Abstract
We have identified the protein MIR16 (for Membrane Interacting protein of RGS16) from a yeast two-hybrid screen by using RGS16 as bait. MIR16 shares strong homology with bacterial glycerophosphodiester phosphodiesterases. It interacts with RGS16 and, more weakly, with several other selected RGS proteins. Analysis of deletion mutants showed that the N-terminal region of the RGS domain in RGS16 is required for its interaction with MIR16. MIR16 is an integral membrane glycoprotein, because it remained associated with membrane fractions after alkaline treatment and because, in some cells, it is sensitive to digestion with endoglycosidase H. By immunofluorescence and immunoelectron microscopy, MIR16 was localized on the plasma membrane in liver and kidney and on intracellular membranes in rat pituitary and cultured pituitary cells. MIR16 represents the only integral membrane protein identified thus far to interact with an RGS domain and, to our knowledge, is the only mammalian glycerophosphodiester phosphodiesterase that has been cloned. The putative enzymatic activity of MIR16 and its interaction with RGS16 suggest that it may play important roles in lipid metabolism and in G protein signaling.
Keywords: G protein, integral membrane protein, lipid metabolism
Heterotrimeric G proteins transduce a variety of extracellular signals from G protein-coupled receptors to downstream G protein effectors (1, 2). Recently, a protein family, the RGS proteins (for Regulator of G protein Signaling), has been discovered; these proteins regulate Gα subunits by acting as GTPase activating proteins (3–5). To date, at least 24 distinct RGS proteins have been found in mammals, but information on the connections of RGS proteins to specific signaling pathways and on the functional roles of RGS proteins in specific biological systems is still limited (3, 4).
We have been interested in defining the G protein-mediated signaling pathways associated with intracellular membranes (6, 7). Previously, we identified GAIP (Gα interacting protein), one of the founding members of the RGS protein family, and localized it to clathrin-coated vesicles in pituitary cells and hepatocytes, among others (6). Besides GAIP, additional RGS proteins have also been found in pituitary, liver, and other secretory cells (3). We have chosen RGS16 (ref. 8; also named RGS-r, ref. 9; and A24-RGS14p, ref. 10) for further study, because our interests focus on secretory cells and because RGS16 was shown to be highly expressed in mouse pituitary and liver (8). RGS16 has been shown to be a GTPase activating protein for members of the Gαi family in vitro (9, 11). However, little information is available concerning the interaction of RGS16 with molecules other than Gα subunits.
To define further signaling pathways involving RGS16, we used the yeast two-hybrid system to identify proteins that interact with RGS16. Herein, we report the discovery of an integral membrane protein MIR16 (for Membrane Interacting protein of RGS16), identified by screening a pituitary cell cDNA library with RGS16 as bait. Sequence analysis indicated that MIR16 shares significant homology with bacterial glycerophosphodiester phosphodiesterases (GP-PDEs). To our knowledge, MIR16 is the only integral membrane protein reported to interact with an RGS domain and is also the only putative mammalian GP-PDE that has been cloned.
Materials and Methods
Yeast Two-Hybrid Screening.
The cDNA encoding mouse RGS16 (amino acids 1–62; ref. 8) was amplified by PCR and inserted into the EcoRI site of pAS2.1 (CLONTECH). The resulting plasmid was used as bait to screen a rat pituitary GC cell cDNA library in the pACT2 vector (12). Approximately 400,000 yeast CG1945 transformants were screened, and positive colonies were scored for β-galactosidase activity by colony lift assay according to the CLONTECH MatchMaker protocol. Prey plasmid DNA from the double-positive (His+/LacZ+) colonies was rescued by electroporation into Escherichia coli HB101 (Life Technologies, Grand Island, NY) and then retransformed back into yeast strain SFY526 containing the original bait plasmid and various control plasmids for one-to-one interactions. Both strands of the DNA inserts from these clones were sequenced by automated sequencing (Center for AIDS Research, DNA sequencing facility, University of California San Diego).
cDNA Cloning.
The insert from one clone, B24 (later named MIR16), was excised and radiolabeled with [α-32P]dCTP by random priming. The probe was then used to screen a rat lung λgt11 cDNA library (CLONTECH) for full-length cDNA clones according to the manufacturer's instructions. The cDNA inserts from selected positive clones were sequenced on both strands by automated sequencing.
Database Searches and Sequence Analysis.
blast searches were performed via the web site of the National Center for Biotechnology Information (13). Protein alignments were carried out with the clustal w 1.7 program and shaded by the macboxshade 2.15 program. macvector 6.5 software (Oxford Molecular, Oxford, U.K.) was used for hydrophobicity analysis and transmembrane region prediction, and signalp 1.1 (http://www.cbs.dtu.dk/services/SignalP/) was used for signal peptide prediction (14).
Northern Blotting Analysis.
Multiple tissue blots of human, rat, and mouse poly(A)+ RNA (CLONTECH) were probed with random-primed 32P-labeled DNA probes containing the coding region of human, rat, and mouse MIR16 (hMIR16, rMIR16, and mMIR16), respectively. Hybridization was carried out at 68°C by using ExpressHyb solution (CLONTECH), and the blots were washed under high-stringency conditions (0.1× SSC/0.1% SDS at 50°C).
