Abstract
Tocopherols are lipophilic antioxidants synthesized exclusively by photosynthetic organisms and collectively constitute vitamin E, an essential nutrient for both humans and animals. Tocopherol cyclase (TC) catalyzes the conversion of various phytyl quinol pathway intermediates to their corresponding tocopherols through the formation of the chromanol ring. Herein, the molecular and biochemical characterization of TCs from Arabidopsis (VTE1 [VITAMIN E 1]), Zea mays (SXD1 [Sucrose Export Deficient 1]) and Synechocystis sp. PCC6803 (slr1737) are described. Mutations in the VTE1, SXD1, or slr1737 genes resulted in both tocopherol deficiency and the accumulation of 2,3-dimethyl-6-phytyl-1,4-benzoquinone (DMPBQ), a TC substrate. Recombinant SXD1 and VTE1 proteins are able to convert DMPBQ to γ-tocopherol in vitro. In addition, expression of maize SXD1 in a Synechocystis sp. PCC6803 slr1737 knockout mutant restored tocopherol synthesis, indicating that TC activity is evolutionarily conserved between plants and cyanobacteria. Sequence analysis identified a highly conserved 30-amino acid C-terminal domain in plant TCs that is absent from cyanobacterial orthologs. vte1-2 causes a truncation within this C-terminal domain, and the resulting mutant phenotype suggests that this domain is necessary for TC activity in plants. The defective export of Suc in sxd1 suggests that in addition to presumed antioxidant activities, tocopherols or tocopherol breakdown products also function as signal transduction molecules, or, alternatively, the DMPBQ that accumulates in sxd1 disrupts signaling required for efficient Suc export in maize.
Tocopherols (α-, β-, γ-, and δ-tocopherol) are lipophilic antioxidants that collectively constitute vitamin E, an essential nutrient for both humans and animals. Tocopherol synthesis has only been observed in photosynthetic organisms (plants, algae, and some cyanobacteria), a distribution that suggests the pathway evolved in cyanobacteria to aid in protecting the cell from reactive oxygen species generated by photosynthesis. Plant tocopherol biosynthetic enzymes are nuclear encoded and were presumably acquired from the endosymbiotic cyanobacteria that gave rise to plastids (Goksoyr, 1967). The localization of tocopherols and most of the tocopherol biosynthetic enzymes in plastid membranes supports the cyanobacterial origins of the pathway in plants (Soll et al., 1980, 1985; Lichtenthaler et al., 1981; Fryer, 1992; Kruk and Strzalka, 1995; Arango and Heise, 1998a).
Although comparatively little is known about tocopherol functions in photosynthetic organisms, the physiological importance of these molecules in human and other animal systems has been studied extensively. The complete absence of dietary tocopherols, for example, results in chronic wasting, death, and fetal reabsorption in rats (Bramley et al., 2000). Less severe tocopherol dietary deficiencies in humans and animal models are associated with numerous degenerative diseases such as atherosclerosis, arthritis, some cancers, vision maladies, weakened immune system, and neuromuscular abnormalities (Brigelius-Flohe and Traber, 1999; Bramley et al., 2000; Ricciarelli et al., 2001).
Among the best characterized functions of tocopherols in cells is their ability to scavenge and quench reactive oxygen species and lipid-soluble byproducts of oxidative stress (Brigelius-Flohe and Traber, 1999; Bramley et al., 2000; Ricciarelli et al., 2001). In addition to being lipophilic, tocopherols are capable of donating a single electron to form the resonance-stabilized tocopheroxyl radical (Liebler, 1993; KamalEldin and Appelqvist, 1996). Tocopherols are unique in this regard to other phenolic antioxidants, such as hydroxyquinones, which must donate two electrons to attain a stable structure. Tocopherols can also donate two electrons, which results in opening of the chromanol ring to form the corresponding tocoquinone derivative. These combined molecular characteristics allow tocopherols to protect polyunsaturated fatty acids from lipid peroxidation by scavenging lipid peroxyl radicals that propagate lipid peroxidation chain reactions in membranes (Burton et al., 1986; Liebler, 1993). Though direct evidence is lacking, tocopherols are thought to play similar roles in protecting the polyunsaturated fatty acid-rich plastid membrane from lipid peroxidation.
Recent studies in mammalian systems have demonstrated additional biological activities of tocopherols that are independent of their antioxidant functions. The underlying mechanisms for these effects are the modulation of signal transduction pathways by specific tocopherols and, in some instances, transcriptional activation of gene expression mediated by tocopherol-binding proteins (Brigelius-Flohe and Traber, 1999; Sen et al., 2000; Chan et al., 2001; Ricciarelli et al., 2001; Yamauchi et al., 2001; Clement et al., 2002; Nobata et al., 2002). Modulation of the protein kinase C signaling cascade and eicosanoid synthesis are two well-characterized examples of the antioxidant-independent effects of tocopherols in mammalian systems (Greenberglevy et al., 1993; Tran et al., 1996; Azzi et al., 2002). Although direct experimental evidence is lacking for antioxidant-independent tocopherol activities in plants, these data raise the possibility that tocopherols may also have roles in plants that extend beyond their proposed antioxidant functions.
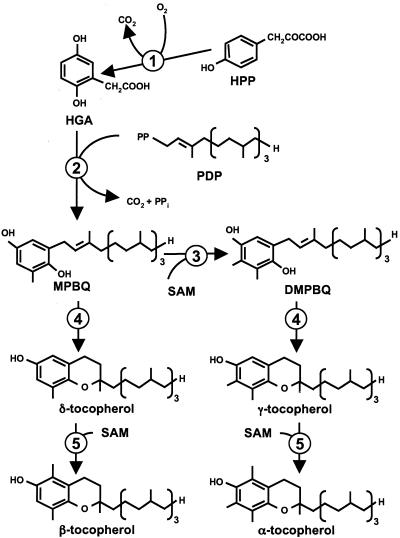
Though the functions of tocopherols in plants remain an open question, much has been learned about tocopherol synthesis and the pathway enzymes during the past 5 years (Norris et al., 1998; Shintani and DellaPenna, 1998; Collakova and DellaPenna, 2001; Schledz et al., 2001; Porfirova et al., 2002; Savidge et al., 2002; Shintani et al., 2002). Tocopherol synthesis draws substrates from two separate metabolic pathways, aromatic amino acid metabolism and isoprenoid synthesis. Homogentisic acid, an intermediate in aromatic amino acid degradation and the head group of tocopherols, is produced from p-hydroxyphenylpyruvate by the cytosolic enzyme p-hydroxyphenylpyruvate dioxygenase (HPPD; Garcia et al., 1997, 1999; Norris et al., 1998; Dahnhardt et al., 2002). The isoprenoid-derived phytyl tail of tocopherols is a product of the plastid-localized 1-deoxy-d-xylulose-5-phosphate pathway (Eisenreich et al., 1998; Lichtenthaler, 1998). The remaining steps in tocopherol synthesis occur within the inner envelope of the chloroplast and include a phytyl transferase, two different methyltransferases, and a ring-producing enzyme, the tocopherol cyclase (TC; Soll et al., 1980, 1985; Arango and Heise, 1998a).
The TC adds a second oxygen-containing ring at the junction between the aromatic head group and phytyl tail to create a two-ring structure known as a chromanol ring (Fig. 1), which is essential for resonance stabilization of tocopheroxyl radicals after single-electron transfer. Previous work has characterized TC activity in chloroplasts and chromoplasts of higher plants and in cyanobacteria (Soll, 1979; Soll and Schultz, 1980; Soll et al., 1985; Stocker et al., 1993, 1994, 1996; Arango and Heise, 1998b). The primary substrate of the TC is reduced (quinol form) 2,3-dimethyl-6-phytyl-1,4-benzoquinone (DMPBQ), which is converted to γ-tocopherol by TC (Soll, 1979; Soll and Schultz, 1980; Soll et al., 1985). However, the enzyme characterized from Anabaena sp. PCC7120 has also been shown to cyclize other 6-prenyl-1,4-benzoquinol substrates in vitro (Stocker et al., 1996). In this report, we describe the isolation and functional characterization of TCs from Arabidopsis, maize (Zea mays), and Synechocystis sp. PCC6803 and discuss the evolutionary implications of tocopherol cyclization for both tocopherol synthesis and function. The identification and characterization of TC from Arabidopsis were recently reported by Porfirova et al. (2002).
Figure 1.
Tocopherol biosynthetic pathway. This figure represents the enzymatic reactions and intermediates that are involved in tocopherol synthesis. 1, HPPD. 2, Homogentisate phytyl transferase (HPT). 3, 2-Methyl-6-phytyl-1,4-benzoquinone (MPBQ) methyltransferase. 4, TC. 5, γ-Tocopherol methyltransferase (γ-TMT). HPP, p-Hydroxyphenylpyruvate; HGA, homogentisic acid; SAM, S-adenosyl l-Met.
