Abstract
Studies with mutants in four members of the five-membered Arabidopsis phytochrome (phy) family (phyA, phyB, phyD, and phyE) have revealed differential photosensory and/or physiological functions among them, but identification of a phyC mutant has proven elusive. We now report the isolation of multiple phyC mutant alleles using reverse-genetics strategies. Molecular analysis shows that these mutants have undetectable levels of phyC protein, suggesting that they are null for the photoreceptor. phyC mutant seedlings were indistinguishable from wild-type seedlings under constant far-red light (FRc), and phyC deficiency had no effect in the phyA mutant background under FRc, suggesting that phyC does not participate in the control of seedling deetiolation under FRc. However, when grown under constant red light (Rc), phyC seedlings exhibited a partial loss of sensitivity, observable as longer hypocotyls and smaller cotyledons than those seen in the wild type. Although less severe, this phenotype resembles the effect of phyB mutations on photoresponsiveness, indicating that both photoreceptors function in regulating seedling deetiolation in response to Rc. On the other hand, phyB phyC double mutants did not show any apparent decrease in sensitivity to Rc compared with phyB seedlings, indicating that the phyC mutation in the phyB-deficient background does not have an additive effect. These results suggest that phyB is necessary for phyC function. This functional dependence correlates with constitutively lower levels of phyC observed in the phyB mutant compared with the wild type, a decrease that seems to be regulated post-transcriptionally. phyC mutants flowered early when grown in short-day photoperiods, indicating that phyC plays a role in the perception of daylength. phyB phyC double mutant plants flowered similarly to phyB plants, indicating that in the phyB background, phyC deficiency does not further accelerate flowering. Under long-day photoperiods, phyA phyC double mutant plants flowered later than phyA plants, suggesting that phyC is able to promote flowering in the absence of phyA. Together, these results suggest that phyC is involved in photomorphogenesis throughout the life cycle of the plant, with a photosensory specificity similar to that of phyB/D/E and with a complex pattern of differential crosstalk with phyA and phyB in the photoregulation of multiple developmental processes.
INTRODUCTION
Plant life depends on light. Plants need light as the source of energy for photosynthesis and also as a crucial environmental signal to ensure survival and reproduction. They constantly monitor the presence, absence, duration, intensity, quality, and direction of light to adjust their growth and development appropriately in a process termed photomorphogenesis. Fluctuations in the light environment are monitored using informational photoreceptors, namely the cryptochromes (Cashmore et al., 1999) and phototropins (Briggs et al., 2001), which are the receptors for UV-A and blue light, and the phytochromes (Neff et al., 2000; Smith, 2000; Fankhauser, 2001), which monitor the red (R) and far-red (FR) regions of the light spectrum. The molecular properties of these photoreceptors enable them to perceive and transduce the light signal to downstream cellular components in a process that culminates in the modulation of the expression of genes responsible for orchestrating photomorphogenesis (Chory and Wu, 2001; Quail, 2002).
The phytochromes are soluble dimeric chromoproteins with two structural domains: a globular photoactive N-terminal half that bears the light-absorbing tetrapyrrole chromophore, and a linear C-terminal half that carries dimerization and regulatory determinants (Quail, 1997). Phytochromes have the unique capacity of existing in two photointerconvertible forms, Pr and Pfr. The molecule is synthesized in the biologically inactive Pr form that absorbs R. Upon R absorption, the Pr form is converted to the biologically active Pfr form that absorbs FR to covert back to Pr. Phytochromes are involved in the control of many major processes during plant development, including germination, seedling deetiolation, synthesis of the photosynthetic machinery, floral induction and tuberization, and responses to competing neighboring plants.
In Arabidopsis, the phytochrome (phy) family is composed of five members, phyA through phyE (Mathews and Sharrock, 1997). Phylogenetic analysis indicates that phytochrome genes in Arabidopsis have evolved by duplication and divergence from a common ancestor (Clack et al., 1994). A PHYA/C and a PHYB/D/E gene ancestor arose relatively early in evolution. The PHYA/C ancestor duplicated fairly early and gave rise to PHYA and PHYC. The PHYB/D/E ancestor duplicated first to give rise to PHYE and again more recently to produce PHYB and PHYD.
phyA is the predominant phytochrome species in etiolated tissue, and phyB is predominant in seedlings grown under light. phyA is highly light labile, whereas the other four phytochromes are much more light stable, although phyB and phyC are less abundant in light-grown seedlings than in the dark. The light-stable phytochromes are present throughout the life cycle of the plant and are present ubiquitously in all organs (Sharrock and Clack, 2002).
Subcellular localization studies using phytochrome:green fluorescent protein (GFP) fusions show that phyA:GFP is localized in the cytoplasm in the dark and translocates to the nucleus in response to R (Kircher et al., 1999). phyB:GFP seems to be predominantly cytoplasmic in the dark and also concentrates in the nucleus upon R irradiation (Kircher et al., 1999; Yamaguchi et al., 1999). By contrast, phyC:GFP, phyD:GFP, and phyE:GFP have been shown to be localized to the nucleus regardless of the light regime (Kircher et al., 2002).
The physiological functions of individual phytochrome species have been studied using different strategies. The most revealing approach has been the isolation and characterization of Arabidopsis mutants deficient in one or more phytochromes (Whitelam and Devlin, 1997). Mutant analysis has revealed that individual members of the family have differential but frequently overlapping functions in the control of plant responses to R and FR. Arabidopsis mutant seedlings lacking phyA do not display deetiolation responses to constant far-red light (FRc) (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993), suggesting that phyA is the only phytochrome that mediates seedling responses to FRc. phyA seedlings have elongated hypocotyls and closed and unexpanded cotyledons when grown in FRc, indistinguishable in appearance from dark-grown, wild-type seedlings. Adult phyA mutant plants show alterations in the induction of flowering, being late flowering under certain photoperiodic conditions (Johnson et al., 1994; Reed et al., 1994; Neff and Chory, 1998). phyA also is involved in seed germination (Shinomura et al., 1994).
Mutants deficient in phyB have reduced sensitivity to R. phyB seedlings display loss of the inhibition of hypocotyl elongation, cotyledon opening and expansion, and chlorophyll synthesis when grown in prolonged R (Koornneef et al., 1980; Reed et al., 1993). phyB also is involved in seed germination (Shinomura et al., 1994). Adult phyB plants are early flowering and have altered leaf morphology in response to low-fluence-rate Rc or a low ratio of R to FR, which provides evidence that phyB is involved in regulating shade-avoidance responses (Goto et al., 1991; Halliday et al., 1994; Smith and Whitelam, 1997).
Mutants deficient in phyD show a slight reduction of the inhibition of hypocotyl elongation and cotyledon expansion when grown under Rc (Aukerman et al., 1997). These seedling phenotypes were more pronounced in a phyB null background. phyD deficiency in adult plants is apparent only in a phyB null background, providing evidence that phyD is involved in mediating the same responses as phyB and that there is some functional redundancy between the two phytochrome species, because phyB phyD double mutants are more elongated and flower earlier than phyB plants (Devlin et al., 1999). Monogenic mutants that lack phyE display a wild-type phenotype. Analysis of mutants deficient in both phyE and phyB revealed that phyE is involved in the regulation of shade-avoidance responses in a partially conditionally redundant manner to phyB (Devlin et al., 1998). phyE also has been shown to be involved in the control of seed germination (Hennig et al., 2002) and to participate in the control of cotyledon expansion under Rc (Franklin et al., 2003).
To date, the isolation of phyC mutants in Arabidopsis using forward genetics has remained elusive. Therefore, we used a reverse-genetics approach to screen the available mutagenized Arabidopsis collections and have identified three different mutant alleles of the PHYC gene. Here, we describe the isolation of these phyC mutants and the physiological and molecular phenotypic effects of phyC deficiency.
RESULTS
Identification of phyC Mutants
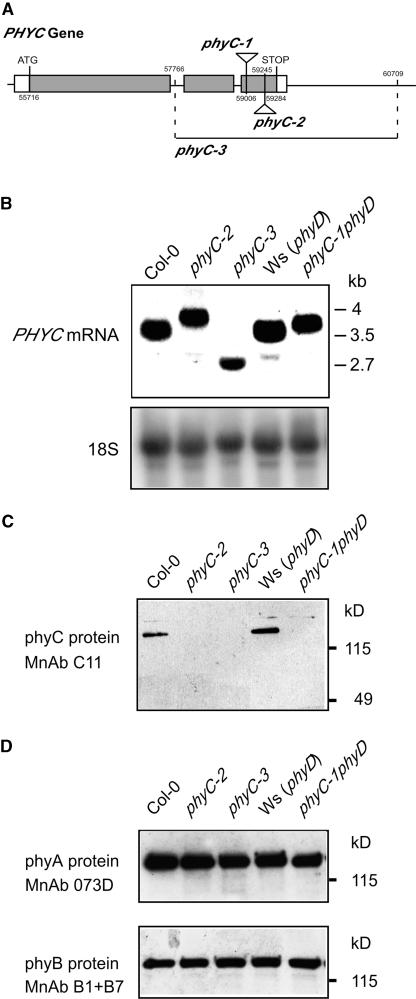
Different pools of Arabidopsis T-DNA insertion lines were screened using a PCR screening strategy, and two mutants were isolated, phyC-1 and phyC-2, each with a T-DNA inserted in the PHYC gene. To determine the exact insertion site, a region containing genomic and T-DNA border sequence was amplified by PCR for each allele. This region was sequenced and compared with the wild-type PHYC gene sequence (Arabidopsis genomic DNA clone MIK22, positions 55716 to 59284). The data indicate that the insertion in phyC-1 is in the 3′ region of the third exon (position 59006; Figure 1A). The T-DNA insertion in phyC-2 also is in the third exon of the PHYC gene at the 5′ end (position 59245; Figure 1A).
Figure 1.
phyC Mutants Express Aberrant PHYC Transcripts and Have Undetectable Levels of phyC Protein.
