Abstract
Arf family GTP-binding proteins are best characterized as regulators of membrane traffic, but recent studies indicate an additional role in cytoskeletal organization. An Arf GTPase-activating protein of the centaurin β family, ASAP1 (also known as centaurin β4), binds Arf and two other known regulators of the actin cytoskeleton, the tyrosine kinase Src and phosphatidylinositol 4,5-bisphosphate. In this paper, we show that ASAP1 localizes to focal adhesions and cycles with focal adhesion proteins when cells are stimulated to move. Overexpression of ASAP1 altered the morphology of focal adhesions and blocked both cell spreading and formation of dorsal ruffles induced by platelet-derived growth factor (PDGF). On the other hand, ASAP1, with a mutation that disrupted GTPase-activating protein activity, had a reduced effect on cell spreading and increased the number of cells forming dorsal ruffles in response to PDGF. These data support a role for an Arf GTPase-activating protein, ASAP1, as a regulator of cytoskeletal remodeling and raise the possibility that the Arf pathway is a target for PDGF signaling.
Cell movement requires that several events occur in the appropriate site and sequence (1–6). The leading edge of the cell forms a ruffle, or lamellipodium, that contains F-actin and regulatory proteins. Adhesion structures containing integrins form when the lamellipodium contacts the substratum. As the adhesions coalesce, they become sites of attachment for stress fibers on the inside of the cell and are known as focal adhesions (FAs). The nucleus and cell body translocate along the stress fibers toward the leading edge, the stress fibers being reinforced under tension. Finally, FAs at the trailing end of the cell are disassembled or detached. The mechanisms ensuring coordination of these events are still being discovered.
Arfs are small GTPases that cycle between GDP and GTP states under the influence of guanine nucleotide exchange factors and GTPase-activating proteins (GAPs). Arfs are best known as regulators of intracellular membrane traffic, apparently controlling the assembly and disassembly of vesicle coat proteins (7, 8). However, they are also involved in remodeling the actin cytoskeleton as cells change shape or move. Thus, Arf6 affects endocytosis and phagocytosis, but also regulates the polymerization of cortical actin and cell spreading (9–16). Arf1 regulates function of the Golgi, endoplasmic reticulum-to-Golgi transport, and recruitment of transport vesicle coat proteins COPI, AP-1, and AP-3, but also affects the recruitment of paxillin to FAs (17–22). The molecular mechanisms by which Arfs affect the cytoskeleton or by which Arfs may coordinate membrane traffic and cytoskeletal changes are not known, but the guanine nucleotide state of Arf is presumed to be critical. Indeed, overexpression of ARNO, an Arf guanine nucleotide exchange factor, induces disassembly of actin stress fibers and, in combination with phorbol ester, induces actin-rich protrusions (13). Overexpression of EFA6, another Arf guanine nucleotide exchange factor, induces ruffles and filopodia (10). Cat1, an Arf GAP first identified as a protein interacting with G protein-coupled receptor kinase (23), was found to be phosphorylated on tyrosine when cells spread on fibronectin (24). An Arf GAP from Saccharomyces cerevisiae affects actin distribution and binds to F-actin (25). A putative Arf GAP, p95, binds to the focal adhesion protein paxillin (26). Two known Arf GAPs, ASAP1 (also called centaurin β4) and PAP (centaurin β3), bind known regulators of cytoskeleton organization, including phosphoinositides and Src family and focal adhesion kinase (FAK) family proteins (27, 28). Furthermore, when transiently expressed, ASAP1 and PAP are found at the edge of transfected cells, where they could influence cortical actin during cell movement (27, 28).