In Vivo Interactions in the Yeast Two-Hybrid System.
To assess the specificity of the interaction between MIR16 and RGS16, MIR16 prey plasmid was cotransformed with pAS2.1 vector containing RGS2, RGS4, GAIP, or Ret-RGS1 (15) into yeast strain SFY526. For mapping the MIR16-interacting region in RGS16, various RGS16 deletion mutants (amino acids 53–180, 1–33, 1–52, 1–62, and 63–201) were made by PCR and inserted into the EcoRI site of the pAS2.1 vector. Sequences of the PCR primers are available on request. All constructs were verified by automated sequencing. One-to-one interactions were examined for β-galactosidase activity by the colony lift assay.
In Vitro Pull-Down Assays.
Full-length RGS16, RGS2, and RGS4 cDNAs and the DNA fragment corresponding to amino acids 1–62 of RGS16 (RGS161–62) were cloned into pGEX-KG (Amersham Pharmacia), and glutathione S-transferase (GST) fusion proteins were expressed in E. coli BL21, purified, and immobilized on glutathione-agarose beads (Sigma) as described in Amersham Pharmacia's instruction manual. 35S-labeled MIR16 products were produced by using the TNT T7 rabbit reticulocyte Quick Coupled Transcription/Translation system (Promega) in the presence of a pCDNA3 construct (Invitrogen) containing MIR16 cDNA and [35S]methionine (1,000 Ci/mmol; 1 Ci = 37 GBq; in vivo cell-labeling grade, Amersham Pharmacia). In vitro translated MIR16 was incubated with ≈5 μg of fusion protein immobilized on glutathione agarose beads in PBS buffer, pH 7.4/0.1% Triton X-100 for 2 h at 4°C with gentle rocking. Beads were then washed three times with the same buffer, and bound proteins were eluted with 25 μl of Laemmli buffer, resolved by SDS/12% PAGE, and visualized by autoradiography with Kodak X-Omat film.
Antibodies.
MIR16 cDNA fragment (amino acids 46–331) was subcloned into the pET28 vector (Novagen), and the resulting plasmid was transformed into E. coli BL21(DE3). Recombinant 6× His-tagged fusion protein was purified on Ni-NTA agarose beads (Qiagen, Chatsworth, CA) according to the manufacturer's instructions and used to immunize New Zealand White rabbits. IgG was purified from antiserum by using Affi-Gel Protein A agarose beads (Bio-Rad) according to the manufacturer's instructions. Purified IgG (1 μg/ml) was able to detect 20 ng of 6× His-tagged MIR16 recombinant protein. Monoclonal antibody 16B12 against the hemagglutinin (HA) epitope was purchased from Babco (Richmond, CA), and rabbit antiserum to calnexin was a gift from J. J. M. Bergeron (McGill University, Montreal). Polyclonal antibodies to RAP were prepared as described (16).
Cell Culture and Transfection.
HEK293 cells, obtained from Joan Heller Brown (University of California San Diego), were maintained in MEMα containing 10% (vol/vol) FCS. A pituitary-derived, luteinizing hormone-secreting cell line, LβT2 (17), was obtained from Pamela Mellon (University of California San Diego) and cultured in DME high glucose medium containing 10% (vol/vol) FCS. GC cells and PC-12 cells, obtained from Michael Karin (University of California San Diego), were cultured in DME high glucose medium containing 12% (vol/vol) horse serum/2.5% (vol/vol) FCS/10% (vol/vol) horse serum/5% (vol/vol) FCS, respectively. Penicillin G (100 units/ml) and streptomycin sulfate (100 μg/ml) were added to all media. Culture media were purchased from GIBCO/BRL, and animal sera products were obtained from HyClone. Transient transfection of HEK293 cells was carried out by using the calcium phosphate precipitation method as described (18).
Membrane Association Assays.
For the in vitro membrane association assay, coupled transcription and translation for MIR16 were carried out as described above, except that 2 μl of canine pancreatic microsomes (Promega) was included in 25 μl of reaction mixture. Microsomes were sedimented by centrifugation at 100,000 × g for 30 min at 4°C. For alkaline extraction, membrane pellets were resuspended in 0.2 M sodium carbonate (pH 11.5) for 30 min on ice and then centrifuged as described above. Proteins associated with the membrane pellet (P100) and supernatant (S100) before and after alkaline treatment were separated by SDS/PAGE followed by autoradiography.
For the in vivo membrane association assay, HEK293 cells were transiently transfected with a pCDNA3 construct containing MIR16 with a C-terminal HA tag. At 48 h after transfection, membrane pellets (≈50 μg) were prepared from postnuclear supernatants by centrifugation at 100,000 × g for 30 min at 4°C as described (6). Alkaline extraction was performed as for the in vitro membrane association assay. Postnuclear supernatant, S100, and P100 fractions were normalized by volume and analyzed by SDS/PAGE followed by immunoblotting with anti-HA IgG (1:1,000).