(A) Mutations in the Arabidopsis PHYC gene (At5g35840). T-DNA inserts in phyC-1 and phyC-2 are located in the third exon of PHYC. The T-DNA insert in phyC-1 is located at position 59006, and phyC-2 carries the T-DNA insert at position 59245. The fast-neutron–induced deletion of the PHYC gene in phyC-3 spans the region between position 57766 in the first intron and position 60709. Numbering of coordinates is based on the Arabidopsis genomic clone MIK22 that contains the Col-0 PHYC gene.
(B) RNA gel blot of RNA extracts of wild-type and phyC mutant seedlings grown in Rc for 4 days probed with a full-length PHYC cDNA probe. As a control, the blot was reprobed to detect 18S RNA.
(C) Immunoblot of protein extracts of wild-type and phyC mutant seedlings grown in the dark for 4 days probed with the phyC-specific monoclonal antibody (MnAb) C11.
(D) Immunoblots of protein extracts of wild-type and phyC mutant seedlings grown in the dark for 4 days probed with monoclonal antibodies 073D specific for phyA and B1 and B7 specific for phyB.
In addition, we also screened pools of Arabidopsis fast-neutron–induced deletion mutants using PCR (Li et al., 2001) and identified a line with a 2.9-kb deletion that affects the PHYC gene. This mutant was designated phyC-3. To determine the exact junction of the deletion in phyC-3, a mutant band encompassing the deleted region was sequenced and compared with the wild type. This analysis indicated that the deletion spans from the first intron in the PHYC gene to ∼1.4 kb downstream of the 3′ untranslated region (positions 57766 through 60709; Figure 1A). We examined whether this deletion might affect any other adjacent gene. Based on the annotated genome, the closest predicted gene downstream of PHYC is gene MIK22.16. Gene MIK22.16 is antiparallel to PHYC, and the predicted open reading frame encodes a protein of 49 amino acids with a predicted stop codon ∼1.3 kb downstream of the deletion junction at position 62072 in the BAC. Therefore, it appears likely that the deletion in phyC-3 affects only the PHYC gene.
phyC-2 and phyC-3 are in the Columbia (Col-0) ecotype, making them monogenic, whereas phyC-1 is in Wassilewskija (Ws), which is naturally null for phyD, rendering phyC-1 a phyC phyD double mutant (Aukerman et al., 1997).
phyC Mutants Express PHYC mRNA of Different Sizes and No Detectable Levels of phyC Protein
The effect of the mutations on the PHYC transcript was assayed by RNA gel blot analysis. RNA was extracted from 4-day-old seedlings grown under Rc and hybridized with a PHYC-specific probe. The T-DNA insertional mutants phyC-1 and phyC-2 expressed higher molecular mass mRNA compared with their respective wild types, Col-0 and Ws (∼4 versus 3.5 kb), whereas the deletion mutant phyC-3 expressed a smaller mRNA compared with the Col-0 wild type (∼2.7 versus 3.5 kb) (Figure 1B). The levels of transcript in phyC-1 and phyC-2 seemed to be reduced slightly compared with that in the wild type. By contrast, the deletion in phyC-3 appeared to have a more severe effect on the stability of the transcript, the levels being reduced by ∼30% with respect to the wild type, Col-0.
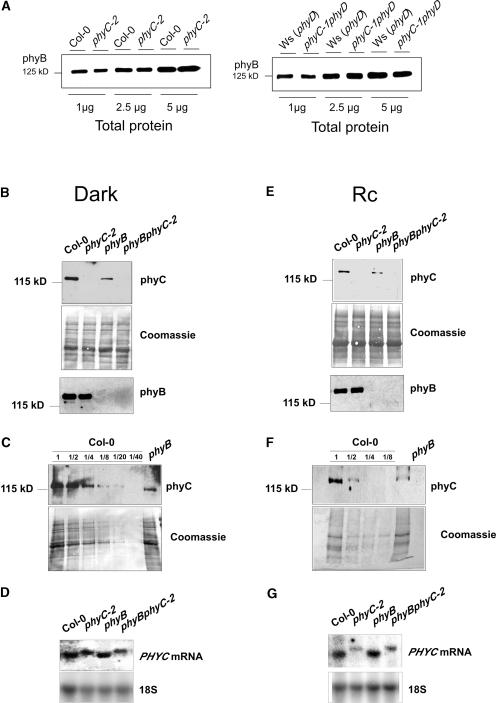
To determine the effect of the phyC mutations on the phyC protein, immunoblot analysis was performed using a monoclonal antibody, C11, that selectively recognizes Arabidopsis phyC (Somers et al., 1991). Monoclonal antibody C11 recognizes an epitope in the N-terminal half of the phyC molecule (J. Martínez-García and E. Monte, unpublished results); thus, it is expected to recognize all three truncated mutant proteins if present. Protein was extracted from 4-day-old seedlings grown in the dark, in which the levels of phyC are highest compared with the levels in seedlings grown under Rc or FRc (Sharrock and Clack, 2002). Figure 1C shows the levels of phyC in the different mutant alleles compared with those of their respective wild types. In phyC-1 and phyC-2, we were unable to detect any band corresponding to phyC even after longer exposure times, indicating that the T-DNA insertions in these mutant alleles cause deficiency in phyC. In the deletion mutant phyC-3, in which no full-length phyC should be present, we detected no band corresponding to either full-length phyC or any truncated derivative, indicating that the deletion in the PHYC gene in phyC-3 causes deficiency for phyC. These immunoblot results suggest that all three of our phyC mutant alleles are likely to be loss-of-function null alleles.
We also tested whether the levels of phyA and phyB are affected as a result of the mutations in PHYC. phyC protein extracts were subjected to immunoblot analysis using monoclonal antibodies specific for phyA and phyB (Somers et al., 1991). Figure 1D shows that the levels of phyA and phyB detected in the phyC mutants do not differ from those of the corresponding wild type.
The phyC Mutation Confers Seedling Hyposensitivity to Rc
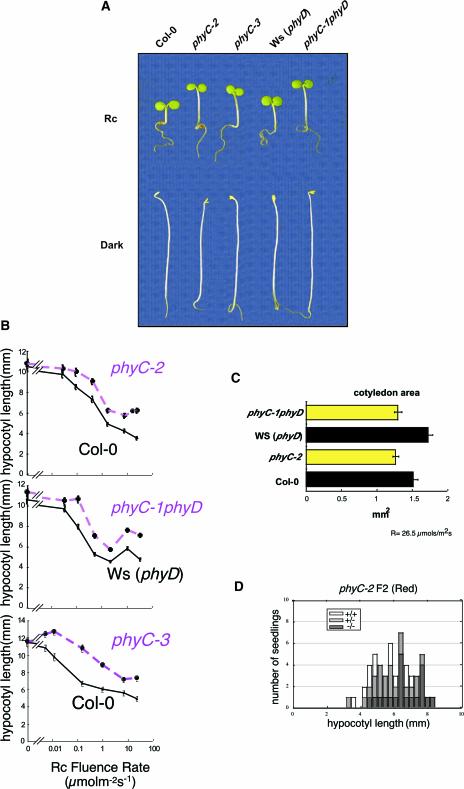
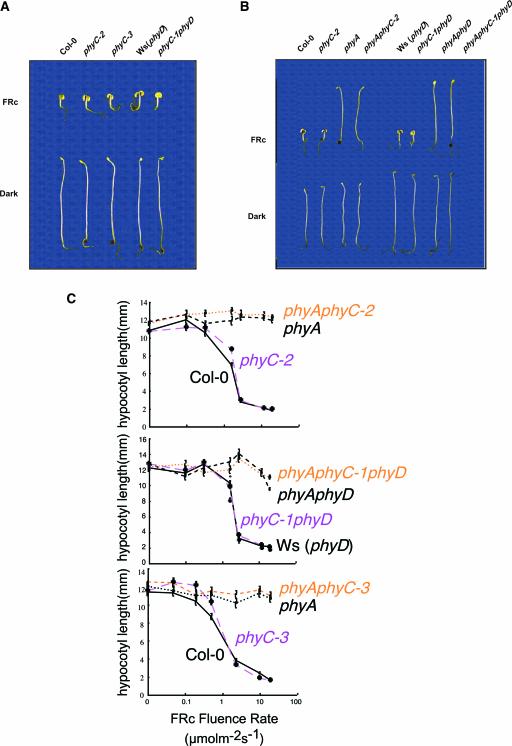
phyC seedlings displayed more elongated hypocotyls compared with wild-type seedlings when grown under Rc (Figure 2A). By contrast, when grown in darkness, the phyC mutant alleles were indistinguishable from the corresponding wild types (Figure 2A). This phenotype was observed for all three mutant alleles over a range of Rc fluence rates (Figure 2B), indicating that phyC mutants are hyposensitive to all Rc fluence rates tested. The loss of sensitivity also was observed in the cotyledons. phyC mutant cotyledons were less expanded than the corresponding wild-type cotyledons in Rc (Figure 2C).
Figure 2.
Deetiolation in phyC Mutants Is Hyposensitive to Rc.
(A) Visual phenotypes of phyC seedlings grown in Rc (18 μmol·m−2·s−1) or darkness for 4 days.
(B) Rc fluence-rate response curves for hypocotyl length in (top to bottom) wild-type Col-0 and phyC-2, Ws (phyD) and phyC-1 phyD, and Col-0 and phyC-3.
(C) Cotyledon area of phyC-1 phyD and phyC-2 seedlings grown under Rc (26.5 μmol·m−2·s−1) for 4 days.
(D) Correlation between hypocotyl length and the presence of T-DNA in phyC-2. Four-day-old seedlings of the segregating F2 population resulting from the cross of phyC-2 to wild-type Col-0 were grown in Rc (1.85 μmol·m−2·s−1), and individual seedlings were measured and genotyped. The distribution of wild-type (+/+), heterozygous mutant (+/−), and homozygous mutant (−/−) seedlings is shown superimposed on hypocotyl length.