ASAP1 was identified both as an Arf GAP, specific for Arf1 and Arf5 but with detectable activity on Arf6, and as a Src SH3-binding protein (27). Src and related tyrosine kinases have been implicated in regulating cell movement (29–32). In addition, ASAP1 contains a C-terminal SH3 domain that binds to the focal adhesion tyrosine kinase FAK (J. T. Parsons, personal communication), whereas the ASAP1 relative PAP binds the FAK relative Pyk2 (28). ASAP1 and PAP also contain ankyrin repeats, a proline-rich region, and a pleckstrin homology (PH) domain. The PH domain of ASAP1 binds phosphatidylinositol 4,5-bisphosphate (PtdInsP2) (33), which is known to regulate the cytoskeletal remodeling that occurs during cell spreading and platelet-derived growth factor (PDGF)-induced membrane ruffling (34–39). The presence of multiple domains capable of binding diverse regulators of the cytoskeleton led us to test whether ASAP1 could regulate cytoskeleton remodeling.
We have found that endogenous ASAP1 is located in peripheral FAs and that it translocates to dorsal ruffles and the edge of the cell during cell movement. Overexpressing ASAP1 blocks FA formation, cell spreading, and PDGF-induced ruffling. These effects are dependent on Arf GAP activity. These data support a role for the Arf GAP ASAP1 as a regulator of the cycle of cytoskeletal changes that are necessary for cell movement.
Materials and Methods
Antibodies.
Peptides conjugated to keyhole limpet hemocyanin through cysteine by using a conjugation kit from Pierce were used to immunize New Zealand White rabbits. Immunizations were performed at Covance Research (Denver, PA). Antiserum 642, raised to the peptide VELAPKPQVGELPPKPGC, had the highest affinity (used at a 1:10,000 dilution for Western blots and 1:2,000 for immunofluorescence) and was used for most of these studies. The antiserum recognized purified ASAP1, recombinant protein, and, as shown in Fig. 1A, a single protein of 130 kDa, the molecular mass of ASAP1, in total lysates of NIH 3T3 cells. The signal, as shown in Fig. 1A, was blocked by incubating the antibody with the peptide to which it was raised. The peptide had no effect on immunofluorescence using antibodies to vinculin (Fig. 1A) or paxillin. Monoclonal antibodies to the indicated proteins or epitopes were from the indicated supplier: FLAG (M5), vinculin (Sigma); myc (9E10; Santa Cruz Biotechnology); paxillin, FAK (Transduction Laboratories, Lexington, KY); phosphotyrosine (4G10; Upstate Biotechnology, Lake Placid, NY). Tetramethylrhodamine B isothiocyanate and FITC-conjugated goat anti-mouse and goat anti-rabbit IgG were from Jackson ImmunoResearch. Horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG were from Amersham Life Sciences. Rhodamine phalloidin was from Molecular Probes.
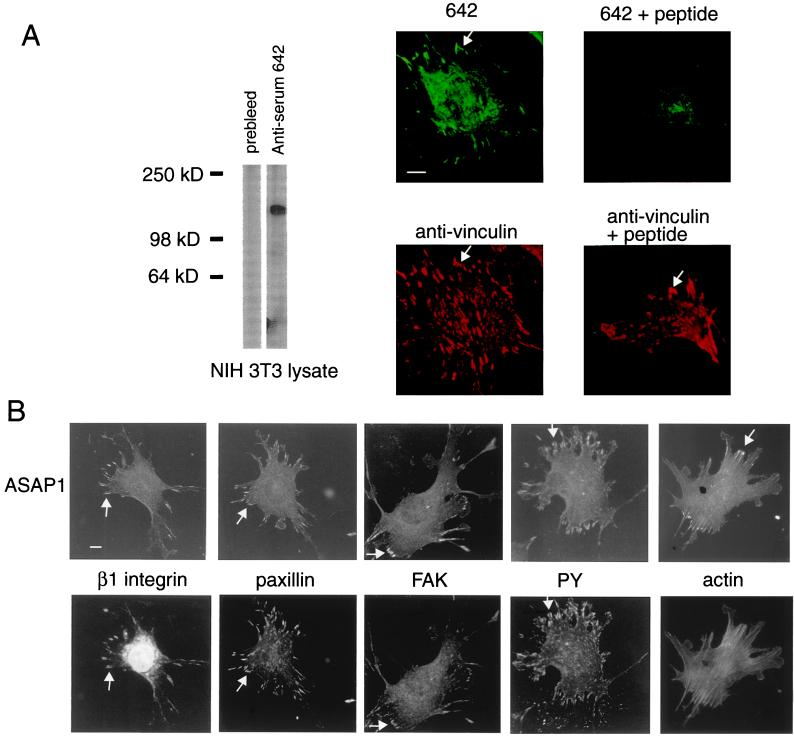
Figure 1.