Immunoblotting.
Proteins were separated by SDS/12% PAGE and transferred to poly(vinylidene difluoride) membranes (Millipore). Membranes were blocked in TBS/5% (vol/vol) calf serum/0.1% Tween 20 and incubated with primary antibodies followed by horseradish peroxidase-conjugated goat anti-rabbit (1:3,000) or anti-mouse IgG (1:5,000; Bio-Rad) and enhanced chemiluminescence detection (Pierce).
Endoglycosidase H (Endo H) Treatment.
Membrane fractions of GC cells, prepared as described above for HEK293 cells, and rat liver lysate (≈50 μg) were incubated with 3 milliunits of endo H (Roche Molecular Biochemicals) in 50 mM citrate buffer (pH 5.5) at 37°C for 3 h (19), after which the proteins were separated by SDS/PAGE followed by immunoblotting with antisera to MIR16 (1:2,000) and RAP (1:5,000).
Immunocytochemistry.
For whole-cell immunofluorescence, GC cells and PC-12 cells were fixed, permeabilized, and incubated in anti-MIR16 IgG (1 h) and Alexa 488 goat anti-rabbit IgG conjugate (Molecular Probes) as described (6). For immunogold or immunofluorescence labeling of cryosections, rat pituitary, kidney, and liver were fixed [4% (vol/vol) paraformaldehyde for 15 min; 8% (vol/vol) paraformaldehyde for 45 min] and processed for semithin or ultrathin cryosectioning as described (6). Ultrathin cryosections were incubated with anti-MIR16 IgG followed by incubation with 5-nm gold, goat anti-rabbit IgG conjugate (Amersham Pharmacia). Sections were stained for 20 min in 2% (vol/vol) neutral uranyl acetate followed by absorption staining and embedding in 0.1% uranyl acetate/0.2% methyl-cellulose/3.2% (vol/vol) polyvinyl alcohol.
Results
Identification of MIR16.
To identify proteins that interact with RGS16, we screened a rat GC pituitary cell cDNA library by using RGS16 as bait. Among ≈400,000 yeast clones screened, 3 clones were confirmed to be His+/LacZ+-positive and required the presence of bait construct for reporter activity. The insert (1,435 bp) from one positive clone, clone B24, was used to obtain a full-length cDNA clone from a rat lung cDNA library. This full-length B24 cDNA encodes an ORF of 331 aa with a calculated molecular mass of 37 kDa and was later named MIR16 (Fig. 1).
Figure 1.
Deduced amino acid sequence of rMIR16. The hydrophobic regions are underlined, and the two putative N-glycosylation sites are indicated by asterisks. Numbers indicate the positions of amino acids.
Kyte–Doolittle analysis predicted that MIR16 contains two hydrophobic regions, one at the N terminus (amino acids 8–45) and the other close to the C terminus (amino acids 243–276; Fig. 1). The N-terminal hydrophobic region is a putative signal peptide (based on a signal peptide prediction program), and the second hydrophobic region is a strong candidate for a membrane-spanning region (based on macvector von Heijne helix transmembrane prediction). Two putative N-glycosylation sites (-N-X-S/T-) were also found (Fig. 1).
MIR16 Has Strong Homology with GP-PDE.
Human and mouse orthologs of rMIR16, including an unknown gene from human chromosome 16 bacterial artificial chromosome clone CIT987SK-327O24 (GenBank accession no. AAC05803), and several mouse and human expressed sequence tags were identified from databases. Two expressed sequence tag clones (accession nos. AA340631 and AA726727) containing human and mMIR16 orthologs, respectively, were obtained from the American Type Culture Collection and sequenced. The deduced human and mMIR16 amino acid sequences showed 94% and 99%, respectively, overall similarity with rMIR16. Four proteins of unknown function (accession nos. AAD14735, CAB01567, AAB52706, and CAB03386) encoded by the Caenorhabditis elegans genome showed significant homology with MIR16, with E values of 8 × 10−32, 2 × 10−24, 6 × 10−22, and 7 × 10−21, respectively. The presence of multiple MIR homologs in C. elegans suggests that MIR may be a member of a gene family sharing similar functions.