RESULTS
Isolation and Characterization of vte1 Mutants
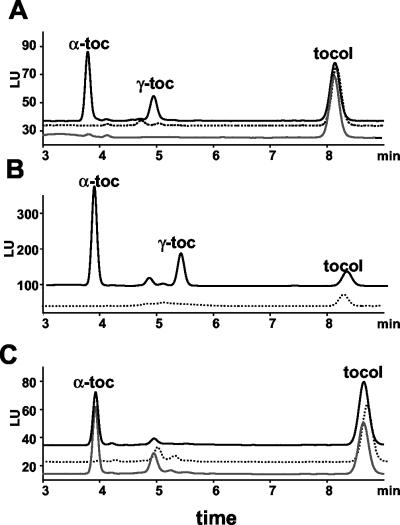
To further understand the tocopherol pathway in higher plants, an HPLC-based screen of Arabidopsis leaf tissue was developed to isolate mutants with tocopherol profiles that differ from wild type. Arabidopsis leaves accumulate approximately 10 ng α-tocopherol mg-1 fresh weight and 0.2 ng γ-tocopherol mg-1 fresh weight under standard growth conditions (see “Materials and Methods”). Numerous mutants were identified from an ethyl methanesulfonate (EMS)-mutagenized population, including two mutants that were devoid of tocopherols in leaf tissue (Fig. 2A). Genetic complementation tests confirmed that the mutants were allelic (data not shown). The mutants were designated vte1-1 and vte1-2 (vitamin e).
Figure 2.
HPLC analysis of tocopherols in wild-type and mutant Arabidopsis, maize, and Synechocystis sp. PCC6803. Tocopherols present in Arabidopsis, maize, and Synechocystis sp. PCC6803 lipid extracts were separated by normal phase HPLC and detected using a fluorescence detector with 290-nm excitation and 325-nm emission. Tocol, a synthetic tocopherol, was used as an internal recovery standard. A, Arabidopsis leaf tissue: solid line, Columbia wild type; dotted line, vte1-1; gray line, vte1-2. B, Maize leaf tissue: solid line, wild type; dotted line, sxd1. C, Synechocystis sp. PCC6803: solid line, wild type; dotted line, Δslr1737 insertional mutant; gray line, SXD1 expressed in the Δslr1737 insertional mutant. Retention times of α-, β-, δ-, and γ-tocopherol and tocol were determined by HPLC analysis of tocopherol standards. LU, Luminescence units.
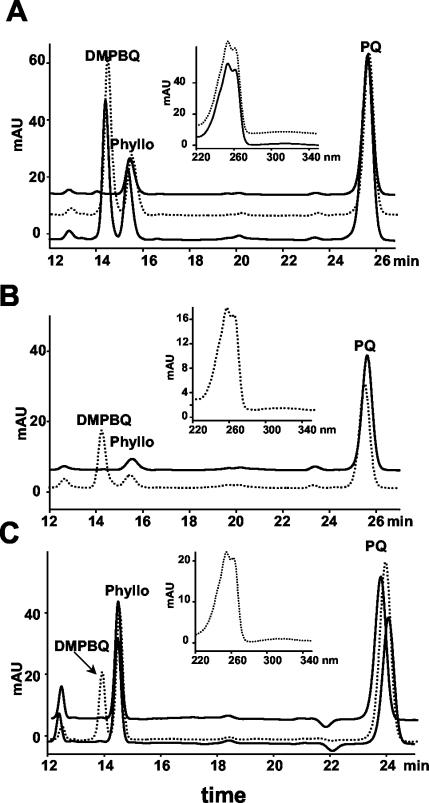
The visible phenotypes of both vte1 mutants did not significantly differ from wild type when grown under normal laboratory conditions (see “Materials and Methods”). Although several possibilities could result in a tocopherol-deficient phenotype, the two most likely are a loss of HPT activity or a loss of TC activity (Fig. 1). Assuming no genetic redundancy, a mutation disrupting either gene would result in a tocopherol-deficient phenotype, but the two classes of mutations should be readily distinguishable by the intermediates that accumulate. A defect in the TC should result in the accumulation of the DMPBQ, whereas a mutation in HPT would not accumulate tocopherol pathway prenyl quinone intermediates (Fig. 1). To understand the biochemical basis of vte1-1 and vte1-2, prenyl quinones were isolated from each mutant and analyzed by HPLC (Fig. 3A). A novel peak with a retention time and spectrum consistent with the prenyl quinone DMPBQ (Hutson and Threlfall, 1980; Marshall et al., 1985; Johnson et al., 2000) was observed in the vte1-1 and vte1-2 mutants but not in wild type (Fig. 3A). This compound was purified by HPLC and a mass of 415 D, the mass of DMPBQ, was determined by fast atom bombardment mass spectroscopy (data not shown). These combined characteristics indicate that the novel compound that accumulates in vte1-1 and vte1-2 mutants is DMPBQ, the substrate of the TC.
Figure 3.
HPLC analysis of the prenyl quinones from wild-type and mutant Arabidopsis, maize, and Synechocystis sp. PCC6803. Lipids were extracted from Arabidopsis, maize, and Synechocystis sp. PCC6803, and total prenyl quinines were isolated by thin-layer chromatography (TLC) and then analyzed by normal phase HPLC (see “Materials and Methods”) A, Arabidopsis. Solid line, Columbia wild type; dotted line, vte1-1; gray line, vte1-2. B, Maize. Solid line, Wild type; dotted line, sxd1. C, Synechocystis sp. PCC6803. Solid line, Wild type; dotted line, Δslr1737 insertional mutant; gray line, SXD1cDNA expressed in the Δslr1737 mutant background. Insets, Spectra of the peak labeled DMPBQ. Phyllo, Phylloquinone; PQ, Plastoquinone.
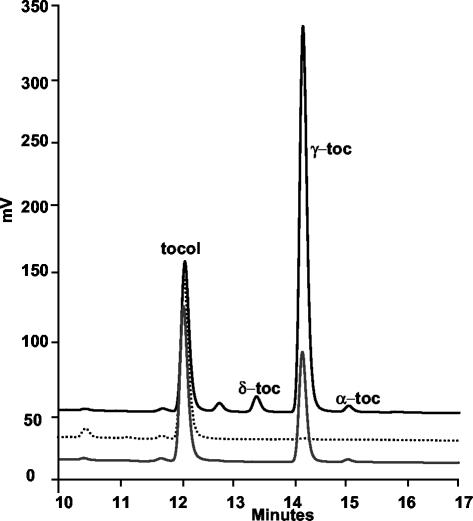
In addition to green tissues, seeds also contain tocopherols, but instead of α-tocopherol predominating as in leaves, γ-tocopherol accumulates due to low γ-TMT activity (Shintani and DellaPenna, 1998). Lipids were extracted from the seeds of wild-type and vte1 mutants, and the tocopherols were analyzed by HPLC (Fig. 4). Tocopherols were absent in vte1-1 seed, but vte1-2 seeds contained approximately 25% of the tocopherol level in wild type (84.9 ± 0.8 versus 322.5 ± 7.4 ng total tocopherols mg-1seed in vte1-2 and wild type, respectively), suggesting vte1-2 is a weaker allele than vte1-1.
Figure 4.
HPLC analysis of seed tocopherols in wild-type Arabidopsis, vte1-1, and vte1-2. Total seed lipids were extracted, and the tocopherols present were separated by reverse phase HPLC and detected using a fluorescence detector; 290-nm excitation and 325-nm emission. Tocol, a synthetic tocopherol, was used as an internal recovery standard. Solid line, Columbia wild type; dotted line, vte1-1; gray line, vte1-2. Retention times of α-, δ-, and γ-tocopherol and tocol were determined by HPLC analysis of tocopherol standards.
A map-based cloning approach was undertaken to isolate the gene encoding the TC from Arabidopsis. vte1-1 was crossed to Landsberg erecta, and 1,100 individuals from an F2 population were used to map the VTE1 locus to a 140-kb interval on the bottom of chromosome 4. Analysis of the genes within this interval identified At4g32770, encoding an unknown protein of 488 amino acids that contains a putative N-terminal chloroplast transit peptide of 68 amino acids. Previously, At4g32770 and the Synechocystis sp. PCC6803 protein slr1737 were identified as homologs of SXD1 (Suc Export Defecient 1) from Maize (Provencher et al., 2001). slr1737 is a protein of unknown function and is in the same operon as slr1736, which encodes HPT (Collakova and DellaPenna, 2001; Schledz et al., 2001; Savidge et al., 2002), another enzyme of the tocopherol biosynthetic pathway (Fig. 1). At4g32770 was fully sequenced in both mutants, and each was found to contain a nonsense mutation. The vte1-1 mutation creates a premature stop codon at amino acid 237 (Trp to stop), whereas the vte1-2 mutation creates a premature stop codon at amino acid 465 (Gln to stop). Both mutations are base transitions, G to A and C to T, respectively, which is consistent with the mutagenic properties of EMS. The identification of At4g32700 as the Arabidopsis TC concurs with a recent report by Porfirova et al. (2002).