To verify that the reduced inhibition of hypocotyl elongation is attributable to the mutation in PHYC, the phenotype was tested for cosegregation with the T-DNA. The hypocotyl length of 4-day-old Rc-grown phyC-2 F2 seedlings segregating for the T-DNA inserted in PHYC was measured and correlated with the copy number of the T-DNA as detected by PCR. The results of this analysis show that the T-DNA was present preferentially in tall seedlings, whereas short seedlings were mostly genotypically wild type (Figure 2D). This correlation was lost when the F2 population was grown in dark conditions (data not shown). These findings provide evidence that the loss of sensitivity to Rc is a consequence of the mutation in the PHYC gene. Also, the fact that the three independent phyC mutant alleles showed the same phenotype provides additional support for the correlation between phyC deficiency and hyposensitivity to Rc.
The analysis of the distribution of hypocotyl length in the F2 segregating population shown in Figure 2D also indicates that the phyC mutation is partially dominant, because the heterozygous population has an intermediate hypocotyl phenotype compared with the wild type and the phyC mutant.
phyC Function in Seedling Deetiolation in Rc Requires the Presence of phyB
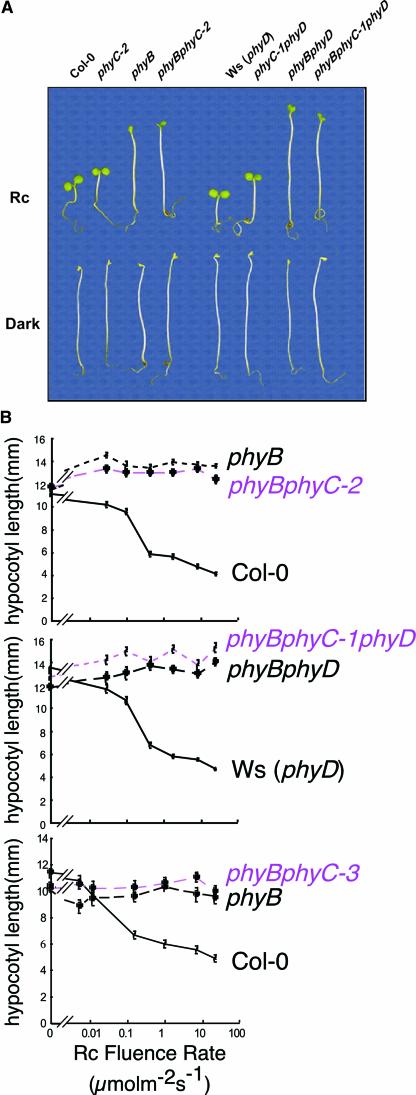
Figure 3A shows that phyB phyC double mutant seedlings were indistinguishable from monogenic phyB mutant seedlings when grown in Rc. This was true for both phyB phyC-2 and phyB phyC-3 mutant alleles in all Rc fluence rates tested (Figure 3B). Analysis of phyB phyC-1 (-phyB phyC phyD triple mutant) also revealed that they had the same hypocotyl length in Rc as the phyB phyD double mutant (Figure 3B). This observation is not unexpected, however, given that the monogenic phyB mutant displayed no detectable hypocotyl responsiveness to Rc under these experimental conditions compared with dark controls (Figure 3B). Therefore, these data indicate that hypocotyl responsiveness to Rc is fully dependent on phyB and that none of the remaining phytochrome family members, including phyC, can substitute functionally for phyB in this response.
Figure 3.
phyC and phyB Hypocotyl Length Hyposensitivity to Rc Are Not Additive.
(A) Visual phenotypes of phyC, phyB, and phyB phyC seedlings grown in Rc (18 μmol·m−2·s−1) or darkness for 4 days.
(B) Rc fluence-rate response curves for hypocotyl length in (top to bottom) phyB and phyB phyC-2, phyB phyD and phyB phyC-1 phyD, and phyB and phyB phyC-3.
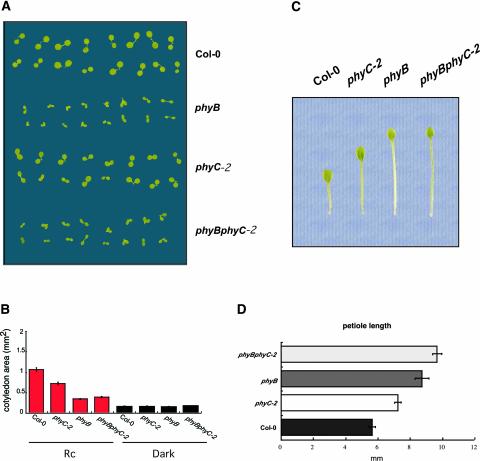
To further characterize phyB phyC seedling deetiolation, the responsiveness of the cotyledons of the phyC-2 mutant to Rc was analyzed and compared with that of the wild type, phyB, and phyB phyC-2. Figures 4A and 4B show that both phyB and phyC monogenic mutants had reduced cotyledon area in Rc compared with the wild type. This reduction was greater in phyB. By contrast, when grown in darkness, the area of the cotyledons was the same in all genotypes tested (Figure 4B). phyB cotyledons expanded more than those of dark controls in response to Rc, indicating that one or more remaining phytochromes is active in this response. However, because the phyB phyC-2 double mutant had the same cotyledon area as the single phyB mutant in Rc, the data suggest that phyC is not responsible for the residual responsiveness of phyB cotyledons to Rc. Thus, for both hypocotyl length and cotyledon expansion, the phenotype of the phyB phyC double mutant was indistinguishable from that of the monogenic phyB, with no additive effect of phyC. Together, these results indicate that phyC function in seedling deetiolation in Rc may require the presence of phyB.
Figure 4.
Cotyledon Area and Petiole Length of phyB, phyC, and phyB phyC Mutant Seedlings.
(A) Visual phenotypes of cotyledons of phyB, phyC-2, and phyB phyC-2 seedlings grown under Rc (18 μmol·m−2·s−1) for 4 days.
(B) Cotyledon area for the different genotypes shown in (A) grown under Rc or in the dark.
(C) Visual phenotypes of primary leaves of phyB, phyC, and phyB phyC rosettes grown under WLc (2.7 μmol·m−2·s−1) for 3 weeks.
(D) Petiole length for the different genotypes shown in (C).
phyC Mutants Exhibit Elongated Petioles and Larger Primary Leaf Area When Grown in Continuous White Light
We grew seedlings under continuous white light (WLc) for 3 weeks from germination and then measured petiole length of the longest leaf in the rosette, which corresponded to the primary leaf in all genotypes tested. Figures 4C and 4D show that the phyC-2 monogenic mutation caused increased petiole elongation compared with that in the Col-0 wild type. These results indicate that phyC plays a role in the control of petiole elongation in WLc. Visually, it was observed that this increase in petiole elongation correlated with a decrease in leaf area (Figure 4C). A role for phyC in primary leaf expansion was proposed by Qin et al. (1997) based on the observation that primary leaves in transgenic Arabidopsis plants overexpressing phyC were larger than those of the corresponding wild type.
As described previously (Reed et al., 1993), the phyB mutant under our conditions also exhibited elongated petioles, longer than either the wild type or phyC (Figures 4C and 4D). Based on the phenotypes of the phyC and phyB single mutants, the contribution of phyC compared with that of phyB seems to be greater in petiole elongation than in hypocotyl growth or cotyledon expansion. Interestingly, the petioles in the phyB phyC-2 double mutant were longer than those in the single phyB mutant (Figure 4D). Thus, although the effect of phyC and phyB deficiency on petiole length and leaf area were not quantitatively additive in phyB phyC-2, the data indicate that phyC is at least partially active in the absence of phyB in regulating this phenotypic response.
phyC Does Not Participate in the Control of Seedling Deetiolation in FRc
When grown under FRc, phyC mutant seedlings were indistinguishable from wild-type seedlings with regard to hypocotyl length over a wide range of FRc fluence rates (Figures 5A and 5C). Also, no significant effect on hypocotyl growth was seen in the double phyA phyC-2 or phyA phyC-1 (-triple phyA phyC phyD) mutant compared with their corresponding monogenic phyA mutant (Figures 5B and 5C). Cotyledons of wild-type and phyC-2 seedlings grown in FRc also were compared, and no difference was detected in area or morphology (data not shown). Nor was any difference seen when the cotyledons of phyA were compared with those of phyA phyC (data not shown). Together, these results indicate that phyC does not play a role in seedling deetiolation under FRc.
Figure 5.
phyC Does Not Participate in the Control of Seedling Deetiolation under FRc.
(A) Visual phenotypes of phyC seedlings grown in FRc (1.9 μmol·m−2·s−1) or darkness for 4 days.
(B) Visual phenotypes of phyC and phyA phyC seedlings grown in FRc (1.9 μmol·m−2·s−1) or darkness for 4 days.
(C) FRc fluence-rate response curves for hypocotyl length in (top to bottom) wild-type Col-0, phyC-2, phyA, and phyA phyC-2; wild-type Ws, phyC-1, phyA phyD, and phyA phyC-1 phyD; and wild-type Col-0, phyC-3, phyA, and phyA phyC-3.
phyA phyC Response under Rc
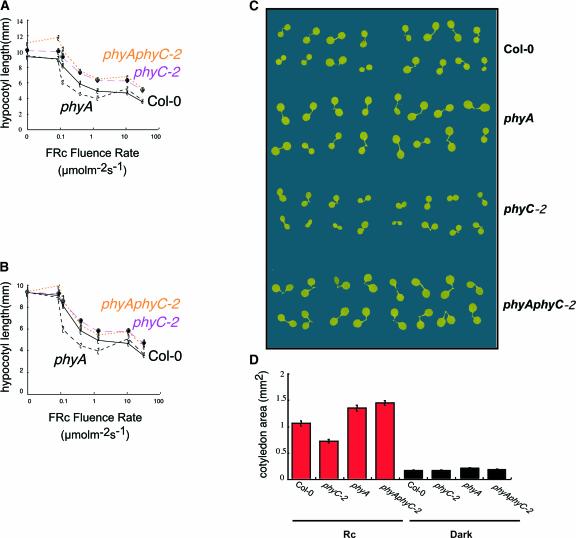
To determine whether phyA might be synergistic to phyC in Rc, the hypocotyl lengths of phyA phyC-2 double mutants were examined over a range of fluence rates of Rc compared with those of the phyC-2 monogenic mutant as well as the wild type and phyA (Figure 6A). To be able to discriminate small effects, the hypocotyl lengths were normalized to the dark Col-0 control values (Figure 6B). We observed that monogenic phyA hypocotyls were shorter than wild-type hypocotyls at low Rc fluence rates but were the same as wild-type hypocotyls at higher fluence rates. When normalized, no significant difference in hypocotyl length was observed between phyC-2 and the phyA phyC-2 double mutant (Figure 6B).