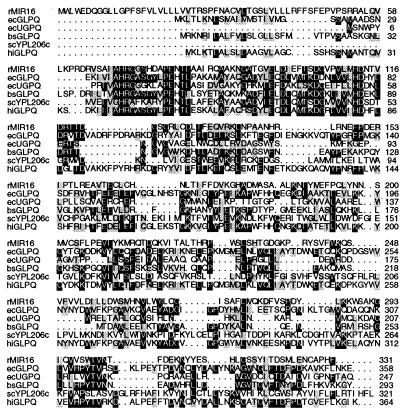
A blast search with the amino acid sequence of MIR16 indicated that MIR16 has significant homology with GP-PDEs (EC 3.1.4.46) from E. coli (20, 21) and Haemophilus influenzae (22), suggesting that MIR16 may have GP-PDE enzymatic activity. GP-PDE hydrolyzes deacylated phospholipid GPs, such as glycerophosphocholine (GPC) and glycerophosphoethanolamine, to sn-glycerol-3-phosphate (G3P) and the corresponding alcohols (22, 23). From the sequence databases, putative GP-PDEs with strong sequence homology to E. coli GP-PDEs can also be found in a number of bacteria (i.e., Bacillus subtilis, Mycobacterium tuberculosis, Mycoplasma pneumoniae, Staphylococcus aureus, Treponema pallidum, and Treponema paraluiscuniculi) and yeast (i.e., Saccharomyces cerevisiae and Schizosaccharomyces pombe). Interestingly, more than one, usually two, putative GP-PDEs are expressed in these organisms. However, the enzymatic activity of the corresponding proteins has not yet been tested. Up to now, no mammalian GP-PDE has been cloned. Fig. 2 shows the alignment of rMIR16 with the E. coli and H. influenzae GP-PDEs together with selected putative GP-PDEs. From the alignment, it can be seen that the N-terminal region of MIR16 (amino acids 70–150) immediately after the putative signal peptide (amino acids 8–45) is highly conserved (40–61% similarity), suggesting that it may contain residues critical for enzymatic activity, i.e., the catalytic site.
Figure 2.
MIR16 protein shares high homology with GP-PDEs. Alignment of rMIR16 protein with GP-PDE proteins. Invariant residues are shaded in black; similar residues are in gray. Numbers indicate the positions of amino acids. Abbreviations for species: r, rat; ec, E. coli; bs, B. subtilis; sc, S. cerevisiae; hi, H. influenzae. The accession numbers of the sequences used in this analysis are as follows: rMIR16, AF212861; ecGLPQ, P09394; ecUGPQ, P10908; bsGLPQ, P37965; scYPL206C, CAA97920; hiGLPQ, Q06282. GlpQ and UGpQ are the two GP-PDEs in E. coli.
MIR16 mRNA Is Widely Expressed in Mammalian Tissues.
Northern blot analysis showed that the MIR16 probe hybridized to a ≈1.8-kilobase transcript in multiple rat tissues (Fig. 3). rMIR16 mRNA was abundantly expressed in heart, brain, liver, kidney, and testis; moderately expressed in lung and skeletal muscle; and not detected in spleen. Similar distribution patterns were also observed for mMIR16 and hMIR16 mRNAs (data not shown).
Figure 3.
Northern blot analysis of rMIR16 mRNA. A multiple tissue blot of rat poly(A)+ mRNA was probed with a cDNA fragment of rMIR16 labeled with [α-32P]dCTP. Tissue sources of the poly(A)+ RNA are shown at the top, and the positions of markers are indicated on the left. A ≈1.8-kilobase (kb) transcript was detected with high expression levels in heart, brain, liver, kidney, and testis and with modest expression levels in lung and skeletal muscle.
MIR16 Selectively Interacts with RGS Proteins.
Using the yeast two-hybrid one-to-one interaction assay, we investigated the specificity of the interaction between MIR16 and several RGS proteins. We found that MIR16 interacted with full-length RGS16 and RGS161–62 and, less strongly, with RGS2, GAIP, and Ret-RGS1, but MIR16 did not interact with RGS4 (Fig. 4A). Next, we performed an in vitro GST pull-down assay and found that 35S-labeled, in vitro-translated MIR16 bound to GST-RGS16 and GST-RGS161–62 but not to GST alone (Fig. 4B). Again, we found that MIR16 was also able to bind weakly to RGS2 but not to RGS4 (Fig. 4B).
Figure 4.
MIR16 selectively interacts with RGS proteins. (A) The interaction between MIR16 and several RGS proteins was tested in the yeast two-hybrid system. A schematic diagram of RGS proteins in the pAS2.1 vector is presented. β-Galactosidase filter assay of yeast transformants was performed on Leu−/Trp− plates, and transformants were scored for the intensity of color after 8 h: ++, strong color; +, intermediate color; −, no color. MIR16 interacts strongly with RGS16 and RGS161–62 and weakly with RGS2, RGS-GAIP, and RET-RGS1 but does not interact with RGS4. The experiment was repeated at least three times. (B) In vitro-translated, 35S-labeled MIR16 was incubated with the indicated GST proteins immobilized on glutathione-agarose beads. Radiolabeled MIR16 bound strongly to GST-RGS16 and more weakly to GST-RGS2 and GST-RGS161–62 but not to GST-RGS4 and GST alone.
Based on the data obtained from the two different interaction assays, we conclude that MIR16 interacts most strongly with RGS16, but it also interacts weakly with selected other RGS proteins.
The N-Terminal Part of the RGS Domain in RGS16 Is Required for Interaction with MIR16.