Sequence Analysis and Evolutionary Origin
BLAST searches revealed that SXD1, VTE1, and slr1737 share a high degree of amino acid sequence similarity (Table I) with other proteins in the nonredundant GenBank database: three proteins of unknown function in the cyanobacteria Anabaena sp. PCC7120, N. punctiforme, and Synechococcus sp. PCC7002. SXD1 is a chloroplast-targeted protein of unknown function that had been identified previously based on a mutation causing a defect in symplastic photosynthate transport near the site of phloem loading within the minor veins of maize leaves (Russin et al., 1996; Provencher et al., 2001). When the SXD1 and VTE1 chloroplast transit peptides are removed, SXD1, VTE1, and the four cyanobacterial proteins share long stretches of amino acid identity.
Table I.
Pairwise comparisons of VTE1 orthologs from plants and cyanobacteria
Pair-wise comparisons were performed using ClustalW and are expressed as percentage amino acid similarity. The predicted mature plant protein sequences were used for alignments. Anabaena, Anabaena sp. PCC7120 (all0245); Nostoc, Nostoc punctiforme (506-74); Synecho, Synechococcus sp. PCC7002.
| SXD1 | VTE1 | Medicago truncatula | Barley (Hordeum vulgare) | slr1737 | Synecho | Nostoc | |
|---|---|---|---|---|---|---|---|
| % | |||||||
| VTE1 | 79 | ||||||
| M. truncatula | 80 | 84 | |||||
| Barley | 92 | 80 | 83 | ||||
| slr1737 | 46 | 46 | 48 | 48 | |||
| Synecho | 47 | 49 | 49 | 48 | 63 | ||
| Nostoc | 55 | 55 | 54 | 55 | 59 | 67 | |
| Anabaena | 55 | 54 | 54 | 55 | 61 | 67 | 85 |
The four cyanobacterial proteins are assumed to be orthologs of VTE1 because the cyanobacterial genomes each contain obvious orthologs of the four other known genes of the tocopherol pathway: HPPD, HPT, MPBQ methyltransferase, and γ-TMT. VTE1 and the cyanobacterial orthologs exist as single genes within their respective sequenced genomes. There are several other cyanobacteria whose genomes also have been sequenced: Prochlorococcus ma- rinus MED4, P. marinus MIT9313, Synechococcus sp. PCC7002 WH8102, and Thermosynechococcus elongatus BP-1. The genomes of these organisms lack obvious VTE1 orthologs and obvious orthologs for HPPD, HPT, and γ-TMT (refer to pathway in Fig. 1). Thus, it appears likely that only a subgroup of cyanobacteria have evolved the ability to synthesize tocopherols.
VTE1, SXD1, and the four cyanobacterial orthologs lack any previously described protein motifs. There are numerous plant expressed sequence tags (ESTs) in the public database that share high similarity with VTE1 and SXD1, and full-length sequences of the M. truncatula and barley VTE1 orthologs were obtained from EST assemblies. These four representative plant sequences are more conserved than the four cyanobacterial sequences (Table I). Sequence alignment of the plant and the cyanobacterial protein sequences identified a highly conserved 30-amino acid carboxyl domain in the plant VTE1 orthologs (starting at Thr-458 of Arabidopsis VTE1) that is absent from the cyanobacteria proteins (Fig. 5). The last five amino acids of this carboxyl domain (KPPGL) are invariant among the plants represented, which include the bryophyte P. patens, monocots, and dicots. With the exception of vascular and nonvascular VTE1 orthologs, this 30-amino acid domain was not found in other proteins in the nonredundant database. Interestingly, the C. reinhardtii VTE1 ortholog has a shortened version of the carboxyl domain (Fig. 5) and lacks the last 12 amino acids (starting at Leu-477 of Arabidopsis VTE1), including the invariant KPPGL motif. The vte1-2 mutation causes premature termination of VTE1 and deletion of 24 amino acids of the conserved carboxyl domain.
Figure 5.
Alignment of the carboxy termini of TC orthologs from plants and cyanobacteria. The asterisk above the At4g32770 protein sequence denotes the position of the vte1-2 mutation. Anabaena, Anabaena sp. PCC7120 (all0245); Chlamy, Chlamydomonas reinhardtii; Medicago, M. truncatula; Nostoc, N. punctiforme (506-74); Physcomitrella, Physcomitrella patens; Synecho, Synechococcus sp. PCC7002. The P. patens, rice (Oryza sativa), and wheat (Triticum aestivum) sequences are partial sequences obtained from ESTs. Dark shading, Amino acid identity; light shading, Amino acid similarity. The threshold for amino acid consensus identity or similarity is 51%.
TC Function in Arabidopsis, Maize, and Synechocystis sp. PCC6803
To confirm that the VTE1 orthologs are required for tocopherol synthesis in plants other than Arabidopsis, lipids were isolated from leaves of the sxd1 mutant and analyzed for tocopherols by HPLC. As with vte1-1 and vte1-2, leaves of the sxd1 mutant lack tocopherols, whereas wild-type maize leaves contain both α- and γ-tocopherols (Fig. 2B). Prenyl quinones from leaves of wild-type maize and the sxd1 mutant were also analyzed by HPLC. This analysis indicated that like the vte1 mutants, sxd1 contained a prenyl quinone that was absent from wild type (Fig. 3B) with an absorbance spectrum and retention time identical to the DMPBQ that accumulated in vte1 mutants.
To show that this gene family has an identical function in cyanobacteria and plants, an insertional mutant, Δslr1737, was created in the slr1737 open reading frame (ORF) of Synechocystis sp. PCC6803 through homologous recombination. The growth rate of Δslr1737 was indistinguishable from wild type (data not shown). Lipids were extracted from Δslr1737 and wild-type cells and analyzed for tocopherol composition by HPLC. Δslr1737 lacked α-tocopherol, similar to the vte1 and sxd1 mutants (Fig. 2). The two unknown peaks detected in Δslr1737 immediately after α-tocopherol are also present in the tocopherol-deficient mutants Δslr1736 and Δslr0090 (Dahnhardt et al., 2002) and do not correspond to tocopherols or tocopherol pathway intermediates. HPLC analysis of prenyl quinones in Δslr1737 and wild type showed that, like vte1 and sxd1, Δslr1737 accumulates DMPBQ (Fig. 3C).
As further proof that the cyanobacterial and plants genes are functionally equivalent, a SXD1 cDNA expression cassette was transformed into the Δslr1737 line. Lipids were isolated from the cells and analyzed by HPLC for tocopherols. The transformed cells contained α-tocopherol (Fig. 2C) and did not accumulate DMPBQ (Fig. 3C). Thus, expression of SXD1 restored a wild-type tocopherol profile to Δslr1737, indicating that SXD1 is able to functionally complement Δslr1737. Hence, these cyanobacterial and plant genes have not only high sequence similarity but also functional equivalency, suggesting a common evolutionary ancestry.
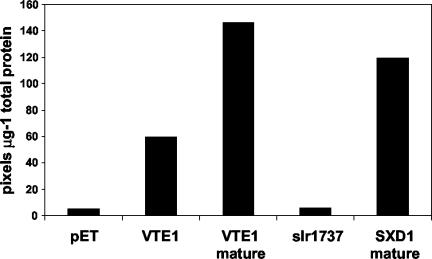
To determine the activity of the VTE1 protein and its maize and cyanobacterial orthologs, we expressed VTE1, SXD1, and slr1737 in Escherichia coli using the pET expression system. Lysates from E. coli expressing either VTE1 or SXD1 were able to convert [14C]2,3-dimethyl-6-phytyl-1,4-benzoquinol into γ-tocopherol (Fig. 6). This result conclusively demonstrates that both genes encode an enzyme with TC activity. Activity was not observed with the slr1737 protein expressed in E. coli for reasons that are unknown.
Figure 6.