Figure 6.
Deetiolation in the phyA phyC Double Mutant under Rc.
(A) Rc fluence-rate response curves for hypocotyl length in wild-type Col-0, phyC-2, phyA, and phyA phyC-2.
(B) Normalized Rc fluence-rate response curves for hypocotyl length in wild-type Col-0, phyC-2, phyA, and phyA phyC-2. Values are normalized to the value for Col-0 in the dark.
(C) Visual phenotypes of cotyledons of phyA, phyC-2, and phyA phyC-2 seedlings grown under Rc (18 μmol·m−2·s−1) for 4 days.
(D) Cotyledon area for the different genotypes shown in (C).
We also examined the cotyledon area and morphology of phyA and phyA phyC-2 seedlings grown in Rc. We observed greater cotyledon area in the phyA mutant than in the wild type (Figures 6C and 6D). The area also was enlarged in phyA phyC-2 double mutant seedlings compared with the wild type, but it was the same as that in the phyA seedlings (Figures 6C and 6D).
Together, these results indicate that phyA does not appear to be synergistic to phyC in the control of hypocotyl length under Rc, and they suggest that phyA might be able to antagonize phyC function in Rc.
phyC Is Involved in the Control of Flowering Time in Arabidopsis
We grew seedlings in long-day (LD) and short-day (SD) photoperiods to determine whether phyC plays a role in the control of flowering time (Figure 7, Table 1). Under LD conditions, the phyC-2 mutant flowers at the same time as the wild type, recorded as days to flowering and number of rosette leaves at bolting (Table 1). phyA flowered at approximately the same time as the wild type, although with a greater number of rosette leaves. In the phyA background, the phyC mutation led to late flowering under LD conditions compared with the wild type and phyA. These results suggest that phyC may play a role in sensing LD photoperiod and that this role is redundant to phyA. As described previously (Goto et al., 1991; Whitelam and Smith, 1991; Bagnall et al., 1995), phyB flowered early also in our LD conditions (Figure 7A, Table 1). In the phyB background, no significant effect of the phyC mutation was observed: the double phyB phyC mutants flowered at the same time as phyB (Figure 7A, Table 1).
Figure 7.
Control of Flowering in Arabidopsis by phyC.
(A) phyB and phyB phyC-2 flower early under LD conditions.
(B) phyC-2 and phyA phyC-2 flower early under SD conditions.
Table 1.
Effect of LD and SD Photoperiods on Flowering Time in phyC-2, phyA phyC-2, and phyB phyC-2 seedlings
| Genotype | Conditions
|
|||
|---|---|---|---|---|
| LDa
|
SDb
|
|||
| Days to Flowering |
Leaf Number at Flowering |
Days to Flowering |
Leaf Number at Flowering |
|
| Col-0 | 25.3 ± 0.3 | 9.2 ± 0.1 | 64.8 ± 2.6 | 20.4 ± 0.5 |
| phyC-2 | 25.2 ± 0.3 | 9.4 ± 0.3 | 42.3 ± 1.5 | 14.7 ± 0.6 |
| phyB | 19.8 ± 0.3 | 6.2 ± 0.2 | 36.5 ± 0.9 | 11.7 ± 0.7 |
| phyA | 26.2 ± 0.3 | 12.1 ± 0.2 | 70.5 ± 0.5 | 24.4 ± 0.6 |
| phyA phyC-2 | 29.5 ± 0.5 | 16.9 ± 0.5 | 44.1 ± 1.3 | 16.1 ± 0.5 |
| phyB phyC-2 | 18.4 ± 0.2 | 5.6 ± 0.1 | 36.4 ± 1.2 | 11.5 ± 0.8 |
Flowering time was recorded as days to flowering after sowing and as number of rosette leaves at time of boiling. Values are averages from 14 plants, and standard errors are shown.
LD conditions are 16 h of white light (166 μmol·m−2·s−1) and 8 h of dark.
SD conditions are 8 h of white light (166 μmol·m−2·s−1) and 16 h of dark.
In SD conditions, phyC was early flowering compared with the wild type (Figure 7B, Table 1), indicating that phyC is required for the perception of SD photoperiod. Thus, in SD conditions, phyC had an inhibitory role in flowering induction. Under our SD conditions, the phyA mutant flowered slightly later than the wild type and with four more leaves on average, suggesting a promotive effect of phyA in SD conditions. Interestingly, the phyA phyC double mutant flowered at the same time as phyC (Figure 7B, Table 1). This result suggests that the promotive effect of phyA on flowering in SD may require phyC. phyB has been described to inhibit flowering in SD (Goto et al., 1991; Whitelam and Smith, 1991; Bagnall et al., 1995). In agreement with this finding, phyB flowered earlier than the wild type in our SD conditions. It also flowered earlier than phyC. Interestingly, we saw no effect of the phyC mutation in the phyB background, because both phyB and phyB phyC flowered at the same time in SD conditions. This result indicates that the effects of the phyB and phyC mutations on flowering time are not additive and that, as we observed for seedling deetiolaton, phyC's function in delaying floral initiation in SD conditions may require the presence of phyB.
phyC Protein Levels Are Lower in the phyB Mutant in Both Dark and Rc, Whereas phyB Levels in the phyC Mutant Are Unaffected
We were intrigued by the nonadditivity of the phenotypic effects of phyC and phyB deficiency observed at both seedling and adult stages. This unexpected result could be explained if the hyposensitive phenotype of the phyC mutant in Rc was an indirect result of a decrease in phyB levels. A coordination of phytochrome levels has been reported previously in phyB mutants (Hirschfeld et al., 1998). By contrast, as shown in Figure 1D, immunoblot analysis revealed no apparent significant difference in the levels of phyB between the wild type and the phyC mutants. To address this issue more quantitatively, an additional dilution series was performed, and the results confirmed that the levels of phyB in the phyC mutant were not altered (Figure 8A). However, when immunoblot analysis was performed to measure phyC in Col-0, phyC-2, phyB, and phyB phyC-2 protein extracts, phyC levels were reduced in the phyB mutant by ∼75% in 4-day-old dark-grown seedlings (Figures 8B and 8C) and by ∼50% in Rc-grown seedlings (Figures 8E and 8F) compared with Col-0 seedlings. These results extend previous data by Hirschfeld et al. (1998), who showed similar reductions in phyC levels in 7-day-old dark- and WLc-grown phyB seedlings compared with Nossen (No-0) and Landsberg erecta wild-type backgrounds. As a result, the difference in phyC levels in the phyB monogenic mutant compared with the phyB phyC-2 double mutant was only ∼25 to 50% of that between Col-0 and phyC-2 (Figures 8B, 8C, 8E, and 8F). PHYC mRNA levels in the phyB mutant appeared normal in both dark- and Rc-grown seedlings (Figures 8D and 8G), suggesting that the alteration of phyC protein levels in phyB likely is post-transcriptional, in accord with the findings of Hirschfeld et al. (1998).
Figure 8.
phyC and phyB Levels in phyC and phyB Mutants.
(A) Immunoblot of a dilution series of protein extracts from wild-type Col-0 and phyC-2 mutant seedlings and wild-type Ws (phyD) and phyC-1 phyD mutant seedlings grown in the dark for 4 days probed with monoclonal antibodies B1 and B7 specific for phyB.
(B) Immunoblot of protein extracts of wild-type Col-0 and phyC-2, phyB, and phyB phyC-2 mutant seedlings grown in the dark for 4 days and probed with the phyC-specific monoclonal antibody C11 (top gel). The Coomassie blue–stained membrane after hybridization is shown (middle gel). The same extracts were probed with the phyB-specific monoclonal antibodies B1 and B7 (bottom gel).
(C) Immunoblot of a dilution series of protein extracts of wild-type Col-0 and phyB mutant seedlings grown in the dark for 4 days probed with monoclonal antibody C11 specific for phyC (top gel). The Coomassie blue–stained membrane after hybridization is shown (bottom gel).
(D) RNA gel blot of RNA extracts from wild-type Col-0 and phyC-2, phyB, and phyB phyC-2 mutant seedlings grown in the dark for 4 days probed with a full-length PHYC cDNA probe. As a control, the blot was reprobed to detect 18S RNA.
(E) Immunoblot of protein extracts from wild-type Col-0 and phyC-2, phyB, and phyB phyC-2 mutant seedlings grown in Rc (8 μmol·m−2·s−1) for 4 days and probed with the phyC-specific monoclonal antibody C11 (top gel). The Coomassie blue–stained membrane after hybridization is shown (middle gel). The same extracts were probed with the phyB-specific monoclonal antibodies B1 and B7 (bottom gel).
(F) Immunoblot of a dilution series of ammonium sulfate–precipitated protein extracts from wild-type Col-0 and phyB mutant seedlings grown in Rc (8 μmol·m−2·s−1) for 4 days probed with monoclonal antibody C11 specific for phyC (top gel). The Coomassie blue–stained membrane after hybridization is shown (bottom gel).
(G) RNA gel blot of RNA extracts from wild-type Col-0 and phyC-2, phyB, and phyB phyC-2 mutant seedlings grown in Rc (18 μmol·m−2·s−1) for 4 days probed with a full-length PHYC cDNA probe. As a control, the blot was reprobed to detect 18S RNA.