Because MIR16 is able to interact with multiple RGS proteins and because RGS domains are conserved among different RGS proteins, we reasoned that the interaction between RGS16 and MIR16 is likely to be mediated by the RGS domain. To test this idea, we analyzed the interaction between RGS16 deletion mutants and MIR16 in the yeast two-hybrid system. As shown in Fig. 5, MIR16 interacted with the RGS domain of RGS16 (amino acids 53–180) but failed to bind to its N terminus (amino acids 1–33 or 1–52). Moreover, it bound to a construct containing the N terminus plus the N-terminal part of the RGS domain (amino acids 1–62) and did not interact with the remainder of RGS16 (amino acids 63–201). These results indicate that the RGS domain of RGS16 is sufficient for its interaction with MIR16, and the first 10 aa of the RGS domain (amino acids 53–62) are critical for this interaction.
Figure 5.
Mapping the MIR16 interacting region in RGS16. Deletion mutants of RGS16, shown schematically, were cloned into pAS2.1 vector and tested for the interaction with MIR16 in the yeast two-hybrid system. β-Galactosidase filter assay of yeast transformants was performed on Leu−/Trp− plates, and scored for the intensity of color after 8 h: +++, very strong color; ++, strong color; −, no color. MIR16 bound to the RGS domain of RGS16 (amino acids 53–180) but not to its N terminus (amino acids 1–33 or 1–52). In addition, it interacted with amino acids 1–62 of RGS16 but not with amino acids 63–201. The experiment was repeated at least three times.
MIR16 Is an Integral Membrane Protein.
The presence of highly hydrophobic regions in MIR16 suggested that it could be an integral membrane protein. To test this possibility, we carried out in vitro translation of 35S-labeled MIR16 in the presence of canine pancreatic microsomes. We found that MIR16 sedimented with microsomes and remained associated with microsomal membranes after alkaline extraction (pH 11.0), a procedure that releases peripheral membrane proteins and content proteins (Fig. 6A).
Figure 6.
Analysis of the membrane association of MIR16 protein. (A) MIR16 protein was translated in vitro in the presence of canine microsomes. Microsomal pellets were sedimented from the reaction mixtures by centrifugation at 100,000 × g. Aliquots of the membrane pellets were treated with 0.2 M sodium carbonate (pH 11.5) for 30 min on ice and recentrifuged. The proteins associated with the membrane (P100) or soluble (S100) fraction with (+) and without (−) alkaline treatment were analyzed by SDS/PAGE. MIR16 sedimented with microsomes in the P100 fraction and was still associated with membranes after alkaline extraction. (B) HA-tagged MIR16 protein was overexpressed in HEK293 cells. S100 and P100 fractions were prepared from postnuclear supernatants (PNS; ≈50 μg) by centrifugation at 100,000 × g. Aliquots of the P100 fractions were subjected to alkaline treatment and centrifuged again at 100,000 × g. Proteins associated with each fraction were separated by SDS/PAGE and immunoblotted with anti-HA IgG. HA-tagged MIR16 is found exclusively in the P100 pellets both before and after alkaline treatment.
Similar results were obtained with HA-tagged MIR16 expressed in HEK293 cells: HA-MIR16 was found exclusively in the membrane pellet (100,000 × g) and remained associated with the membrane pellet after alkaline extraction (Fig. 6B). These results from both in vitro and in vivo membrane association assays show that MIR16 is an integral membrane protein. The precise membrane topology of MIR16 remains to be determined.
MIR16 Is a Glycoprotein.
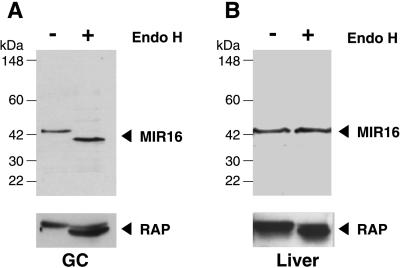
As mentioned earlier, the primary sequence of MIR16 contains two putative N-glycosylation sites (Fig. 1). To determine whether MIR16 undergoes glycosylation, we carried out digestion with endo H on membrane preparations from pituitary-derived GC cells followed by immunoblotting for MIR16. As indicated in Fig. 7A, before endo H treatment, the anti-MIR16 antibody recognized a protein of apparent molecular mass of 43 kDa. Treatment with endo H caused a shift in the molecular mass to ≈37 kDa, which is the predicted molecular mass of MIR16. Similar results were obtained with membranes prepared from LβT2 pituitary cells (data not shown). The positive control RAP, an endo H-sensitive glycoprotein (16), also showed a slight shift in mobility after treatment. These results indicate that, in two pituitary-derived cell lines, MIR16 is an endo H-sensitive, N-linked glycoprotein with high mannose oligosaccharides.
Figure 7.
MIR16 is a N-linked glycoprotein. P100 membrane fractions (50 μg) from GC cells (A) or rat liver lysates (B) were treated (+) or not (−) with endo H (3 milliunits) for 3 h and immunoblotted with antibodies to MIR16 or RAP. In GC cells, MIR16 undergoes a mobility shift from 43 to 37 kDa after endo H digestion. In contrast, no mobility shift is detected for MIR16 from rat liver, but a change is seen in the mobility of RAP used as a positive control.