TC activity of proteins expressed in E. coli. E. coli cell lysates from cells overexpressing the empty pET vector or pET engineered to express TC proteins from Arabidopsis, maize, and Synechocystis sp. PCC6803 were incubated with radiolabeled 2,3-methyl-6-phytyl-1,4-benzonequinol (3 methyl 14C) for 4 h as described in “Materials and Methods.” Total lipids were extracted, separated by TLC, and radiolabeled products were detected by phosphor imager analysis. Products were identified by comigration with standards. The 14C incorporation into γ-tocopherol was quantified densitometrically and expressed as pixels per microgram of total protein.
Carbohydrate Assimilation in vte1
Although SXD1 and VTE1 have similar enzymatic activities (Fig. 6) and primary biochemical phenotypes (tocopherol deficiency and DMPBQ accumulation, Figs. 2 and 3), sxd1 was initially isolated because of a secondary phenotype, a Suc transport defect (Russin et al., 1996; Provencher et al., 2001). To determine whether vte1 caused a similar Suc transport defect, Glc, Suc, and starch levels were analyzed in mature leaves from 4-week-old vte1-1 plants. The leaves were sampled at the end and at the beginning of the photoperiod. There were no significant differences between wild-type and vte1-1 leaves for Glc, Suc, and starch at the beginning or end of the photoperiod (Table II). Thus, unlike the sxd1 mutant, carbohydrate metabolism in mature leaves appears unaffected by the vte1 mutation. The molecular and biochemical similarities of the VTE1 and SXD1 proteins suggest the difference in sxd1 and vte1 carbohydrate metabolism phenotypes in maize and Arabidopsis reflects additional roles of tocopherols or the tocopherol pathway beyond antioxidant chemistry, rather than simply a difference in the enzymatic activity of the SXD1 and VTE1 proteins.
Table II.
Analysis of Glc, Suc, and starch in wild-type Arabidopsis and vte1-1
Glc, Suc, and starch levels were analyzed spectrophotometrically using the enzyme-coupled assays described in “Materials and Methods.” Glc and Suc are expressed as nanomoles per milligram fresh wt (n = 4).
| Beginning of Photoperiod
|
End of Photoperiod
|
|||
|---|---|---|---|---|
| Carbohydrates | Columbia | vte1-1 | Columbia | vte1-1 |
| Glc | 2.32 ± 0.62 | 2.46 ± 0.58 | 2.36 ± 0.79 | 2.63 ± 0.83 |
| Suc | 0.07 ± 0.07 | 0.16 ± 0.16 | 1.56 ± 0.60 | 1.75 ± 0.28 |
| Starcha | 13 ± 3 | 13 ± 4 | 86 ± 14 | 85 ± 11 |
Starch is expressed as nanomoles of Glc monomers per milligram fresh wt
DISCUSSION
In this report, we have shown that the three proteins, VTE1, SXD1, and slr737 from a dicot, monocot, and cyanobacterium, respectively, function as TCs. Mutations in the TC gene from each organism result in identical primary biochemical phenotypes, a block in tocopherol synthesis, and accumulation of DMPBQ, the endogenous substrate for the TC. In addition, the SXD1 and VTE1 proteins expressed in E. coli were able to convert DMPBQ to γ-tocopherol. Finally, expression of maize SXD1 was sufficient to complement the tocopherol-deficient phenotype of the Synechocystis sp. PCC6803 slr1737 deletion mutant (Δslr1737). This result demonstrates that slr1737 and SXD1 are functionally equivalent and that the biochemical activity of TCs has been evolutionarily conserved between plants and cyanobacteria. Our finding that At4g32770 encodes a functional TC in Arabidopsis concurs with a recent report by Porfirova et al. (2002).
Sequence Analysis and Evolutionary Implications of the TC Family
The TCs (VTE1, SXD1, and slr1737) share significant amino acid similarity with each other and define an evolutionarily conserved gene family that includes putative orthologs in a large number of other plants and cyanobacteria. VTE1 orthologs were not identified in databases of fungal, animal, or nonphotosynthetic bacterial species, none of which are known to produce tocopherols. Full-length sequences of two additional VTE1 orthologs from plants (barley and M. truncatula) and three from cyanobacteria (N. punctiforme, Anabaena sp. PCC7120, and Synechococcus sp. PCC7002) were identified in the public databases. Although all VTE1 proteins share a high degree of amino acid similarity, they are devoid of any previously described protein motifs, with the exception of ubiquitous phosphorylation and myristolation motifs. All VTE1 orthologs are hydrophobic proteins with low pIs and a high number of conserved Trp residues (Provencher et al., 2001). These characteristics are consistent with the TC activity characterized in Anabaena variabilis being membrane associated (Stocker et al., 1993, 1994, 1996).
Although plant and cyanobacterial TCs exhibit a high degree of protein sequence similarity, plant orthologs have additional N- and C-terminal domains that are absent in the four cyanobacterial TCs. The N-terminal domains of plant VTE1 orthologs are poorly conserved and are predicted to encode chloroplast transit peptides that would target each protein to the chloroplast. The N-terminal sequence of SXD1 has been demonstrated experimentally to be required for import into plastids (Provencher et al., 2001). HPT and γ-TMT, two other tocopherol biosynthetic enzymes, are also predicted to be chloroplast targeted (Shintani and DellaPenna, 1998; Collakova and DellaPenna, 2001; Schledz et al., 2001; Savidge et al., 2002), and chloroplastic localization of the TC is consistent with the reported localization of TC activity and tocopherol synthesis in plastids (Soll et al., 1985; Arango and Heise, 1998a).
In contrast to the N-terminal domain of plant TCs, the 30-amino acid C-terminal domain is highly conserved between angiosperms and the moss P. patens. This evolutionary conservation suggests an important function for this domain in tocopherol synthesis in plants, whereas the absence of the sequence from the four cyanobacterial VTE1 orthologs suggests that the domain is not an absolute requirement for TC enzymatic activity per se. The restriction of this C-terminal domain to vascular and nonvascular plants suggests it arose relatively recently in progenitors of land plants rather than in the endosymbiotic cyanobacteria that gave rise to plastids (Goksoyr, 1967). The shortened C-terminal domain present in the C. reinhardtii TC further suggests evolution took place in an ancestor common to C. reinhardtii and plants (i.e. before the split of Chlorophyta and Charophyta; Karol et al., 2001). The vte1-2 mutation causes deletion of the majority of the conserved C-terminal domain, and tocopherols fail to accumulate in vte1-2 leaf tissue but reach 25% of wild-type levels in seeds, indicating the truncated protein retains at least partial activity in vivo. The vte1-2 phenotype suggests the C-terminal domain plays a more significant role in TC activity/function in leaf chloroplasts than in the plastids of seeds. The complementation of Δslr1737 by SXD1 suggests that the presence of the C-terminal domain does not affect TC activity and function in cyanobacteria. The relevance of this highly conserved carboxyl domain for TC function in plants requires further investigation.
The sxd1 and vte1 Phenotypes
A surprising phenotype of the sxd1 mutants is a block in Suc export from leaves and an accumulation of anthocyanins and starch in leaf blades (Russin et al., 1996). This pleiotropic phenotype results from aberrant plasmodesmata at the interface between the bundle sheath cells and the vascular parenchyma cells surrounding the minor veins. These defective plasmodesmata block symplastic transport of Suc to the phloem and, hence, cause the Suc export defect. Unlike sxd1, vte1-1 and vte1-2 do not accumulate anthocyanins in leaves, do not accumulate starch in cells surrounding the leaf veins, and do not have stunted growth (data not shown). In addition, the similar Suc, Glc, and starch levels in mature leaves of vte1-1 and wild type (Table II) indicate that a functional Suc export pathway is present in vte1. However, this most obvious difference in phenotype between sxd1 and vte1 is not entirely unexpected considering the anatomical and physiological differences between C3 and C4 plants. Maize bundle sheath cells contain chloroplasts that differ morphologically and physiologically from the chloroplasts of mesophyll cells (Evert et al., 1977a; Fahn, 1990). Maize bundle sheath plastids have few grana, little PSII activity, high NADP-malate decarboxylase activity, and accumulate starch (Evert et al., 1977a, 1977b, 1978; Fahn, 1990). Arabidopsis lacks a physiologically equivalent cell type to the C4 bundle sheath cell. Chloroplasts within the analogous cells surrounding the minor veins in Arabidopsis do not differ significantly from the chloroplasts of mesophyll cells (Haritatos et al., 2000). Mutants of the Arabidopsis SUC2 gene, which encodes a Suc-H+ symporter required for apoplastic phloem loading, share striking similarities to sxd1 mutants (Gottwald et al., 2000). suc2 mutants accumulate anthocyanins and starch in their cotyledons, have stunted growth, and are seedling lethal in soil (Gottwald et al., 2000). Thus, the absence of the sxd1 Suc export phenotype in vte1 suggests that disruption of VTE1 in Arabidopsis either does not affect Suc export, most likely because of the fundamentally different mechanisms of Suc export in maize and Arabidopsis, or that the effect is too small to be observable as an analogous whole-plant phenotype in Arabidopsis.