ASAP1 associates with FAs. Arrows point to FAs. (A) Antibody specific for ASAP1. Antiserum 642 was used for Western blotting (Left) at a 1:10,000 dilution and for immunofluorescence (Right) at a dilution of 1:2,000. Cell lysate (1 μg) was used for Western blotting. The peptide (500 μM) to which the antibody was raised was included where indicated. (B) Colocalization of ASAP1 with markers of FAs. PY, protein phosphotyrosine. (Bars = 10 μm.)
Plasmids.
All ASAP1 constructs, including mutants, were derived from the isotype ASAP1b (GenBank accession no. AF075462). A mammalian expression vector for FLAG-tagged ASAP1b and a bacterial expression vector for His-tagged (residues 325–724) ASAP1 (referred to as PZA for PH, zinc finger, and ANK repeat domains) have been described (27). Point mutations were introduced as described (27). Mutations were confirmed by sequencing using an ABI 373 (Applied Biosystems) and Perkin–Elmer reagents.
Assays of Cytoskeletal Remodeling.
NIH 3T3 fibroblasts, obtained from Douglas Lowy (National Cancer Institute, Bethesda, MD), were used unless otherwise indicated. Where indicated, cells were transfected with Fugene (Roche Molecular Biochemicals) and used 24 h later. Cells were removed from culture plates by using trypsin/EDTA, washed, and incubated on fibronectin-coated coverslips in serum-free medium (Optimem; GIBCO/BRL). To follow spreading, cells were fixed at the indicated times in PBS containing 2% formaldehyde. To examine FAs and to determine responses to PDGF, the cells were incubated 5–7 h in Optimem before use. Cells were prepared for immunofluorescence as described (40).
Miscellaneous.
A protein consisting of residues 325 to 724 of ASAP1, referred to as PZA, and [R497K]PZA were expressed in and purified from bacteria as described (27). Far-UV circular dichroism spectra of purified PZA and [R497K]PZA were determined as described (33), and recombinant Arf1 was prepared as described (41). Arf GAP activity was assayed as described (42). Coverslips were coated with fibronectin from bovine plasma (Sigma) (43). Enhanced chemiluminescence was performed using reagents from New England Nuclear. Nonlinear regressions were performed with figp (Biosoft, Milltown, NJ). ANOVA with Tukey–Kramer post tests was performed with graphpad prism software.
Results
ASAP1 Associates with FAs.
We localized endogenous ASAP1 in NIH 3T3 fibroblasts by immunofluorescence using an antiserum raised to a peptide from a unique sequence in the protein (see Materials and Methods). Consistent with FA association, ASAP1 was concentrated in linear structures at the end of actin stress fibers immediately behind the edge of the cell (Fig. 1B) and colocalized with several markers of FAs, including vinculin, β1 integrin, paxillin, FAK, and phosphotyrosine (Fig. 1 A and B, arrows). Peripheral FAs were specifically labeled. Little or no staining of central FAs was detected, which was most evident when counterstaining for vinculin.