Together, these data show that the morphological phenotype of phyB mutants likely is the result not only of phyB deficiency but also of a deficiency of ∼50 to 75% in the levels of phyC. The data also show that a further reduction of the remaining ∼25 to 50% content of phyC in phyB phyC compared with phyB does not have a detectable phenotypic effect on hypocotyl elongation, cotyledon expansion, or flowering time under the conditions tested. By contrast, this ∼25 to 50% reduction in phyC content seems to have an effect on petiole elongation in the phyB phyC double mutant compared with phyB, suggesting that this percentage reduction might have a higher impact on processes in which the contribution of phyC compared with phyB is more important. Our findings also show that whereas phyB deficiency implies an accompanying reduction in phyC levels, phyC deficiency does not compromise phyB protein levels. Thus, our results suggest that this is a one-way dependence of phyC on phyB.
phyC Contributes Marginally to the Rc-Imposed Repression of ATHB-2
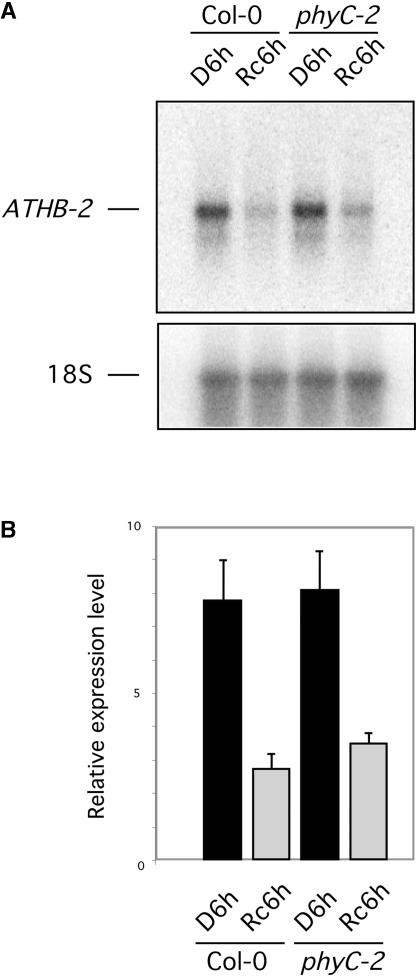
To determine whether the visible deetiolation phenotype observed in phyC mutants is accompanied by the reduced photoresponsiveness of light-regulated genes, we examined the expression of the ATHB-2 gene in wild-type and phyC-2 seedlings in Rc. This gene was selected as a marker because Franklin et al. (2003) recently reported that the ATHB-2 gene in the phyA phyB phyD phyE quadruple mutant still retained some photoresponsiveness in white light–grown seedlings, suggesting a role for phyC in regulating its expression. The ATHB-2 transcript has been shown to accumulate in the dark and in response to end-of-day FR-enriched light and is downregulated rapidly by light (Carabelli et al., 1993, 1996; Steindler et al., 1997; Tepperman et al., 2001). We have found that in dark-grown seedlings, robust repression of ATHB-2 transcript accumulation occurred within 1 h of Rc treatment, and levels were very low after 6 h of Rc (J. Tepperman and P. Quail, unpublished data). ATHB-2 transcript levels were examined in the phyC-2 monogenic mutant after 4 days of growth in the dark followed by 6 h of Rc (Rc6h) or an additional 6 h of dark as controls (D6h) and were compared with the Rc6h and D6h levels in Col-0 (Figure 9).
Figure 9.
phyC Contributes Marginally to the Rc Repression of ATHB-2 Expression.
(A) RNA gel blot of RNA extracts from wild-type Col-0 and phyC-2 after 4 days plus 6 h of growth in the dark (D6h lanes) or after 4 days of growth in the dark followed by 6 h of Rc irradiation (7 μmol·m−2·s−1; Rc6h lanes). As a control, the blot was reprobed to detect 18S RNA.
(B) Histogram of the relative expression levels of ATHB-2 mRNA in wild-type Col-0 and phyC-2 normalized to 18S rRNA levels. Mean values and standard errors (bars) were obtained by quantification of triplicate RNA gel blots (including the one shown in [A]), prepared from three separate seedling batches.
This experiment was performed in triplicate using separately grown seedling batches to enable us to detect any small differences confidently. A quantification of the transcript levels showed that the downregulation of ATHB-2 in Rc was slightly less pronounced in the phyC mutant than in the wild type (Figure 9B). Application of the statistical t test showed that the difference between the means of the ATHB-2 transcript levels in Col-0 and phyC-2 seedlings after 6 h of Rc was statistically significant, with a confidence P value of 0.03. We calculated the quantitative contribution of phyC to the Rc-imposed repression of ATHB-2 as a percentage of the total using the following formula:
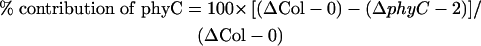 |
where ΔCol-0 = (Col-0 Rc6h) − (Col-0 D6h) and ΔphyC = (phyC-2 Rc6h) − (phyC-2 D6h). The values used are the mean values from the three experiments. Based on this calculation, phyC contributes ∼9% of the total Rc-imposed repression of ATHB-2 in Col-0. These results indicate that phyC may participate, albeit marginally, in the repression of ATHB-2 expression driven by Rc during seedling deetiolation.
DISCUSSION
Phytochrome mutants have been crucial in determining the functional roles of different members of the photoreceptor family (Quail, 1998; Whitelam et al., 1998). Mutants deficient in phyA (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993), phyB (Koornneef et al., 1980; Reed et al., 1993), and phyE (Devlin et al., 1998) have been isolated in forward-genetics screens designed to identify mutants with reduced sensitivity to light, and a naturally occurring mutation was identified in the PHYD gene of the Ws ecotype (Aukerman et al., 1997). However, although these forward screens have been numerous, none had yielded a phyC mutant. Based on the available data from higher order mutants (Halliday et al., 1994; Reed et al., 1994; Devlin et al., 1996, 1998, 1999; Shalitin et al., 2002), it was inferred that phyC likely contributed marginally to photomorphogenesis. Studies with Arabidopsis phyC overexpressors showed a marginal hypocotyl phenotype in Rc and a possible role for phyC in primary leaf expansion (Qin et al., 1997). Very recently, Franklin and collaborators reported that a phyA phyB phyD phyE quadruple mutant still retains some weak responses to Rc, specifically in cotyledon development (Franklin et al., 2003). Thus, it was expected that the phyC monogenic mutant likely had a very subtle phenotype that would be difficult to detect reliably in a forward-genetics screen.
The availability of a number of implementable reverse-genetics strategies in Arabidopsis in recent years has offered a new opportunity to approach this problem in a targeted manner. The two different strategies used here have yielded three phyC mutant alleles. By screening two collections of mutants produced by T-DNA insertion (http://signal.salk.edu; Sussman et al., 2000) and one collection of mutants produced by fast-neutron irradiation (Li et al.., 2001), we have isolated three apparently null alleles, one from each collection. The parallel behavior of these differently derived alleles provides mutually reinforcing evidence that the phenotypes are specific to the PHYC locus and that all three are similarly null. These phyC mutations provide evidence that phyC plays a role in Arabidopsis in a diversity of light-regulated developmental processes throughout the life cycle, including seedling deetiolation, vegetative architecture, and floral initiation.
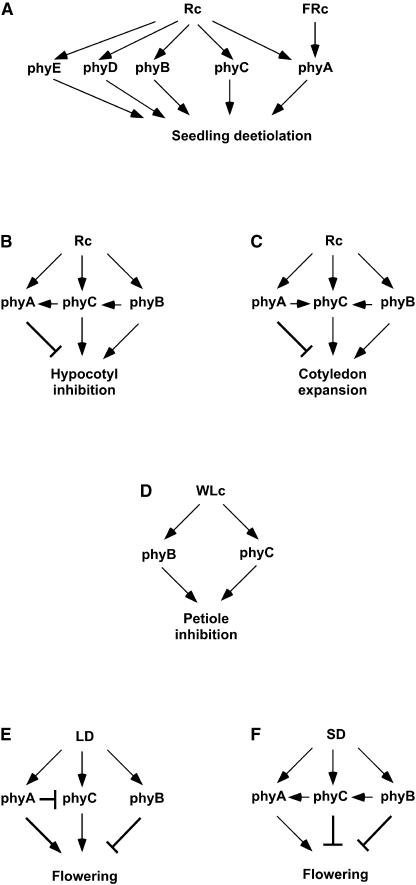
We have used the well-characterized seedling deetiolation process (Quail et al., 1995; Quail, 2002) as a system to investigate the photosensory specificity of phyC and the functional interactions with other members of the family. The data show that phyC is involved in the responses of the seedling to Rc but does not participate in processes regulated by FRc. Thus, phyC has a photosensory specificity that is similar to that of phyB and phyD and that is different from that of phyA. This result confirms that phyA is the only phytochrome responsible for the seedling responses to FRc. Also, the finding that phyC is involved in Rc sensing indicates that the seedling has four different phytochromes (phyB, phyC, phyD, and phyE) dedicated to mediating responses exclusively to Rc. phyA also has been shown to be able to function in Rc in addition to its role in FRc perception, specifically in cotyledon expansion (Franklin et al., 2003). The different photosensory specificities of each phytochrome species in seedling deetiolation are represented in Figure 10A.
Figure 10.
Differential Crosstalk among Phytochromes in the Photoregulation of Multiple Processes throughout the Life Cycle.
(A) Rc and FRc photosensory specificity of phyA, phyB, phyC, phyD, and phyE in seedling deetiolation.
(B) Functional interaction of phyC, phyA, and phyB in seedling hypocotyl inhibition under Rc.
(C) Functional interaction of phyC, phyA, and phyB in seedling cotyledon expansion under Rc.
(D) Functional interaction of phyC and phyB in petiole elongation under WLc.
(E) Functional interaction of phyC, phyA, and phyB in floral initiation under LD conditions.
(F) Functional interaction of phyC, phyA, and phyB in floral initiation under SD conditions.
The crosstalk indicated by the arrows in (B) through (F) are based on the data presented here.