In sxd1 mutants, the link between the production of aberrant plasmodesmata in the BS parenchyma cells and the defect in symplastic transport of Suc was straightforward and easy to rationalize (Provencher et al., 2001). However, the mechanistic link between the disruption of a gene encoding a chloroplast protein of unknown function (SXD1) and defective Suc transport was not so obvious. Provencher et al. (2001 raised the possibility that the sxd1 mutation exerts its pleiotropic phenotype by disrupting or altering a signal from the chloroplast to the nucleus. The nature of this signal was unknown, and cloning of the SXD1 locus did not provide further insight. Although the precise nature of this signal remains unclear, the demonstration that SXD1 encodes a TC and determination of the primary sxd1, vte1, and Δslr1737 biochemical phenotypes now greatly limits the possibilities.
The absence of tocopherols in vte1 and Δslr1737 does not cause a pleiotropic phenotype analogous to sxd1. vte1 mutants are indistinguishable from wild type under normal growth conditions, and Δslr1736 and Δslr1737 mutants grow at rates identical to wild type (Collakova and DellaPenna, 2001). These data suggest that the sxd1 phenotype is not simply due to the absence of tocopherols as lipophilic antioxidants because similar affects would be observed in the vte1, Δslr1737, and Δslr1736 mutants, which also lack tocopherols. In addition, the DMPBQ that accumulates in vte1, sxd1, and Δslr1737 mutants can still act as an antioxidant by donating a pair of electrons and then being recycled by reduction back to the quinol form of DMPBQ (Kruk et al., 1994; Kruk and Strzalka, 1995; Liebler and Burr, 2000). Thus, even in the absence of tocopherols, the sxd1 mutant is not entirely deficient in membrane-associated antioxidants.
Influencing membrane fluidity is another potential function of tocopherols, and this could be related to the plasmodesmatal defect in sxd1 mutants. However, the consensus from cell fractionation studies is that tocopherols are localized exclusively in plastid membranes (Lichtenthaler et al., 1981; Fryer, 1992; Kruk and Strzalka, 1995), and although it cannot be excluded, it is unlikely that tocopherols or DMPBQ would be present in plasma membranes to directly impact plasmodesmatal development. However, the bundle sheath cell is a specialized cell type, and direct physical connections between plasmodesmata and chloroplasts through the endoplasmic recticulum have been reported in maize bundle sheath cells (Evert et al., 1977a, 1977b). This potential association between the chloroplast and plasmodesmata would provide a means for tocopherols or prenyl quinones to impact membrane fluidity at the plasma membrane. Still, it is difficult to envision a mechanism whereby membrane fluidity would alter plasmodesmata development. If diminished antioxidant capacity or altered membrane fluidity is not the cause of the sxd1 phenotype, the question still remains: How does disruption of TC activity result in the pleiotropic sxd1 phenotype?
In addition to the well-defined role of tocopherols as antioxidants, specific tocopherols, tocotrienols, and their oxidized products have been demonstrated to have biological activities in mammalian systems that are independent of their antioxidant functions. The unifying theme for these antioxidant-independent activities is the modification or modulation of various signal transduction pathways (Brigelius-Flohe and Traber, 1999; Ricciarelli et al., 2001; Clement et al., 2002). The effects of tocopherols on the protein kinase C signaling cascade and the synthesis of eicosanoids in mammals have been well characterized. α-Tocopherol posttranslationally inhibits the activity of protein kinase C in several mammalian systems (Chan et al., 2001; Azzi et al., 2002; Clement et al., 2002). Plant genomes also contain protein kinase C homologs and other components of this signaling pathway. Tocopherols also have been shown to posttranslationally inhibit the activity of phospholipase A2 (Tran et al., 1996; Chandra et al., 2002), cyclooxygenase (COX-2; Jiang et al., 2000; Wu et al., 2001), and lipoxygenase-5 (Greenberglevy et al., 1993; Wang et al., 2000; Ricciarelli et al., 2002). These enzymes are involved in the production of eicosanoid signaling molecules (prostaglandins, prostacyclins, thromboxanes, and leukotrienes) from polyunsaturated fatty acids in mammals (Ohuchi and Levine, 1980; Tran et al., 1996; Kim et al., 2001). Like eicosanoids in animals, jasmonic acid and other oxylipins in plants are synthesized from polyunsaturated lipids by the action of lipoxygenase(s) and phospholipase(s) (Blee, 2002; Howe and Schilmiller, 2002). A third example of α-tocopherol-dependent, antioxidant-independent signal transduction in mammals is the transcriptional activator TAP (tocopherol-associated protein). Upon the binding of α-tocopherol by TAP, the complex is translocated into the nucleus, where it has been shown to activate the transcription of transgenes (Yamauchi et al., 2001).
Although tocopherols have not yet been shown to affect protein kinase C signaling, transcriptional regulation, or the synthesis of jasmonic acid or other oxylipins in plants, studies in mammalian systems suggest plausible mechanisms whereby the absence of TC activity (and, hence, tocopherols) could affect signaling and result in the pleiotropic sxd1 phenotype. Thus, it is possible that many tocopherol functions will be universal, including the roles tocopherols play in modulating signal transduction pathways or acting as signals themselves. Although the downstream events of signal transduction pathways would not necessarily be evolutionarily conserved between plants and mammals, many of the core components and biochemical motifs of signal transduction pathways are. We suggest that the sxd1 phenotype is the first evidence that tocopherols act as signaling molecules or modulators of signaling in plants. Tocopherols, tocopherol derivatives, or tocopherol pathway intermediates may provide or modulate signals required for the development of maize bundle sheath vascular parenchyma plasmodesmata, analogous to the effects of tocopherols in mammalian signaling. Alternatively, the DMPBQ that accumulates in sxd1 may interfere with an endogenous signaling pathway required for the process. Several groups have provided evidence that the redox status of the chloroplast, which is monitored through the plastoquinone (PQ) pool, regulates nuclear-encoded photosynthetic gene expression (Pfannschmidt et al., 1999a, 1999b, 2001; Alfonso et al., 2000; Allen and Pfannschmidt, 2000; Kujat and Owttrim, 2000; Li and Sherman, 2000; Pursiheimo et al., 2001; Trebitsh and Danon, 2001; Yang et al., 2001). DMPBQ has the same 2,3-dimethyl-1,4-benzoquinone head group as PQ and could interfere with redox signaling through the PQ pool. Another more remote possibility is that the SXD1 protein has an unknown substrate in addition to DMPBQ, and this product is required for signal transduction in maize. Although none of these models can be excluded based on the present data, they do provide a framework for future study.
The observation that Arabidopsis vte1 mutants do not exhibit phenotypes analogous to sxd1 suggests that the downstream signal transduction events impacted by tocopherol deficiency differ between monocots and dicots. The pathways leading to maize bundle sheath vascular parenchyma plasmodesmata formation may either be absent or not equivalent in Arabidopsis, or the effects are too subtle to be observed at the whole-organism level as in sxd1. Experiments to assess the whole-genome responses of Arabidopsis vte1 mutants are under way.
MATERIALS AND METHODS
Growth Conditions and Seed Stocks
Arabidopsis plants were grown at 22°C under a 12-h photoperiod (120 μE) in a vermiculite and potting soil mixture. M3 EMS-mutagenized Arabidopsis seeds (Columbia ecotype) were purchased from Lehle Seed (Round Rock, TX). vte1-1 was backcrossed to wild type three times, and vte1-2 was backcrossed twice. Maize (Zea mays) plants were grown under greenhouse conditions in the same soil mixture and fertilized biweekly with 20-20-20 fertilizer. The maize sxd1-2 allele used in this publication was isolated through the Trait Utility System for Corn (Pioneer Hybrids, Johnston, IA). The sxd1-2 Mu insertion site and mutant phenotype were described previously (Provencher et al., 2001). Synechocystis sp. PCC6803 was grown on BG-11 media photoautotrophically or photomixotrophically (BG11 media containing 15 mm Glc) on plates or in liquid culture at 30°C and 50 to 70 μE light.