We also performed endo H digestion on rat liver lysates. Unlike in GC or LβT2 cells, no shift in the mobility was seen after endo H treatment of rat liver lysates (Fig. 7B). Anti-MIR16 IgG recognizes the same 43-kDa band, presumably the N-glycosylated form, before and after endo H treatment. These results suggest that, in rat liver, the majority of MIR16 is endo H insensitive, suggesting that it is localized in a post-Golgi compartment.
Subcellular Distribution of MIR16.
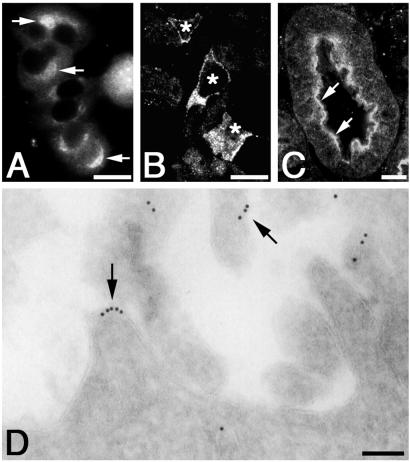
To determine the precise cellular distribution of MIR16, we carried out immunofluorescence on GC cells, PC-12 cells, and semithin cryosections of rat pituitary, kidney, and liver. MIR16 was localized in the juxtanuclear region of GC cells (Fig. 8A) and PC-12 cells (not shown). This localization and its endo H sensitivity suggest that MIR16 is present in a pre-mid-Golgi compartment (ERGIC, cis-Golgi) in these cell types. Staining was also seen on intracellular membranes of selected cells in semithin cryosections of rat pituitary (Fig. 8B). In rat kidney, MIR16 was mainly detected along the apical domain of proximal tubule cells (Fig. 8C), and in rat liver, MIR16 was distributed around the sinusoids (not shown). At the electron microscopic level, MIR16 was found on the sinusoidal domain of the plasma membrane (PM) of hepatocytes (Fig. 8D) and proximal tubule cells (not shown). The PM distribution of MIR16 in liver is in agreement with its endo H insensitivity. We conclude that MIR16 is found on intracellular membranes in the rat pituitary, in several cultured cell lines, and on the PM in rat liver and kidney.
Figure 8.
Subcellular localization of MIR16. (A) In GC cells, staining for MIR16 is found in the juxtanuclear region (arrows) by immunofluorescence. (B) In semithin cryosections of rat pituitary, MIR16 is localized on intracellular membranes in selected cells (marked with asterisks). (C) In semithin cryosections of rat kidney, MIR16 is concentrated along the apical domain of cells of the proximal tubule. (D) After immunogold labeling of ultrathin cryosections prepared from rat liver, gold particles, indicating the distribution of MIR16, are found on the sinusoidal domain of the PM of hepatocytes (arrows). GC cells and rat pituitary and kidney tissues were fixed and prepared as described in Materials and Methods and incubated with protein A-purified MIR16 IgG followed by Alexa 488 goat anti-rabbit IgG conjugate (A–C) or 5 nm gold, goat anti-rabbit IgG conjugate (D). (Bars = 10 μm in A–C and 100 nm in D.)
Discussion
In this study we have identified, characterized, and localized MIR16, a protein that interacts with RGS16. MIR16 binds to RGS16 as well as to several other RGS proteins in the yeast two-hybrid and GST pull-down assays. Moreover, we also have shown that MIR16 is an integral membrane glycoprotein mainly localized on the PM in rat liver and kidney and on intracellular membranes in rat pituitary, PC-12 cells, and several pituitary cell lines.
The primary sequence of MIR16 protein shows high homology with bacterial GP-PDEs (EC 3.1.4.46), suggesting that MIR16 is a putative GP-PDE. The two E. coli GP-PDEs GlpQ and UgpQ play important roles in the uptake and metabolism of GPs as well as the degradation product G3P, which is a major carbon and phosphate source (21, 24). In mammals, no GP-PDE has been cloned up to now. However, GP-PDE enzymatic activity with GPC (GPC-PDE, EC 3.1.4.2), glycerophosphoethanolamine, or glycerophosphoinositol (glycerophosphoinositol PDE, EC 3.1.4.44) as substrates has been observed in a number of mammalian tissues including liver (25, 26), brain (27), and kidney (28). Preliminary studies on the biochemical properties of the enzyme suggest that multiple enzymes may be responsible for the activity (29, 30). The MIR16 protein identified in this study may represent one of these enzymes.
Studies on the cellular roles of mammalian GP-PDEs are very limited; however, in mammals, GP-PDEs serve important metabolic roles. For example, GPC, the substrate of GPC-PDE, is an intermediate product of phosphocholine catabolism (31). The two esterified fatty acid moieties in the sn-1 and sn-2 position of phosphocholine can be removed by phospholipase A2 and lysophospholipase or phospholipase B alone to produce GPC. GPC-PDE subsequently hydrolyzes GPC to G3P, which can be reused in phospholipid synthesis and the G3P-NADH shuttle (28). Interestingly, GPC and other GPs have been suggested to regulate membrane phospholipid composition which in turn modifies membrane fluidity and activity of membrane-bound proteins (32). It has been proposed that lipid microdomains, such as caveolae and lipid rafts, may exist as organization centers for signal transduction (33, 34). Thus, it is conceivable that GP-PDEs play roles in modulation of signal transduction through altering the concentration of GPs. Assuming that the enzymatic activity of MIR16 can be confirmed, the identification and characterization of a putative mammalian GP-PDE should facilitate delineation of the physiological roles of GP-PDEs.