Our data from studies of the monogenic phyC mutants indicate that phyC is functional in multiple developmental processes throughout the life cycle. However, the behavior of the various double and triple mutants suggests a matrix of complex interactions between phyC and other members of the phytochrome family in regulating these processes. In seedling photomorphogenesis, phyC monogenic mutants are hyposensitive to Rc, exhibiting elongated hypocotyls and small cotyledons compared with those of the wild type. Thus, phyC is required for normal seedling deetiolation and is involved in regulating the responsiveness of both organs to Rc. Although less severe, the phenotype of the phyC mutants resembles that of the Arabidopsis phyB mutants under Rc (Koornneef et al., 1980; Reed et al., 1993). This observation indicates that both phyC and phyB function in promoting seedling hypocotyl inhibition and cotyledon expansion in response to Rc, but with phyB having the predominant role (Figures 10B and 10C).
For hypocotyl inhibition, phyB mutants appear to have lost all responsiveness to Rc, because phyB seedlings are as tall in Rc as in the dark. This finding implies that phyB alone accounts for all of the hypocotyl responsiveness to Rc and therefore that no other phytochrome is expected to contribute to hypocotyl deetiolation in Rc in the presence of phyB. However, the phyC monogenic mutant exhibits a partial loss of Rc responsiveness, with elongated hypocotyls compared with the wild type. This apparent contradiction could be explained if part of the loss of hypocotyl inhibition in either phyC or phyB was caused by a reduction in the levels of phyB or phyC, respectively. We have shown that the levels of phyB in the phyC monogenic mutant are not affected but that phyC levels in the phyB mutant are decreased by 50 to 75%. This finding is consistent with the interpretation that part of the loss of hypocotyl responsiveness to Rc in the phyB mutant is caused by a decrease in phyC levels and reconciles our finding that phyC is required for hypocotyl inhibition with the observation that the phyB mutant seems to have lost all hypocotyl responsiveness to Rc.
Accordingly, phyB phyC double mutants are indistinguishable from phyB, and phyB phyC phyD triple mutants are indistinguishable from phyB phyD. Alternatively, intrinsic phyC activity (as distinct from level) in hypocotyl inhibition might depend on phyB, with the result that phyC deficiency would not be additive to phyB deficiency. Regardless, the data indicate one-way crosstalk between phyB and phyC (Figure 10B). Previous work has suggested that both phyA phyB (Reed et al., 1994) and phyB phyD (Aukerman et al., 1997) double mutants have more elongated hypocotyls than phyB under Rc. However, the reported hypocotyl lengths in Rc for both phyA phyB and phyB phyD in Rc are longer than those of the corresponding dark controls. This observation is conceptually difficult to explain. The authors interpreted it as Rc-stimulated growth by the activity of a remaining phytochrome (Aukerman et al., 1997).
For cotyledon expansion, the phenotype of the phyB mutant reveals that phyB is an important contributor to cotyledon responsiveness to Rc. However, because the cotyledons of dark-grown seedlings are smaller than those of Rc-grown phyB, the cotyledons still respond to Rc in the absence of phyB. We have shown with the monogenic phyC mutant that phyC also contributes to this response (Figure 10C). However, phyB phyC double mutants have the same cotyledon area as phyB. This nonadditive effect could be explained as a consequence of the reduction of phyC levels in the phyB mutant. Alternatively, intrinsic phyC activity in regulating cotyledon expansion might depend on phyB. As was the case for hypocotyl inhibition, these data suggest crosstalk between phyB and phyC. phyC was shown recently to function to a small extent in cotyledon expansion in the absence of the other four phytochrome members (Franklin et al., 2003). However, the possibility cannot be excluded that the functional interactions of phyC vary depending on the active phytochrome species present in the plant.
In examining the response of phyA phyC double mutants to Rc compared with that of phyA and phyC, we found that phyA is hypersensitive to some Rc fluence rates, exhibiting shorter hypocotyls than the wild type, suggesting an inhibitory role for phyA in hypocotyl deetiolation (Figure 10B). This finding is in contrast to the conclusions of previous reports on phyA from this and other laboratories, in which it was concluded that the phyA response to Rc was indistinguishable from that of the wild type (Dehesh et al., 1993; Reed et al., 1994; Quail et al., 1995). However, closer inspection of the data presented in those reports, and a direct comparison with our present data for comparable fluence rates, reveals that phyA previously showed a measurably shorter hypocotyl phenotype in this same Rc fluence-rate range than did the wild type (Dehesh et al., 1993; Reed et al., 1994; Quail et al., 1995). The extent of this difference is smaller than that observed in the present work, but it is statistically significant, at least in one of the reports (Dehesh et al., 1993). This discrepancy might reflect differences among the ecotypes used in these studies: Col-0 (used here), RLD (used by Dehesh et al., 1993), and Landsberg (used by Reed et al., 1994). Consistent with this possibility, another recent study from this laboratory using the Col-0 ecotype (Fairchild et al., 2000) also revealed clear hypersensitivity of the phyA mutant to Rc, comparable in magnitude to that reported here. This inhibitory activity of phyA in Rc appears to require phyC (Figure 10B), because the phyA phyC double mutant is indistinguishable from the phyC monogenic mutant in Rc. For cotyledon expansion, phyA mutants have larger cotyledons than wild-type plants when grown under Rc, suggesting a similar inhibitory role for phyA in cotyledon expansion (Figure 10C). Interestingly, the cotyledon area of the phyA phyC double mutant is larger than that of phyC and the same as that of phyA. This finding suggests that the promotive activity of phyC in this response requires phyA. Thus, the direction of the crosstalk between phyA and phyC appears to be organ dependent (Figures 10B and 10C).
The phyC mutations also affect the vegetative architecture of the plant, as indicated by the production of elongated petioles by the phyC mutants when grown under WLc. This function also overlaps with the previously described role of phyB (Reed et al., 1993), indicating that both phyC and phyB promote petiole growth inhibition (Figure 10D). phyD also has been shown to participate in this response in a phyB-deficient background (Aukerman et al., 1997). Because phyB phyC double mutants have more elongated petioles than monogenic phyB, it appears that these two phytochromes have an additive role in regulating this response. Thus, in contrast to the seedling deetiolation responses referred to above, phyC appears to function at least partially independently of phyB in inhibiting petiole elongation.
Previously, it was shown that phyA, phyB, phyD, and phyE are involved in the regulation of flowering time (for review, see Lin, 2000). Under LD conditions, we observed that the phyA phyC double mutant flowered significantly later than the monogenic phyA mutant, a late-flowering mutant (Johnson et al., 1994; Neff and Chory, 1998). This result indicates that phyC is able to promote flowering under 16-h photoperiods in the absence of phyA and, therefore, that phyA inhibits this promotive function of phyC (Figure 10E). Under SD conditions, the monogenic phyC mutant flowered earlier than the wild type, indicating that phyC inhibits the initiation of flowering in SD conditions (Figure 10F). The acceleration of flowering in phyC is not as pronounced as it is in the phyB monogenic mutant, indicating that the contribution of phyC to the inhibition of flowering in SD conditions is less important than that of phyB. Interestingly, the phyB phyC double mutant flowered at the same time as the monogenic phyB. Therefore, the effects of phyC and phyB deficiency in floral initiation under SD conditions are not additive.
These observations are similar to our findings on the control of seedling deetiolation under Rc and indicate that the functional interaction of phyC with phyB is similar in both responses. If so, this would suggest that phyC function in the flowering response also may require phyB (Figure 10F). This finding would be consistent with the possibility that phyC protein levels are downregulated in the phyB mutant at the adult stage, as has been observed for seedlings. However, similar phyC levels in phyA phyB phyD and the wild type at the adult stage were reported recently by Halliday et al. (2003), suggesting that another crosstalk mechanism is operative. This nonadditivity of phyB and phyC is in contrast to previous reports that monogenic phyD and phyE mutants display a wild-type flowering-time phenotype but that both phyB phyD (Aukerman et al., 1997; Devlin et al., 1999) and phyB phyE (Devlin et al., 1999) flower earlier than phyB under all conditions tested. In our study, the phyA monogenic mutant displayed a late-flowering-time phenotype under SD conditions. However, the phyA phyC double mutant flowered as early as the monogenic phyC mutant, suggesting that phyA promotive activity may require phyC (Figure 10F). This effect also has been described for the phyA phyB double mutant, which flowers as early as the phyB monogenic mutant (Reed et al., 1994).
Our flowering data indicate that phyC is required for the control of floral initiation under SD conditions, with an inhibitory effect that may require phyB. By contrast, the data suggest that phyC has a promotive effect on flowering under LD conditions in a manner redundant with phyA. Therefore, phyC seems to have opposite effects on floral initiation under SD and LD conditions. Based on the information available, antagonistic interactions in flowering regulation have been proposed for phyB/D/E and phyA (Lin, 2000). However, opposite effects in two photoperiodic conditions have not been described for any other phytochrome and may reflect a unique property of phyC.
The finding that the phyC monogenic mutants are impaired in their responses to Rc implies that phyC is involved in modulating the expression of Rc-regulated genes that are responsible for orchestrating photomorphogenesis. The degree of attenuation of the Rc-imposed repression of ATHB-2 in the phyC mutant during deetiolation indicates that phyC contributes somewhat to the regulation of its expression. In etiolated seedlings, phyA has been shown to be responsible for the downregulation of ATHB-2 by a FR pulse, and a novel phytochrome(s) other than phyA or phyB has been shown to be responsible for the downregulation of ATHB-2 by a R pulse (Carabelli et al., 1996). phyC appears to be partly responsible for this downregulation by Rc. However, the relatively minor contribution of phyC to the Rc-imposed repression of ATHB-2 in etiolated seedlings suggests that phyD and/or phyE dominates in repressing ATHB-2 expression in Rc during the early deetiolation process.
