase sequences were retrieved from the functional genomic databases YPD (Costanzo et al., 2001) and PlantsP (Gribskov et al., 2001), respectively. Yeast proteins (122) were categorized as protein kinase in YPD. However, three proteins (CTK2, CTK3, and SSK1) were excluded from this analysis because CTK2 and CTK3 are non-catalytic regulatory subunits, and SSK1 is a non-catalytic response regulator of the phosphate relay system. Protein kinases (1,019) from Arabidopsis were identified in the PlantsP database. They include conventional protein kinases as well as unconventional protein kinases (His kinases and kinases whose catalytic domains are more closely related to His kinase catalytic domain but exhibit non-His kinase activity). Also included are the ATM-like protein kinases, such as AtRAD3. The catalytic domains of these kinases are more closely related to that of phosphatidylinositol kinase, but exhibit protein kinase activity (Mallory and Petes, 2000). These sequences constitute the sequence set {S} used in this study.
Determination of Inter-Protein Distances
BLAST version 2.09 (Altschul et al., 1997) was used in an all-against-all comparison to generate a distance matrix between the protein sequences in the sequence set {S} mentioned above. The BLAST program was run using default parameters but with sequence filtering turned off. Inter-protein distances were expressed as BLAST E values. BLAST detects all conserved regions shared by two sequences. The E value takes into consideration all high-scoring regions (Karlin and Altschul, 1993) and, therefore, is a good measure of full-length-based protein-protein distances. The resulting distance matrix {S} × {S} was the initial input for the clustering program discussed below.
Hierarchical Clustering
The clustering was performed in multiple steps. Each step used a sequentially less stringent clustering threshold than the previous step. This procedure is similar to that used by Yona et al. (1999) in the construction of the protoMap system. The output of each step was used as input for the next step, until the least stringent clustering threshold was reached.
The threshold E values were determined based on inspection of the distribution of the E values in the {S} × {S} BLAST comparison. Clusters were observed. Selected thresholds correspond to minima in this distribution. Such thresholds have been used successfully by Rost (2002). We used these threshold E values of: E = 10-180, E = 10-110, E = 10-80, E = 10-35, and E = 1.0.
A maximum linkage clustering program written in C performed clustering at each step. This program, cluster.c, is available from the authors upon request. Inputs to this program are a clustering threshold and a distance matrix, initially the most stringent threshold and the distance matrix {S} × {S} mentioned above. This program loops over the following three steps: (a) sort the distances and identify the shortest one (smallest E value), (b) merge the corresponding pair of clusters/sequences, and (c) update the distances from each sequence (or previously existing cluster) to the new cluster as the maximum of the distances from it to the two clusters/sequences being merged (hence the term maximum linkage). The loop stops when the shortest distance identified in step one exceeds the threshold and results in a cluster set {C}.
Distances between generated clusters are then recalculated as the geometric mean of the distances between the members of the clusters. This resulted in a new distance matrix {C} × {C}, which is used as input for the next round of maximum linkage clustering.
This work was supported by the National Science Foundation (grant no. DBI–9975808) and was assisted by the facilities of the National Biomedical Computation Resource (through grant no. P41–RR08605 from the National Institutes of Health National Center for Research Resources).