Tocopherol Analysis
For tocopherol analyses, total lipids were extracted from 30 to 35 mg of Arabidopsis or maize leaf tissue or 15 to 20 mg of plate-grown Synechocystis sp. PCC6803 cells (Bligh and Dyer, 1959; Collakova and DellaPenna, 2001), and dissolved in 100 μL of methanol or hexane. Methanol extracts (50 μL) were subject to HPLC (Agilent 1100 series, Agilent, Wilmington, DE) on a Spherisorb ODS-2 5-μm, 250-× 4.6-mm reverse phase column (Column Engineering, Ontario, CA) at 28°C with a flow rate of 2 mL min-1 with 95% (v/v) methanol and 5% (v/v) isopropanol. Hexane extracts (50-μL volume) were subjected to HPLC on a ReliaSil Silica 5-μm, 250-× 4.6-mm normal phase column (Column Engineering) at 42°C with a flow rate of 2 mL min-1 with 83% (v/v) hexane and 17% (v/v) isopropyl ether. Tocopherols were detected by fluorescence using 290-nm excitation and 325-nm emission.
Analysis of Prenyl Quinones
One gram of Arabidopsis or maize leaf tissue and a 500-mL culture of Synechocystis sp. PCC6803 (OD730 = 0.8) were harvested and total lipids extracted (Collakova and DellaPenna, 2001). Prenyl quinones were purified by TLC as described by Pennock (1985) and eluted with diethyl ether. After drying, the samples were resuspended in 500 μL of hexane. A 100-μL aliquot was dried, resuspended in 75 μL of isopropanol, and subjected to HPLC on a reverse phase column (described above) under conditions previously described by Johnson et al. (2000). Alternatively, 60 μL of hexane extract was subjected to HPLC, on a normal phase column (described above), at 30°C with a 1 mL min-1 flow rate using 0.1% (v/v) dioxane in hexane. Prenyl quinones were detected by A254 using a diode array detector.
Map-Based Cloning of vte1
PCR-based markers were designed using INDEL or SNP from the Cereon Arabidopsis Polymorphism and Landsberg erecta Sequence Collection (Cereon Genomics LLC, Cambridge, MA; Jander et al., 2002). DNA was extracted from 1- to 2-mm developing leaves using Plant DNazol (Invitrogen, Carlsbad, CA) following the protocol of the manufacturer. Alternatively, DNA was isolated from 1- to 2-mm developing leaves in 1.1-mL tubes arrayed in a 96-well format. Two 4-mm glass beads were added to tubes along with 200 μL of 10 mm Tris (pH 8.0) and 200 μL of chloroform. The tubes were shaken with a commercial paint shaker for 5 min and centrifuged at 3,750 rpm for 10 min. One microliter of the aqueous phase or resuspended DNA from DNazol extractions was used in a 20-μL PCR reaction.
Sequence Analysis
All DNA sequences other than vte1 mutant alleles were obtained from public databases using BLAST: wheat (Triticum aestivum; BQ619591), rice (Oryza sativa; AU031770), Physcomitrella patens (BJ164574), M. truncatula (BF641171 and TC48011), barley (Hordeum vulgare; TC33553 and TC32886), Arabidopsis (AF302188), and maize (AF302187). A TC prefix denotes sequences obtained from The Institute for Genomic Research. All others have GenBank accession numbers. The cyanobacteria and algae sequences were obtained from their respective genome sequencing projects. Synechocystis sp. PCC6803 and Anabaena sp. PCC7120 sequences were obtained from Cyanobase (http://www.kazusa.or.jp/cyano/cyano.html). Chlamydomonas reinhardtii and Nostoc sequences were obtained from the Joint Genome Institute (http://www.jgi.doe.gov). The Synechococcus sp. PCC7002 sequence was obtained from National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Alignments were performed using MacVector 7.0 (Oxford Publishing, London), which includes the ClustalW algorithm.
Construction of slr1737, SXD1, and VTE1 Protein Expression Vectors
Primers 5′-CATATGACCCCTAATTTATCTTCCTTTG-3′ (F1), 5′-CATATGGACAAGATCTCCGTTAAACCTG-3′ (F2), 5′-CTCGAGTTACAGACCCGGTGGCTTG-3′ (R1), and turbo Pfu (Stratagene, La Jolla, CA) were used to PCR amplify the VTE1 cDNA from a seed cDNA library (a gift from Dr. John Ohlrogge, Michigan State University, East Lansing). F1 was used to amplify the full-length cDNA, and F2 was used to amplify a truncated version of the cDNA that encodes a version of the protein lacking chloroplast transit peptide. PCR products were cloned into the SmaI site of pBluescript II SK-. The two versions of the VTE1 cDNA were cloned as NdeI-XhoI fragments (sites underlined in primers) into NdeI and XhoI sites of the pET30b expression vector (Novagen, Madison, WI).
Primers 5′-CATATGAAATTTCCGCCCCACAGTGGTTAC-3′ (F3) and 5′-GGATCCTAACGAATCAAAACAAGGC-3′ (R2), and turbo Pfu were used to amplify the slr1737 gene from Synechocystis sp. PCC6803 genomic DNA. The PCR product was cloned into the EcoRV site of pBluescript II SK-. The slr1737 gene was cloned as NdeI-BamHI fragments (sites underlined in primers) into NdeI and BamHI sites of the pET30b expression vector (Novagen).
Primers 5′-TTCATATGGCAACGCCGCATAGCGGGTACCAC-3′ (F4) and 5′-TTGCGGCCGCTTCATTGTGACATTCGTTGG (R3) were used to amplify the SXD1 cDNA without a chloroplast transit peptide. The PCR product was cloned into NdeI and NotI sites (sites underlined in primers) of pET24d (Novagen). The fidelity of all constructs was confirmed by sequencing.
Generation of Δslr1737
Primers 5′-CTGTGTATTCTGACGGTGC-3′ (F5) and 5′-GGAGATTGAGAGAATTTATGATGC-3′ (R4) and Pfu polymerase (Stratagene) were used to PCR amplify the 5′ region flanking slr1737 from Synechocystis sp. PCC6803 genomic DNA. Primers 5′-ATAAATTCTCTCAATCTCCGTACGGAATAACACTGCCTTGTTTTG-3′ (F6) and 5′-ACCTGTTCTTCTAACCACTTG-3′ (R5) and Pfu polymerase (Stratagene) were used to PCR amplify the 3′ region flanking slr1737 from Synechocystis sp. PCC6803 genomic DNA (underlined nucleotides indicate an added BsiWI restriction site). The 5′- and 3′-flanking PCR products were joined through re-amplification with Pfu polymerase using Primers F5 and R5 to generate a contiguous 1,041-bp fragment containing a BsiWI site separating the 5′- and 3′-flanking portions. The PCR product was cloned into the pPCR-Script AMP vector (Stratagene) to generate pSLR1737-5′-3′flank. The aadA gene encoding spectinomycin adenyltransferase was inserted as a BsiWI fragment into the corresponding site of pSLR1737-5′-3′flank. The resulting plasmid was then used to replace ORF slr1737 by homologous recombination (Williams, 1988). Recombinant lines were selected by spectinomycin resistance, and replacement of the slr1737 ORF with the aadA gene was confirmed by PCR.
Complementation of Δslr1737 by Expression of the Maize SXD1 cDNA
A vector, designated pSynExp-2, was used to express SXD1 in Synechocystis sp. PCC6803. pSynExp-2 was derived from pPCR-Script AMP (Stratagene) and contained the Synechocystis sp. PCC6803 psbA2 promoter linked by a multicloning site to Tn9, which encodes chloramphenicol acetyltransferase. To facilitate homologous recombination, the promoter and multicloning site were flanked by 512- and 429-bp sequences from the slr2699locus.
Primers 5′-TTTTTTTTGCTAGCACGCCGCATAGCGGGTACCAC-3′ (F7) and 5′-TTTTTTGCTAGCTGATCACATTCGTTGGTGATCCTATAG-3′ (R6) and Pfu polymerase were used to PCR amplify the coding sequence of the mature maize SXD1 polypeptide from the SXD1 cDNA. The PCR product was cloned into the NheI and BclI restriction sites of the multicloning site behind the psbA2 promoter. The resulting plasmid was used to transform Δslr1737 through homologous recombination of the slr2699 locus. Recombination events were selected by chloramphenicol resistance, and the introduction of the maize SXD1 cDNA into slr1737 knockout mutants was confirmed by PCR analysis of genomic DNA isolated from chloramphenicol- and spectinomycin- (selection for Δslr1737) resistant cell lines.