On the other hand, the observed contribution of phyC to the regulation of ATHB-2 expression during deetiolation suggests that phyC also might participate in the shade-avoidance responses in fully green plants, because ATHB-2 is a homeobox gene proposed to be involved in mediating the shade-avoidance syndrome (Steindler et al., 1999). In green plants, ATHB-2 responds strongly to shade-avoidance–inducing signals (Carabelli et al., 1993) and is regulated by changes in R/FR ratio through the action of phyB and a novel phytochrome(s) other than phyA or phyB (Carabelli et al., 1996). Recently, phyE was suggested to play a role (Franklin et al., 2003). However, given that the phyA phyB phyD phyE quadruple mutant still retains a certain degree of responsiveness to low R/FR ratios in fully green plants (Franklin et al., 2003), it is plausible that phyC may play a more prominent role in regulating ATHB-2 expression under shade-avoidance conditions in such plants than in etiolated seedlings. Therefore, the combined evidence from the present work and studies from other laboratories suggests that phyB, phyC, and phyE each may contribute partially to the Rc-imposed repression of ATHB-2.
Together, our data provide evidence for the existence of a potentially complex web of interdependent interactions between members of the phytochrome family in a manner that can vary in direction and magnitude from one developmental process to another. Such crosstalk between signaling pathways is an emerging theme in plant regulatory systems (Eckardt, 2002).
METHODS
Isolation of phyC Mutant Alleles
The collections of Arabidopsis thaliana mutants screened by PCR for disruption of the PHYC gene were as follows: the Ecker/Alonso collection of T-DNA insertional mutants (http://signal.salk.edu), consisting of a population of Columbia (Col-0) lines transformed with pROK2 plasmid (Baulcombe et al., 1986); the BASTA collection of T-DNA insertional mutants from the Arabidopsis Knockout Facility at the University of Wisconsin (Sussman et al., 2000), consisting of a population of Wassilewskija (Ws) lines transformed with pSK1015 plasmid (Weigel et al.., 2000); and the Maxygen collection of fast-neutron deletion mutants in the Col-0 ecotype (Li et al., 2001).
phyC-1 was detected by DNA gel blot analysis of PCR-amplified products in DNA superpool 17 of the BASTA collection using primers phyC-R (EMO2, 5′-GAAGACTTTCAAAAACACCACACTTATTC-3′) and JL202 (EMO115, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′). PCR reagents were 1× Takara Ex-Taq polymerase buffer (Takara Mirus Bio, Madison, WI), 0.2 mM deoxynucleotide triphosphates (dNTPs), 0.24 pmol/μL phyC-R primer, 0.24 pmol/μL JL202 primer, and 0.05 unit/μL Takara Ex-Taq polymerase. PCR conditions were 96°C for 5 min and 36 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 4 min. phyC-2 was detected by DNA gel blot analysis of PCR-amplified products in the 40K set of DNAs of the Ecker/Alonso collection using primers phyC-F (EMO51, 5′-CGGTCTAGCACTGCAAAACAGATCAAGTGG-3′) and JMLB1 (EMO49, 5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′). PCR reagents were 1× Takara Ex-Taq polymerase buffer, 0.2 mM dNTPs, 0.5 μM phyC-F primer, 0.5 μM JMLB1 primer, and 0.025 unit/μL Takara Ex-Taq polymerase. PCR conditions were 94°C for 2 min and 35 cycles of 94°C for 15 s, 56°C for 15 s, and 72°C for 3 min.
phyC-3 was detected by size analysis of PCR-amplified products in the Maxygen fast-neutron mutagenized collection using the procedures of Li et al. (2001). PCR was performed using the following primers. For the primary screen on megapools, first round, 53060F (5′-GAAGAATAGAAGTAGATGCTCATGGGCAGG-3′) and 62095R (5′-TGTTAAAGAAAGGAAGGCCAGCTGGAACTC-3′); second round, 53112F (5′-GTATCAGTCACAGCGAGGAGATATGGTTTC-3′) and 62002R (5′-ATTGATATTTCCTCTACGCGCACAAACCAC-3′). Subsequent secondary screens on superpools and pools were as follows: first round, 53060F and 62095R; second round, 55448F (5′-TCGTAGTCCGTACTTAAACCCGCCACGTAG-3′) and 62002R. PCR reagents were 1× Takara Ex-Taq polymerase buffer, 0.2 mM dNTPs, 0.3 μM forward (F) primer, 0.3 μM reverse (R) primer, and 0.025 unit/μL Takara Ex-Taq polymerase. PCR conditions for the primary screen and the first round of secondary screens were as follows: 94°C for 2 min and 40 cycles of 94°C for 15 s, 65°C for 30 s, 68°C for 90 s, and 68°C for 5 min. PCR conditions for the second round of secondary screens were 94°C for 2 min and 40 cycles of 94°C for 15 s, 65°C for 30 s, 68°C for 50 s, and 68°C for 5 min.
Seeds corresponding to the identified hits were grown and DNA was extracted from leaves (Edwards et al., 1991) for genotyping and sequencing of the mutant lesions. For phyC-1, primers EMO22 (5′-CATAGAGAAGCTTTTATTTGGCT-3′) and EMO2 were used to detect the PHYC wild-type copy, and primers EMO2 and EMO115 were used to detect the presence of the T-DNA in the PHYC gene. PCR conditions were 94°C for 2 min and 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. The wild-type band was ∼1.2 kb, and the T-DNA band was 0.5 kb. For phyC-2, primers EMO24 (5′-CATCAATGAGATTGTATGGAGAC-3′) and EMO53 (5′-GACGACCGTTAACTGGGATGTC-3′) were used to detect the PHYC wild-type copy, and primers EMO24 and EMO49 were used to detect the presence of the T-DNA in the PHYC gene. PCR conditions were as described above for phyC-1. The wild-type band was ∼1.0 kb, and the T-DNA band was ∼0.5 kb. PCR products were separated on agarose gels, and individual segregating plants for phyC-1 and phyC-2 were genotyped based on the presence or absence of wild-type and T-DNA bands. A T-DNA PCR product was purified and sequenced to map the T-DNA insertion site. For phyC-3, primers JO23 (5′-AAATGGATGCAATTAATTCTC-3′) and 62002R were used to detect in the same reaction the PHYC wild-type copy and the copy carrying the deletion in the PHYC gene. PCR conditions used were 94°C for 2 min and 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 5 min. The wild-type band was ∼4.5 kb, and the deleted band was ∼1.6 kb. PCR products were separated on agarose gels, and individual plants for phyC-3 were genotyped based on the presence or absence of wild-type and deleted bands. A PCR band corresponding to the deleted product was used to sequence the deletion junction.
Homozygous phyC-1 and phyC-2 mutant plants were outcrossed once to their respective wild types (phyC-1 to Ws and phyC-2 to Col-0). The resulting F2 segregating population was analyzed by PCR as described above, and individual homozygous mutant plants were selected. Wild-type F2 siblings also were selected to use as controls. Various F2 segregating phyC-1 mutants were tested for BASTA resistance, and only those exhibiting a clear 3:1 (resistant:sensitive) segregation indicative of a single insertion locus were kept for further analysis. Various F2 phyC-2 mutants were analyzed to determine the number of T-DNA inserts. Based on DNA gel blot analysis, phyC-2 carries a single T-DNA insertion locus. phyC-3 mutants were outcrossed twice to Col-0 before phenotypic analysis.
Construction of Double and Triple Mutants
phyA phyC double mutants were obtained by crossing the phyC-1 mutant to phyA-5 (a gift from G. Whitelam, University of Leicester, UK) and crossing phyC-2 to phyA-211 (derived from line pOCA107-2 carrying a transgenic cab promoter construct; Reed et al., 1994). phyB phyC double mutants were obtained by crossing phyC-1 to phyB-10 (Reed et al., 1993) and crossing phyC-2 to phyB-9 (Reed et al., 1993). Selection of double phyA phyC mutants was performed by a sequential selection of F2 segregants by PCR to select for homozygosity for phyC followed by analysis under constant far-red light (FRc) of the corresponding F3 progeny to select for long hypocotyls, indicative of homozygosity for phyA. Selection of phyB phyC double mutants was performed by a sequential selection of F2 segregants for long hypocotyls under constant red light (Rc), indicative of homozygosity for phyB, followed by PCR analysis to select for homozygosity for phyC.
Seedling and Plant Growth and Measurements
Seeds were surface-sterilized in 20% bleach and 0.03% Triton X-100 for 10 min and plated on GM medium (Valvekens et al., 1988) without sucrose, stratified at 4°C in the dark for 4 days, exposed to white light for 3 h to induce germination, and placed in a growth chamber at 21°C in darkness for 21 h. For Rc and FRc treatments, seedlings were moved to Rc or FRc growth chambers at 21°C for 3 days. Dark control seedlings were kept in darkness. Hypocotyl length and cotyledon area of at least 20 seedlings were determined after the light treatments. For flowering-time experiments, plants were grown after stratification in short-day (8 h of white light + 16 h of dark) or long-day (16 h of white light + 8 h of dark) photoperiods in chambers at 21°C (white light = 166 μmol·m−2·s−1), and the number of rosette leaves of 16 plants was determined when the floral stem was 1 cm long. Petiole measurements were performed as described by Wagner et al. (1997). Stratified seeds were germinated directly in continuous white light (2.7 μmol·m−2·s−1) growth chambers at 21°C. After 3 weeks, petioles were measured on the longest leaves of 16 plants. Fluence rates were measured with a spectroradiometer (LI-1800; Li-Cor, Lincoln, NE). Hypocotyl length, cotyledon area, and petiole length were determined using NIH Image software (public domain; National Institutes of Health, Bethesda, MD).
DNA Analysis
DNA gel blot analysis of PCR products was performed by separating the PCR products on 1% agarose gels and transferring the products to a nylon membrane after denaturing. A PHYC probe corresponding to the full-length cDNA (Sharrock and Quail, 1989) was labeled by random priming, and hybridization was performed in Church buffer (Church and Gilbert, 1984) at 65°C.
To test for the number of T-DNA insertion loci in phyC-2, DNA was extracted from leaves with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). A total of 0.5 μg was digested with XbaI, run on a 0.8% agarose gel, and transferred to a nylon membrane after denaturing. A T-DNA right border probe was amplified by PCR from pROK2 (Baulcombe et al., 1986) using primers EMO154 (5′-TTCGTCGAAGGCGTCTATCGC-3′) and EMO155 (5′-GGCGCTTTACTGGCACTTCAGG-3′). The probe was labeled by random priming, and hybridization was performed in Church buffer (Church and Gilbert, 1984) at 65°C.