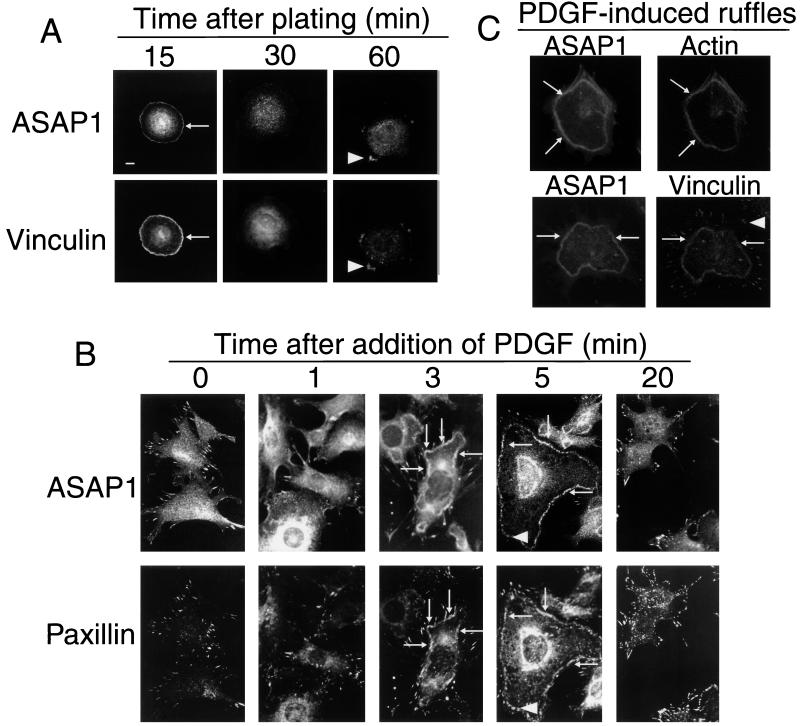
Further evidence that endogenous ASAP1 is in FAs was provided by examining cells under conditions in which FAs are remodeled. First, cells were allowed to attach to fibronectin, and localization of ASAP1 during cell spreading and FA assembly was monitored (Fig. 2A). At 15 min, ASAP1 was found in the cell edge (arrows). Within 30 min, ASAP1 staining at the edge began to decrease, and within 1 h, ASAP1 colocalized with paxillin (data not shown) and vinculin (Fig. 2A) in FAs (arrowheads). Second, cells were stimulated with PDGF (Fig. 2 B and C). Within 1 min, ASAP1 staining within FAs decreased and cytosolic staining increased. By 3 min, ASAP1 was associated with dorsal ruffles (arrows), colocalizing with paxillin (Fig. 2B), actin (Fig. 2C; ref. 33), and vinculin (Fig. 2C). By 5 min, ASAP1 within the dorsal ruffles began to organize into circular and linear structures (Fig. 2B, arrowheads) and to associate with forming FAs. The rate of ASAP1 association with the FAs was slower than paxillin association (Fig. 2B). At 5 min, paxillin was forming linear FAs (arrowheads) under the ASAP1-rich dorsal ruffle. Similarly, whereas vinculin and ASAP1 colocalized in dorsal ruffles, a population of FAs was observed that contained vinculin but not ASAP1 (Fig. 2C, arrowhead). By 20 min, ASAP1 was in the newly formed FAs, and no protein was detected in the cytoplasm or at the cell edge. PDGF at 1 ng/ml was sufficient to induce this response. These results suggest that ASAP1 associates with FAs that already contain paxillin and vinculin, and dissociates and moves to the cell edge when FA remodeling is stimulated by PDGF.
Figure 2.
ASAP1 cycles between FAs, the cytoplasm, and plasma membrane when cells are stimulated to move. (A) Cell spreading. NIH 3T3 fibroblasts were removed from plates with trypsin/EDTA and incubated in serum-free medium with fibronectin-coated glass coverslips for the indicated times. (Bars = 10 μm.) (B) PDGF-induced cytoskeletal remodeling: time course. PDGF (10 ng/ml) was added to cells on fibronectin-coated coverslips in serum-free medium. (C) PDGF-induced dorsal ruffles contain actin and vinculin. Cells were treated for 5 min with 10 ng/ml PDGF. Actin was visualized by using phalloidin conjugated to rhodamine. Arrows, dorsal ruffles; arrowheads, FAs.