Besides binding to Gα proteins through their diagnostic RGS domains, RGS proteins have been found to interact with a variety of other proteins. Some of these RGS-interacting proteins, such as Gβ5 (35) and G protein-coupled receptors (36), are components of G protein signaling pathways. In other cases, functional domains in RGS proteins, including the RGS domain, provide protein–protein interaction links to other signaling molecules, such as small GTPases, protein kinase A, and components of Wnt signaling pathways (3). Another interesting RGS-interacting protein is retinal guanylyl cyclase, a critical enzyme in phototransduction. RGS9 was shown to bind to retinal guanylyl cyclase and inhibit its activity in a dose-dependent manner (37).
What could be the functional relationship between MIR16 and RGS16? There are at least three possibilities. First, the putative enzymatic activity of MIR16 suggests that, as in the case of guanylyl cyclase and RGS9, MIR16 could be a downstream effector of RGS16 and that the activity of GP-PDE might be regulated by RGS proteins and/or heterotrimeric G proteins. Secondly, MIR16 might serve as a regulator of RGS16's GTPase activating protein activity. Our finding that MIR16 interacts with the RGS domain of RGS16, which also mediates the interaction between RGS16 and Gα subunits, suggests that MIR16 could have competitive or cooperative effects on the interaction between Gα proteins and RGS16. Finally, there is a possibility that MIR16 could function as a membrane anchoring protein for RGS16. When overexpressed in HEK293 cells (unpublished results) and in yeast cells (38), RGS16 was shown to exist in two pools, cytosolic and membrane-associated. The finding that MIR16 is an integral membrane makes it a candidate for an anchoring protein.
It is noteworthy that MIR16 not only binds RGS16 but also selectively binds other RGS proteins. The mammalian RGS family has recently been classified into six subfamilies (A–F; ref. 39). We have tested the interaction between MIR16 and several RGS proteins from mammalian RGS subfamilies A and B. Whether MIR16 can interact with members of other RGS subfamilies remains to be seen. The N-terminal 10 aa of the RGS domain of RGS16 are required for the interaction, but comparison of the 10 aa in RGS proteins that bind MIR16 vs. RGS4, which does not bind, failed to reveal any common feature that could explain the selectivity.
MIR16 is apparently found on both intracellular membranes and on the PM in different cell types. It was localized on intracellular membranes in rat pituitary and in pituitary-derived GC cells and in PC-12 cells but was found on the PM in liver and kidney. In preliminary experiments, we localized RGS16 in rat liver by using a polyclonal antibody against the N terminus of RGS16 and found labeling for RGS16 on the sinusoidal PM of hepatocytes, suggesting overlap in the distribution of MIR16 and RGS16 (unpublished results). Currently, it is not possible to colocalize MIR16 and RGS16 because of lack of suitable antibodies.
In summary, we have discovered a RGS16-interacting membrane protein, MIR16, which is a putative GP-PDE and is localized on the PM and intracellular membranes. The future challenge will be to characterize the enzymatic activity of MIR16 and to understand its functional connection with heterotrimeric G protein signaling.
Acknowledgments
We thank Tammy McQuistan for technical assistance with the immunofluorescence and immunoelectron microscopy. This work was supported by National Institutes of Health Grants DK17780 and CA58689 (to M.G.F.). B.Z. and D.C. are members of the Molecular Pathology Graduate Program, University of California San Diego, and B.Z. is supported by the Huang Memorial Scholarship.