References
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA 88: 8602-8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322-5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Aravind L, Sherlock G, Ball CA, Koonin EV, Dwight SS, Harris MA, Dolinsky K, Mohr S, Smith T et al. (1998) Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science 282: 2022-2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Crawford ME, Hirschman JE, Kranz JE, Olsen P, Robertson LS, Skrzypek MS, Braun BR, Hopkins KL, Kondu P et al. (2001) YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res 29: 75-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covic L, Lew RR (1996) Arabidopsis thaliana cDNA isolated by functional complementation shows homology to serine/threonine protein kinases. Biochim Biophys Acta 1305: 125-129 [DOI] [PubMed] [Google Scholar]
- Dorow DS, Devereux L, Tu GF, Price G, Nicholl JK, Sutherland GR, Simpson RJ (1995) Complete nucleotide sequence, expression, and chromosomal localisation of human mixed-lineage kinase 2. Eur J Biochem 234: 492-500 [DOI] [PubMed] [Google Scholar]
- Fankhauser C (2001) The phytochromes, a family of red/far-red absorbing photoreceptors. J Biol Chem 276: 11453-11456 [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhao Y, Bottino PJ, Kung SD (1993) Cloning and characterization of a novel member of protein kinase family from soybean. Biochim Biophys Acta 1172: 200-204 [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, Villarroel R, Van Montagu M, Inze D (1991) The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3: 531-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M et al. (1996) Life with 6000 genes. Science 274: 546-563 [DOI] [PubMed] [Google Scholar]
- Gribskov M (2002) A systematic classification of plant protein kinases. Plant Physiol (in press)
- Gribskov M, Fana F, Harper J, Hope DA, Harmon AC, Smith DW, Tax FE, Zhang G (2001) PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Res 29: 111-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735-3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPKs: a kinase for every Ca2+. Trends Plant Sci 5: 154-159 [DOI] [PubMed] [Google Scholar]
- Hunter T, Plowman GD (1997) The protein kinases of budding yeast: six score and more. Trends Biochem Sci 22: 18-22 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Shinozaki K (1997) ATMRK1, an Arabidopsis protein kinase related to mammal mixed-lineage kinases and Raf protein kinases. Plant Sci 130: 171-179 [Google Scholar]
- Jouannic S, Hamal A, Leprince AS, Tregear JW, Kreis M, Henry Y (1999) Plant MAP kinase kinase kinases structure, classification and evolution. Gene 233: 1-11 [DOI] [PubMed] [Google Scholar]
- Jouannic S, Champion A, Segui-Simarro JM, Salimova E, Picaud A, Tregear J, Testillano P, Risueno MC, Simanis V, Kreis M et al. (2001) The protein kinases AtMAP3Kepsilon1 and BnMAP3Kepsilon1 are functional homologues of S. pombe cdc7p and may be involved in cell division. Plant J 26: 637-649 [DOI] [PubMed] [Google Scholar]
- Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudi J, P (2000) CDK-related protein kinases in plants. Plant Mol Biol 43: 607-620 [DOI] [PubMed] [Google Scholar]
- Karlin S, Altschul SF (1993) Applications and statistics for multiple high-scoring segments in molecular sequences. Proc Natl Acad Sci USA 90: 5873-5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427-441 [DOI] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127: 949-962 [PMC free article] [PubMed] [Google Scholar]
- Lance GN, Williams WT (1967) A general theory of classificatory sorting strategies: I. Hierarchical systems. Computer J 9: 373-380 [Google Scholar]
- Mallory JC, Petes TD (2000) Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USA 97: 13749-13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R, McCombie WR (2001) The first plant genome. Cell 105: 571-574 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K (1998) Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett 437: 56-60 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 765-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Kamada H, Shinozaki K (1993) Cloning and characterization of two cDNAs encoding casein kinase II catalytic subunits in Arabidopsis thaliana. Plant Mol Biol 21: 279-289 [DOI] [PubMed] [Google Scholar]
- Mizunuma M, Hirata D, Miyaoka R, Miyakawa T (2001) GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J 20: 1074-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Banno H, Kawahara E, Irie K, Machida Y (1997) Possible involvement of differential splicing in regulation of the activity of Arabidopsis ANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs). Plant J 12: 39-48 [DOI] [PubMed] [Google Scholar]
- Nozawa A, Koizumi N, Sano H (2001) An Arabidopsis snf1-related protein kinase, atsr1, interacts with a calcium-binding protein, atcbl2, of which transcripts respond to light. Plant Cell Physiol 42: 976-981 [DOI] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I (1999) An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol 119: 1527-1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B (2002) Enzyme function less conserved than anticipated. J Mol Biol 318: 595-608 [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischman W et al. (2000) Comparative genomics of the eukaryotes. Science 287: 2204-2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519-1521 [DOI] [PubMed] [Google Scholar]
- Schaller GE (2000) Histidine kinases and the role of two-component system in plants. Adv Bot Res 32: 109-148 [Google Scholar]
- Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 28: 1409-1438 [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL (1995) The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol 15: 5716-5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu W, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392-400 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear JW, Jouannic S, Schwebel-Dugue N, Kreis M (1996) An unusual protein kinase displaying characteristics of both the serine/threonine and tyrosine families is encoded by the Arabidopsis thaliana gene ATN1. Plant Sci 117: 107-119 [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743-1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Cooper JA (1995) Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol 7: 798-805 [DOI] [PubMed] [Google Scholar]
- Weng S, Dong Q, Balakrishnan R, Christie K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, Hong E et al. (2003) Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res 31: 216-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona G, Linial N, Linial M (1999) ProtoMap: automatic classification of protein sequences, a hierarchy of protein families, and local maps of the protein space. Proteins 15: 360-378 [PubMed] [Google Scholar]