Overexpression of ASAP1 Affects FA Assembly and Cellular Events Dependent on Reorganization of the Actin Cytoskeleton.
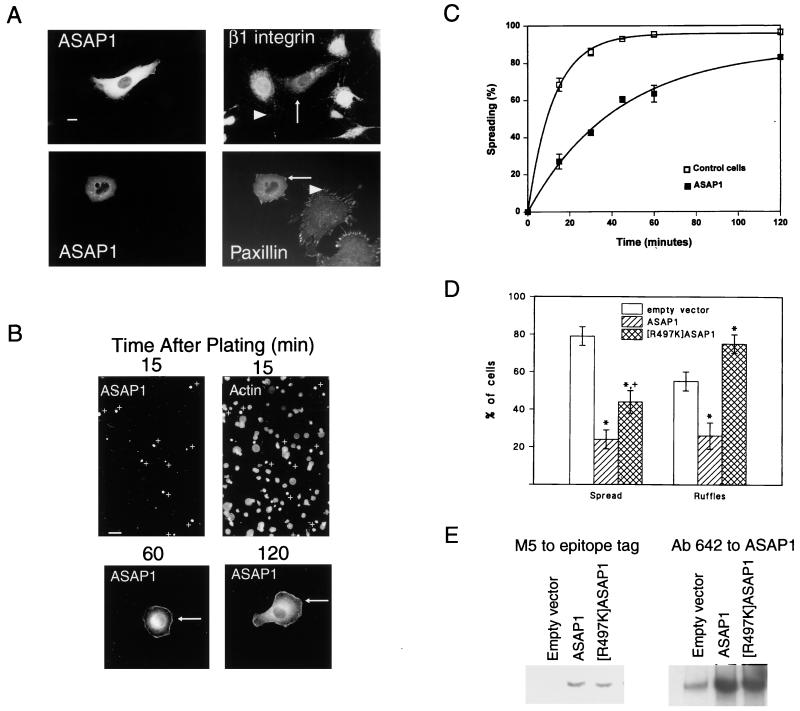
To test whether ASAP1 may regulate events associated with FA assembly or cell movement, the effects of overexpressing ASAP1 were examined (Fig. 3A). Two markers were used, β1 integrin and paxillin. In nontransfected cells, β1 integrin and paxillin were both apparent in the linear FAs (Fig. 3A, arrowheads). In transfected cells overexpressing epitope-tagged ASAP1, β1 integrin and paxillin were associated with punctate structures at the edge of the cell (arrows) rather than linear FAs. In addition, paxillin was found throughout the cytoplasm. Overexpression of ASAP1 had no effect on the Golgi marker β COP (27). The punctate structures are similar to those seen in cells in which Rho is inactivated and Rac is activated (44) and have been referred to as focal complexes. These observations suggest that high-level expression of ASAP1 can interfere with formation or stabilization of FAs and can induce focal complexes.
Figure 3.