Abbreviations
- endo H
endoglycosidase H
- G3P
sn-glycerol-3-phosphate
- GPC
glycerophosphocholine
- GP
glycerophosphodiester
- PDE
phosphodiesterase
- GST
glutathione S-transferase
- HA
hemagglutinin
- r-
m-, and hMIR16, rat, mouse, and human MIR16
- PM
plasma membrane
Footnotes
References
- 1.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 3.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar M G. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Hepler J R. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- 5.De Vries L, Farquhar M G. Trends Cell Biol. 1999;9:138–144. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 6.De Vries L, Elenko E, McCaffery J M, Fischer T, Hubler L, McQuistan T, Watson N, Farquhar M G. Mol Biol Cell. 1998;9:1123–1134. doi: 10.1091/mbc.9.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson B S, Komuro M, Farquhar M G. Endocrinology. 1994;134:233–244. doi: 10.1210/endo.134.1.8275939. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Zheng B, Han J, Lin S C. J Biol Chem. 1997;272:8679–8685. doi: 10.1074/jbc.272.13.8679. [DOI] [PubMed] [Google Scholar]
- 9.Chen C K, Wieland T, Simon M I. Proc Natl Acad Sci USA. 1996;93:12885–12889. doi: 10.1073/pnas.93.23.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckbinder L, Velasco-Miguel S, Chen Y, Xu N, Talbott R, Gelbert L, Gao J, Seizinger B R, Gutkind J S, Kley N. Proc Natl Acad Sci USA. 1997;94:7868–7872. doi: 10.1073/pnas.94.15.7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Druey K M, Ugur O, Caron J M, Chen C K, Backlund P S, Jones T L. J Biol Chem. 1999;274:18836–18842. doi: 10.1074/jbc.274.26.18836. [DOI] [PubMed] [Google Scholar]
- 12.Lin P, Le-Niculescu H, Hofmeister R, McCaffery J M, Jin M, Hennemann H, McQuistan T, De Vries L, Farquhar M G. J Cell Biol. 1998;141:1515–1527. doi: 10.1083/jcb.141.7.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S, Gish W, Miller W, Myers E, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.De Vries L, Lou X, Zhao G, Zheng B, Farquhar M G. Proc Natl Acad Sci USA. 1998;95:12340–12345. doi: 10.1073/pnas.95.21.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlando R A, Farquhar M G. Proc Natl Acad Sci USA. 1994;91:3161–3165. doi: 10.1073/pnas.91.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas P, Mellon P L, Turgeon J, Waring D W. Endocrinology. 1996;137:2979–2989. doi: 10.1210/endo.137.7.8770922. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Shraga-Levine Z, Sokolovsky M. Biochem Biophys Res Commun. 1998;246:495–500. doi: 10.1006/bbrc.1998.8646. [DOI] [PubMed] [Google Scholar]
- 20.Kasahara M, Makino K, Amemura M, Nakata A. Nucleic Acids Res. 1989;17:2854. doi: 10.1093/nar/17.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tommassen J, Eiglmeier K, Cole S T, Overduin P, Larson T J, Boos W. Mol Gen Genet. 1991;226:321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- 22.Munson R S, Jr, Sasaki K. J Bacteriol. 1993;175:4569–4571. doi: 10.1128/jb.175.14.4569-4571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson T J, Ehrmann M, Boos W. J Biol Chem. 1983;258:5428–5432. [PubMed] [Google Scholar]
- 24.Brzoska P, Boos W. FEMS Microbiol Rev. 1989;5:115–124. doi: 10.1016/0168-6445(89)90015-6. [DOI] [PubMed] [Google Scholar]
- 25.Dawson R M C. Biochem J. 1956;62:689–696. doi: 10.1042/bj0620689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd-Davies K A, Michell R H, Coleman R. Biochem J. 1972;127:357–368. doi: 10.1042/bj1270357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster G R, Marples E A, Thompson R H S. Biochem J. 1957;65:374–377. doi: 10.1042/bj0650374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baldwin J J, Cornatzer W E. Biochim Biophys Acta. 1968;164:195–204. doi: 10.1016/0005-2760(68)90146-x. [DOI] [PubMed] [Google Scholar]
- 29.Spanner S, Ansell G B. Biochem J. 1982;208:845–850. doi: 10.1042/bj2080845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross B M, Sherwin A L, Kish S J. Lipids. 1995;30:1075–1081. doi: 10.1007/BF02536607. [DOI] [PubMed] [Google Scholar]
- 31.Burg M B. Am J Physiol. 1995;268:F983–F996. doi: 10.1152/ajprenal.1995.268.6.F983. [DOI] [PubMed] [Google Scholar]
- 32.Burt C T, Ribolow H J. Biochem Med. 1984;31:21–30. doi: 10.1016/0006-2944(84)90054-1. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson K, Dietrich C. Trends Cell Biol. 1999;9:87–91. doi: 10.1016/s0962-8924(98)01495-0. [DOI] [PubMed] [Google Scholar]
- 34.Anderson R G. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 35.Snow B E, Krumins A M, Brothers G M, Lee S F, Wall M A, Chung S, Mangion J, Arya S, Gilman A G, Siderovski D P. Proc Natl Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snow B E, Hall R A, Krumins A M, Brothers G M, Bouchard D, Brothers C A, Chung S, Mangion J, Gilman A G, Lefkowitz R J, et al. J Biol Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- 37.Seno K, Kishigami A, Ihara S, Maeda T, Bondarenko V A, Nishizawa Y, Usukura J, Yamazaki A, Hayashi F. J Biol Chem. 1998;273:22169–22172. doi: 10.1074/jbc.273.35.22169. [DOI] [PubMed] [Google Scholar]
- 38.Chen C, Seow K T, Guo K, Yaw L P, Lin S C. J Biol Chem. 1999;274:19799–19806. doi: 10.1074/jbc.274.28.19799. [DOI] [PubMed] [Google Scholar]
- 39.Zheng B, De Vries L, Farquhar M G. Trends Biochem Sci. 1999;14:411–415. doi: 10.1016/s0968-0004(99)01474-7. [DOI] [PubMed] [Google Scholar]