TC activity from Escherichia coli-Expressed Proteins
C43 (DE3) cells containing the relevant pET vectors engineered to express TCs from the three organisms were grown at 30°C in 50 mL of Luria-Broth culture (50 μg mL-1 kanamycin) to mid-log phase (0.4–0.6 OD600 nm), then 1 mm isopropylthio-β-galactoside was added and grown at 15°C for 18 h (Miroux and Walker, 1996). Cells were harvested by centrifugation and washed with 100 mm KHPO4 (pH 7.8) and 4 mm MgCl2. The cells were resuspended in 1 mL of 100 mm KHPO4 (pH 7.8), 4 mm MgCl2, 4 mm dithiothreitol, and 4 mm glutathione, and sonicated six times for a duration of 10 s at 40 Hz. Dodecyl maltoside was added to a final concentration of 0.08 mm, and the cell lysate was shaken gently for 1 h at 4°C. Radiolabeled DMPBQ substrate was prepared by enzymatic labeling of MPBQ with 14C-S-adenosyl l-Met (Amersham, Piscataway, NJ) using heterologously expressed Synechocystis sp. PCC6803 MPBQ methyl transferase (Shintani et al., 2002). The labeled DMPBQ was purified from the reaction by TLC (Pennock, 1985). Each 100-μL TC assay contained 4.5 × 10-4 μCi of 2,[14C]3-dimethyl-6-phytyl-1,4-benzoquinol. The substrate was reduced with 2 mg of NaBH4 immediately before the assay. Each reaction contained 100 mm KHPO4 (pH 7.8), 4 mm MgCl2, 0.8 mm dithiothreitol, 0.8 mm glutathione, 0.8 mm dodecyl maltoside, 20 mm ascorbic acid, 1 mm cyclodextrin, and 20 μL of cell lysate from E. coli expressing SXD1, VTE1, or slr1737. The dodecyl maltoside and cyclodextrin were added first to solubilize the substrate. The reactions were sparged with nitrogen gas and gently shaken for 4 h at room temperature. Total lipids were extracted in extraction buffer containing 1 mg mL-1 butylated hydroxytoluene and unlabeled 20 μg mL-1 γ-tocopherol as a carrier. The prenyl lipids were separated by TLC, and the presence of the unlabeled prenyl quinone and tocopherol standards was determined by staining with Emmeric-Engel reagent (Pennock, 1985). 14C-labeled compounds were detected by a 4-d exposure to a low-energy PhosphorImage screen (Molecular Dynamics, Piscataway, NJ) and analyzed using a PhosphorImager (Molecular Dynamics).
Analysis of Glc, Suc, and Starch in Leaves
Mature leaves from 4-week-old Arabidopsis plants were harvested at the end and the beginning of the photoperiod. Sugars were extracted from leaf tissue in 80% (v/v) ethanol at 80°C for 30 min. Starch was also extracted from the cleared leaf tissue using 0.2 m KOH at 95°C for 45 min and neutralized with 1 m acetic acid to pH 5.0. The sugars and starch levels were measured enzymatically through the conversion of NAD to NADH by Glc-6-phosphate dehydrogenase and observed at 340 nm as described (Stitt et al., 1989).
Acknowledgments
We would like to acknowledge Chris Cook and Hiroshi Maeda for technical assistance in the map-based cloning of vte1, Eva Collakova for help with the analysis of prenyl quinones, Dr. Zigang Cheng for technical assistance in preparation of MPBQ substrate, and Maria Magallenes-Lundback for assistance with the HPLC analysis. We are also thankful to members of the Dr. DellaPenna laboratory for reviewing this manuscript and for their helpful discussions.
This work was supported in part by the Michigan State University Center for Novel Plant Products.
References
- Alfonso M, Perewoska I, Kirilovsky D (2000) Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803: involvement of the cytochrome b(6)/f complex. Plant Physiol 122: 505-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Pfannschmidt T (2000) Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond Ser B Biol Sci 355: 1351-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango Y, Heise KP (1998a) Localization of alpha-tocopherol synthesis in chromoplast envelope membranes of Capsicum annuum L. fruits. J Exp Bot 49: 1259-1262 [Google Scholar]
- Arango Y, Heise KP (1998b) Tocopherol synthesis from homogentisate in Capsicum annuum L. (yellow pepper) chromoplast membranes: evidence for tocopherol cyclase. Biochem J 336: 531-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A, Ricciarelli R, Zingg JM (2002) Non-antioxidant molecular functions of alpha-tocopherol (vitamin E). FEBS Lett 519: 8-10 [DOI] [PubMed] [Google Scholar]
- Blee E (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7: 315-322 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911-917 [DOI] [PubMed] [Google Scholar]
- Bramley PM, Elmadfa I, Kafatos A, Kelly FJ, Manios Y, Roxborough HE, Schuch W, Sheehy PJA, Wagner KH (2000) Vitamin E. J Sci Food Agric 80: 913-938 [Google Scholar]
- Brigelius-Flohe R, Traber MG (1999) Vitamin E: function and metabolism. FASEB J 13: 1145-1155 [PubMed] [Google Scholar]
- Burton GW, Cheng SC, Webb A, Ingold KU (1986) Vitamin E in young and old human red blood cells. Biochim Biophys Acta 860: 84-90 [DOI] [PubMed] [Google Scholar]
- Chan SS, Monteiro HP, Schindler F, Stern A, Junqueira VBC (2001) Alpha-tocopherol modulates tyrosine phosphorylation in human neutrophils by inhibition of protein kinase C activity and activation of tyrosine phosphatases. Free Radic Res 35: 843-856 [DOI] [PubMed] [Google Scholar]
- Chandra V, Jasti J, Kaur P, Betzel C, Srinivasan A, Singh TP (2002) First structural evidence of a specific inhibition of phospholipase A(2) by alpha-tocopherol (vitamin E) and its implications in inflammation: crystal structure of the complex formed between phospholipase A(2) and alpha-tocopherol at 1.8 angstrom resolution. J Mol Biol 320: 215-222 [DOI] [PubMed] [Google Scholar]
- Clement SA, Tan CC, Guo JL, Kitta K, Suzuki YJ (2002) Roles of protein kinase C and alpha-tocopherol in regulation of signal transduction for GATA-4 phosphorylation in HL-1 cardiac muscle cells. Free Radic Biol Med 32: 341-349 [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D (2001) Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 127: 1113-1124 [PMC free article] [PubMed] [Google Scholar]
- Dahnhardt D, Falk J, Appel J, van der Kooij TAW, Schulz-Friedrich R, Krupinska K (2002) The hydroxyphenylpyruvate dioxygenase from Synechocystis sp. PCC 6803 is not required for plastoquinone biosynthesis. FEBS Lett 523: 177-181 [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A (1998) The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol 5: R221-R233 [DOI] [PubMed] [Google Scholar]
- Evert RF, Eschrich W, Heyser W (1977a) Distribution and structure of plasmodesmata in mesophyll and bundle sheath cells of Zea mays L. Planta 136: 77-89 [DOI] [PubMed] [Google Scholar]
- Evert RF, Eschrich W, Heyser W (1978) Leaf structure in relation to solute transport and phloem loading in Zea mays L. Planta 138: 279-294 [DOI] [PubMed] [Google Scholar]
- Evert RF, Eschrich W, Neuberger DS, Eichhorn SE (1977b) Tubular extensions of plasmalemma in leaf cells of Zea mays L. Planta 135: 203-205 [DOI] [PubMed] [Google Scholar]
- Fahn A (1990) Plant Anatomy. Butterworth-Heinemann Ltd., Oxford
- Fryer MJ (1992) The antioxidant effects of thylakoid vitamin E (alphatocopherol). Plant Cell Environ 15: 381-392 [Google Scholar]
- Garcia I, Rodgers M, Lenne C, Rolland A, Sailland A, Matringe M (1997) Subcellular localization and purification of a p-hydroxyphenylpyruvate dioxygenase from cultured carrot cells and characterization of the corresponding cDNA. Biochem J 325: 761-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Rodgers M, Pepin R, Hsieh TF, Matringe M (1999) Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from Arabidopsis in transgenic tobacco. Plant Physiol 119: 1507-1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksoyr J (1967) Evolution of eukaryotic cells. Nature 214: 1161. [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979-13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberglevy SH, Budowski P, Grossman S (1993) Lipoxygenase and other enzymes of arachidonic acid metabolism in the brain of chicks affected by nutritional encephalomalacia. Int J Biochem 25: 403-409 [DOI] [PubMed] [Google Scholar]
- Haritatos E, Medville R, Turgeon R (2000) Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 211: 105-111 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230-236 [DOI] [PubMed] [Google Scholar]