RNA Analysis
For PHYC mRNA detection (Figures 1B, 8D, and 8G), total RNA was extracted from 4-day-old Rc-grown seedlings with the RNeasy Plant Mini Kit (Qiagen). Five micrograms was loaded per lane and then transferred to a nylon membrane. PHYC and 18S probes were labeled by random priming, and hybridization was performed in Church buffer (Church and Gilbert, 1984) at 65°C. The PHYC probe corresponds to the full-length cDNA (Sharrock and Quail, 1989). The probe to detect 18S RNA was as described by Cantón and Quail (1999).
For ATHB-2 mRNA detection (Figure 9), 4-day-old dark-grown seedlings were irradiated with Rc for 6 h (7 μmol·m−2·s−1) or kept in darkness as a control. Material was harvested from three different replicates, and total RNA was isolated as described by Tepperman et al. (2001). Five micrograms of total RNA was loaded per lane and then transferred to a nylon membrane. The ATHB-2 probe was amplified by PCR from Arabidopsis Col-0 DNA with specific primers EMO195 (5′-GGAGGTAGACTGCGAGTTC-3′) and EMO196 (5′-AACTACATGCATATCTGGTCC-3′). The ATHB-2 probe was labeled by random priming, and hybridization was performed in Church buffer (Church and Gilbert, 1984) at 65°C. Hybridization signal was quantified with the Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and normalized to 18S rRNA levels.
Protein Analysis
For total protein detection (Figures 1C, 1D, 8A, 8B, 8C, and 8E), protein extracts were prepared from 4-day-old seedlings by briefly grinding ∼200 seedlings in 0.4 mL of extraction buffer [100 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.6, 40 mM β-mercaptoethanol, 10% glycerol, 100 mM NaCl, 2 mM phenylmethylsulfonyl fluoride, 20 mM iodoacetamide, 2 μg/mL apoprotein, 0.7 μg/mL pepstatin, and 5 μg/mL leupeptin] on ice in a 1.5-mL tube and pestle and centrifuging for 5 min at 14,000 rpm in a microcentrifuge at 4°C. The protein concentration in the supernatant was determined by the method of Bradford (1976). The remaining supernatant was mixed with the appropriate volume of 5× SDS-PAGE sample buffer (Laemmli, 1970). Samples were heated for 3 min at 95°C and loaded on 8% SDS-polyacrylamide gels according to Laemmli (1970). Proteins were electroblotted to nitrocellulose membranes, blocked in 2% nonfat dry milk in TBS-T (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween), incubated with the primary monoclonal antibodies and secondary anti-mouse antibody conjugated to horseradish, and detected with enhanced chemiluminescence reagents (Pierce, Rockford, IL). For dark-grown seedlings, 20 μg of total protein was loaded per lane to detect phyC and 10 μg was loaded to detect phyB (Figure 8B). The dark dilution series starts with 30 μg of total protein (Figure 8C). For Rc-grown seedlings, 30 μg of total protein was loaded per lane to detect phyC and 10 μg was loaded to detect phyB (Figure 8E).
For ammonium sulfate precipitation (Figure 8F), larger amounts of seedlings were ground in extraction buffer on ice with a mortar and pestle. The extracts were centrifuged for 5 min at 14,000 rpm in a microcentrifuge at 4°C, and a sample of the supernatant was taken for protein quantitation using the method of Bradford (1976). A total of 0.20 g of (NH4)2SO4 was added to 1 mL of supernatant in a 1.5-mL tube and stirred at 4°C for 30 min. The extracts were centrifuged for 20 min at 14,000 rpm in a microcentrifuge at 4°C. Pellets were solubilized in 60 μL of extraction buffer, and the protein concentration was determined by the method of Bradford (1976). An appropriate volume of 5× SDS-PAGE sample buffer (Laemmli, 1970) was added to the supernatants. The Rc dilution series starts with 15 μg of ammonium sulfate precipitation (Figure 8F), corresponding to ∼300 μg of initial total protein. C11, B1 and B7, and 073D mouse monoclonal antibodies were used at a 1:500 dilution to detect phyC, phyB, and phyA proteins, respectively (Somers et al., 1991; Hirschfeld et al., 1998).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Peter H. Quail, quail@nature.berkeley.edu.
Accession Numbers
GenBank accession numbers for the sequences mentioned are as follows: Z32538 (Arabidopsis PHYC gene; At5g35840) and AB005236 (Arabidopsis genomic clone MIK22).
Acknowledgments
Seeds for the phyA-5 mutant were a kind gift from Garry Whitelam. We thank Michael Sussman and Patrick Krysan for assistance at the University of Wisconsin Biotechnology Center. We are grateful to Wendy Fong and Barbara Simpson for excellent technical assistance and to David Hantz and the Plant Gene Expression Center greenhouse staff for plant care. We also thank Stephen Grigg and our laboratory members for helpful discussions and support. This work was supported by a postdoctoral fellowship from the Spanish Ministry of Science and Technology to E.M. and by Department of Energy Grant DE-FG03-87ER13742 and U.S. Department of Agriculture Agricultural Research Service Current Research Information System 5335-21000-017-00D to P.H.Q.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012971.
References
- Aukerman, M.J., Hirschfeld, M., Wester, L., Weaver, M., Clack, T., Amasino, R.M., and Sharrock, R.A. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J., King, R.W., Whitelam, G.C., Boylan, M.T., Wagner, D., and Quail, P.H. (1995). Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 108, 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C., Saunders, G.R., Bevan, M.W., Mayo, M.A., and Harrison, B.D. (1986). Expression of biologically-active viral satellite RNA from the nuclear genome of transformed plants. Nature 321, 446–449. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantón, F.R., and Quail, P.H. (1999). Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 121, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli, M., Morelli, G., Whitelam, G., and Ruberti, I. (1996). Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc. Natl. Acad. Sci. USA 93, 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli, M., Sessa, G., Baima, S., Morelli, G., and Ruberti, I. (1993). The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J. 4, 469–479. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Chory, J., and Wu, D. (2001). Weaving the complex web of signal transduction. Plant Physiol. 125, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Dehesh, K., Franci, C., Parks, B.M., Seeley, K.A., Short, T.W., Tepperman, J.M., and Quail, P.H. (1993). Arabidopsis HY8 locus encodes phytochrome A. Plant Cell 5, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., Halliday, K.J., Harberd, N.P., and Whitelam, G.C. (1996). The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: Novel phytochromes control internode elongation and flowering time. Plant J. 10, 1127–1134. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., Patel, S.R., and Whitelam, G.C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., Robson, P.R.H., Patel, S.R., Goosey, L., Sharrock, R.A., and Whitelam, G.C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119, 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt, N.A. (2002). Specificity and cross-talk in plant signal transduction: January 2002 Keystone Symposium. Plant Cell 14 (suppl.), S9–S14. [DOI] [PMC free article] [PubMed]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C. (2001). The phytochromes, a family of red/far-red absorbing photoreceptors. J. Biol. Chem. 276, 11453–11456. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Praekelt, U., Stoddart, W.M., Billingham, O.E., Halliday, K.J., and Whitelam, G.C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, N., Kumagai, T., and Koornneef, M. (1991). Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plant. 83, 209–215. [Google Scholar]
- Halliday, K.J., Koornneef, M., and Whitelam, G.C. (1994). Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 104, 1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Hennig, L., Stoddart, W.M., Dieterle, M., Whitelam, G.C., and Schäfer, E. (2002). Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 128, 194–200. [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld, M., Tepperman, J.M., Clack, T., Quail, P.Q., and Sharrock, R.A. (1998). Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E., Bradley, M., Harberd, N.P., and Whitelam, G.C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognár, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Ádám, E., Schäfer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li, X., Song, Y., Century, K., Straight, S., Ronald, P., Dong, X., Lassner, M., and Zhang, Y. (2001). A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 27, 235–242. [DOI] [PubMed] [Google Scholar]
- Lin, C. (2000). Photoreceptors and regulation of flowering time. Plant Physiol. 123, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, S., and Sharrock, R.A. (1997). Phytochrome gene diversity. Plant Cell Environ. 20, 666–671. [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, M., Kuhn, R., Moran, S., and Quail, P.H. (1997). Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J. 5, 1163–1172. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (1997). An emerging molecular map of the phytochromes. Plant Cell Environ. 20, 657–665. [Google Scholar]
- Quail, P.H. (1998). The phytochrome family: Dissection of functional roles and signalling pathways among family members. Philos. Trans. R. Soc. Lond. 353, 1399–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 14, 180–188. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417, 763–767. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Clack, T. (2002). Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Chory, J., and Furuya, M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 104, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. (2000). Phytochromes and light signal perception by plants: An emerging synthesis. Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Smith, H., and Whitelam, G.C. (1997). The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20, 840–844. [Google Scholar]
- Somers, D.E., Sharrock, R.A., Tepperman, J.M., and Quail, P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3, 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler, C., Carabelli, M., Borello, U., Morelli, G., and Ruberti, I. (1997). Phytochrome A, phytochrome B and other phytochrome(s) regulate ATHB-2 gene expression in etiolated and green Arabidopsis plants. Plant Cell Environ. 20, 759–763. [Google Scholar]
- Steindler, C., Matteucci, A., Sessa, G., Weimar, T., Ohgishi, M., Aoyama, T., Morelli, G., and Ruberti, I. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126, 4235–4245. [DOI] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Hoecker, U., and Quail, P.H. (1997). RED1 is necessary for phytochrome B–mediated red light–specific signal transduction in Arabidopsis. Plant Cell 9, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., and Devlin, P.F. (1997). Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 20, 752–758. [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., Patel, S., and Devlin, P.F. (1998). Phytochromes and photomorphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. 353, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., and Smith, H. (1991). Retention of phytochrome-mediated shade avoidance responses in phytochrome-deficient mutants of Arabidopsis, cucumber and tomato. J. Plant Physiol. 139, 119–125. [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]