- Hutson KG, Threlfall DR (1980) Synthesis of plastoquinone-9 and phytylplastoquinone from homogentisate in lettuce chloroplasts. Biochim Biophys Acta 632: 630-648 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Elson-Schwab I, Courtemanche C, Ames BN (2000) gammatocopherol and its major metabolite, in contrast to alpha-tocopherol, inhibit cyclooxygenase activity in macrophages and epithelial cells. Proc Natl Acad Sci USA 97: 11494-11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TW, Shen G, Zybailov B, Kolling D, Reategui R, Beauparlant S, Vassiliev IR, Bryant DA, Jones AD, Golbeck JH et al. (2000) Recruitment of a foreign quinone into the A(1) site of photosystem I: I. Genetic and physiological characterization of phylloquinone biosynthetic pathway mutants in Synechocystis sp. pcc 6803. J Biol Chem 275: 8523-8530 [DOI] [PubMed] [Google Scholar]
- KamalEldin A, Appelqvist LA (1996) The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 31: 671-701 [DOI] [PubMed] [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF (2001) The closest living relatives of land plants. Science 294: 2351-2353 [DOI] [PubMed] [Google Scholar]
- Kim KR, Kim MJ, Rhee SJ (2001) Effects of vitamin E on arachidonic acid cascade in platelets and aorta of acute cadmium-poisoned rats. Nutr Res 21: 657-665 [Google Scholar]
- Kruk J, Schmid GH, Strzalka K (1994) Antioxidant properties of plastoquinol and other biological prenylquinols in liposomes and solution. Free Radic Res 21: 409-416 [DOI] [PubMed] [Google Scholar]
- Kruk J, Strzalka K (1995) Occurrence and function of alpha-tocopherol quinone in plants. J Plant Physiol 145: 405-409 [Google Scholar]
- Kujat SL, Owttrim GW (2000) Redox-regulated RNA helicase expression. Plant Physiol 124: 703-713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sherman LA (2000) A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182: 4268-4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK (1998) The plants' 1-deoxy-d-xylulose-5-phosphate pathway for biosynthesis of isoprenoids. Fett Lipid 100: 128-138 [Google Scholar]
- Lichtenthaler HK, Prenzel U, Douce R, Joyard J (1981) Localization of prenylquinones in the envelope of spinach chloroplasts. Biochim Biophys Acta 641: 99-105 [DOI] [PubMed] [Google Scholar]
- Liebler DC (1993) The role of metabolism in the antioxidant function of vitamin E. Crit Rev Toxicol 23: 147-169 [DOI] [PubMed] [Google Scholar]
- Liebler DC, Burr JA (2000) Antioxidant reactions of alpha-tocopherolhydroquinone. Lipids 35: 1045-1047 [DOI] [PubMed] [Google Scholar]
- Marshall PS, Morris SR, Threlfall DR (1985) Biosynthesis of tocopherols: a re-examination of the biosynthesis and metabolism of 2-methyl-6-phytyl-1,4-benzoquinol. Phytochemistry 24: 1705-1711 [Google Scholar]
- Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260: 289-298 [DOI] [PubMed] [Google Scholar]
- Nobata Y, Urakaze M, Temaru R, Sato A, Nakamura N, Yamazaki K, Kishida M, Takata M, Kobayashi M (2002) alpha-tocopherol inhibits IL-8 synthesis induced by thrombin and high glucose in endothelial cells. Hormone Metab Res 34: 49-54 [DOI] [PubMed] [Google Scholar]
- Norris SR, Shen X, DellaPenna D (1998) Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol 117: 1317-1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi K, Levine L (1980) Alpha-tocopherol inhibits 12–0-tetradecanoylphorbol-13-acetate-stimulated deacylation of cellular lipids, prostaglandin production, and changes in cell morphology of Madin-Darby canine kidney Cells. Biochim Biophys Acta 619: 11-19 [DOI] [PubMed] [Google Scholar]
- Pennock JF (1985) Biosynthesis of plastoquinone. Methods Enzymol 110: 313-319 [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF (1999a) Photosynthetic control of chloroplast gene expression. Nature 397: 625-628 [Google Scholar]
- Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF (1999b) Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. Iubmb Life 48: 271-276 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Schutze K, Brost M, Oelmuller R (2001) A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem 276: 36125-36130 [DOI] [PubMed] [Google Scholar]
- Porfirova S, Bergmuller E, Tropf S, Lemke R, Dormann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99: 12495-12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher LM, Miao L, Sinha N, Lucas WJ (2001) Sucrose export defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13: 1127-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursiheimo S, Mulo P, Rintamaki E, Aro EM (2001) Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J 26: 317-327 [DOI] [PubMed] [Google Scholar]
- Ricciarelli R, Zingg JM, Azzi A (2001) Vitamin E: protective role of a Janus molecule. FASEB J 15: 2314-2325 [DOI] [PubMed] [Google Scholar]
- Ricciarelli R, Zingg JM, Azzi A (2002) The 80th anniversary of vitamin E: beyond its antioxidant properties. Biol Chem 383: 457-465 [DOI] [PubMed] [Google Scholar]
- Russin WA, Evert RF, Vanderveer PJ, Sharkey TD, Briggs SP (1996) Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell 8: 645-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B, Weiss JD, Wong YH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE (2002) Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 129: 321-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledz M, Seidler A, Beyer P, Neuhaus G (2001) A novel phytyltransferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett 499: 15-20 [DOI] [PubMed] [Google Scholar]
- Sen CK, Khanna S, Roy S, Packer L (2000) Molecular basis of vitamin E action: tocotrienol potently inhibits glutamate-induced pp60(c-Src) kinase activation and death of HT4 neuronal cells. J Biol Chem 275: 13049-13055 [DOI] [PubMed] [Google Scholar]
- Shintani D, DellaPenna D (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282: 2098-2100 [DOI] [PubMed] [Google Scholar]
- Shintani DK, Cheng Z, DellaPenna D (2002) The role of 2-methyl-6-phytylbenzoquinone methyltransferase in determining tocopherol composition in Synechocystis sp. PCC6803. FEBS Lett 511: 1-5 [DOI] [PubMed] [Google Scholar]
- Soll J (1979) Comparison of geranylgeranyl and phytyl substituted methylquinols in the tocopherol synthesis of spinach chloroplasts. Biochem Biophys Res Commun 91: 715-720 [DOI] [PubMed] [Google Scholar]
- Soll J, Kemmerling M, Schultz G (1980) Tocopherol and plastoquinone synthesis in spinach chloroplast subfractions. Arch Biochem Biophys 204: 544-550 [DOI] [PubMed] [Google Scholar]
- Soll J, Schultz G (1980) 2-Methyl-6-phytylquinol and 2,3-dimethyl-5-phytylquinol as precursors of tocopherol synthesis in spinach chloroplasts. Phytochemistry 19: 215-218 [Google Scholar]
- Soll J, Schultz G, Joyard J, Douce R, Block MA (1985) Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch Biochem Biophys 238: 290-299 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174: 518-552 [Google Scholar]
- Stocker A, Fretz H, Frick H, Ruttimann A, Woggon WD (1996) The substrate specificity of tocopherol cyclase. Bioorg Med Chem 4: 1129-1134 [DOI] [PubMed] [Google Scholar]
- Stocker A, Netscher T, Ruttimann A, Muller RK, Schneider H, Todaro LJ, Derungs G, Woggon WD (1994) The reaction mechanism of chromanol ring formation catalyzed by tocopherol cyclase from Anabaena variabilis Kutzing (cyanobacteria). Helv Chim Acta 77: 1721-1737 [Google Scholar]
- Stocker A, Ruttimann A, Woggon WD (1993) Identification of the tocopherol cyclase in the blue-green algae Anabaena variabilis Kutzing (cyanobacteria). Helv Chim Acta 76: 1729-1738 [Google Scholar]
- Tran K, Wong JT, Lee E, Chan AC, Choy PC (1996) Vitamin E potentiates arachidonate release and phospholipase A(2) activity in rat heart myoblastic cells. Biochem J 319: 385-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Danon A (2001) Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc Natl Acad Sci USA 98: 12289-12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CX, Zien CA, Afitlhile M, Welti R, Hildebrand DF, Wang XM (2000) Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12: 2237-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167: 766-778 [Google Scholar]
- Wu DY, Hayek MG, Meydani SN (2001) Vitamin E and macrophage cyclooxygenase regulation in the aged. J Nutr 131: 382S-388S [DOI] [PubMed] [Google Scholar]
- Yamauchi J, Iwamoto T, Kida S, Masushige S, Yamada K, Esashi T (2001) Tocopherol-associated protein is a ligand-dependent transcriptional activator. Biochem Biophys Res Commun 285: 295-299 [DOI] [PubMed] [Google Scholar]
- Yang DH, Andersson B, Aro EM, Ohad I (2001) The redox state of the plastoquinone pool controls the level of the light-harvesting chlorophyll a/b binding protein complex II (LHC II) during photoacclimation: cytochrome b(6)f deficient Lemna perpusilla plants are locked in a state of high-light acclimation. Photosynth Res 68: 163-174 [DOI] [PubMed] [Google Scholar]