Overexpression of ASAP1 affects cytoskeletal remodeling. NIH 3T3 fibroblasts were transfected with plasmids for FLAG-tagged ASAP1. (A) FA morphology. For double staining for β1 integrin, ASAP1 was detected by using a monoclonal antibody to the FLAG epitope. For paxillin, ASAP1 was detected with the polyclonal antibody 642. In this case, endogenous ASAP1 was apparent in FAs in nontransfected cells, visible in photographs in which the transfected cells were overexposed. (Bar = 10 μm.) (B) Comparison of transfected and nontransfected cells during spreading. A low-power field photographed from experiments used to derive the data for C is shown. (Bar = 40 μm.) At higher magnification, two transfected cells, fixed 1 and 2 h after plating, are shown. (C) Effect of ASAP1 overexpression on cell spreading rate. NIH 3T3 fibroblasts were seeded on fibronectin-coated coverslips. At the indicated times, cells were removed and fixed. Transfected cells were detected by using a monoclonal antibody to the FLAG epitope and were counterstained with rhodamine-phalloidin. A cell was considered spread when the total diameter was 2-fold or greater than the nuclear diameter. The data are the means and range for two experiments. (D) Effect of ASAP1 and [R497K]ASAP1 overexpression on cell spreading and PDGF-induced dorsal ruffles. Cells were transfected with plasmids encoding FLAG epitope-tagged ASAP1 or [R497K]ASAP1. Spreading was determined at 15 min. To examine ruffling, cells were treated for 4 min with 10 ng/ml PDGF. Dorsal ruffles were visualized by using rhodamine-phalloidin. Data are shown as mean ± SD for four experiments. ∗, Different from control, P < 0.001; +, different from wild type, P < 0.01. (E) Expression levels of FLAG-tagged ASAP1 and FLAG-tagged [R497K]ASAP1. Cells were transfected in parallel to those used for spreading and ruffling assay and lysed in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). ASAP1 in 1 μg of cell lysate protein was detected with an antibody to the FLAG epitope (Left) and in 0.1 μg of cell lysate protein with antibody 642 (Right).
To test for a role for ASAP1 as a regulator of other cytoskeletal changes associated with cell movement, the effects of ASAP1 overexpression on cell spreading and PDGF-induced cell ruffling were examined. Cell spreading was studied during adhesion to fibronectin. Like the endogenous protein, the ectopically expressed ASAP1 was at the edge of the cell during cell spreading. However, the overexpressed ASAP1 persisted at the edge for a longer period, remaining evident at 1 to 2 h (Fig. 3B, arrows), and slowed spreading in both NIH 3T3 (Fig. 3 B, C, and D) and HeLa (data not shown) cells. In the low-power field shown in Fig. 3B, none of the transfected cells, marked by +, had spread after 15 min, whereas more than half of the nontransfected cells had spread. As shown in Fig. 3C, the transfected cells spread at about 1/3 the rate of the nontransfected cells, and at 15 min, 79 ± 5% of control cells and 24 ± 5% (n = 4; mean ± SD) of ASAP1-overexpressing cells had spread (Fig. 3D). The effect of ASAP1 overexpression on dorsal ruffles formed in response to a 4-min treatment to PDGF was examined (Fig. 3D). Of the nontransfected cells, 55 ± 5% (n = 4; mean ± SD) had dorsal ruffles in contrast to 26 ± 7% of cells overexpressing ASAP1. Thus, ectopically expressed ASAP1 can interfere both with the formation of FAs during cell spreading and with a response to PDGF, dorsal ruffle formation.
Effect of ASAP1 on PDGF-Induced Dorsal Ruffles Depends on Arf GAP Activity.
To determine whether the effects of ASAP1 on the cytoskeleton were related to Arf GAP activity, different mutations were introduced into ASAP1. Arginine at position 497, highly conserved among proteins known to have Arf GAP activity (45), was changed to lysine. The effect of the mutation on GAP activity was assessed in bacterially expressed proteins consisting of the PH, Arf GAP, and ANK repeat domains (PZA). PZA has been shown previously to be the minimal structure required for GAP activity (27, 33). No differences in the far-UV circular dichroism spectra of wild-type PZA and [R497K]PZA were detected, indicating no gross differences in folding. However, [R497K]PZA had <0.001% the activity of wild-type PZA. In contrast, mutation of conserved cysteine 472 to serine interfered with both PZA structure and activity.
The effects of [R497K]ASAP1 on cell spreading and PDGF-induced dorsal ruffles were compared with those of wild-type ASAP1 to determine the role of the Arf GAP activity in these events. [R497K]ASAP1 was only half as effective as wild-type ASAP1 in inhibiting cell spreading (Fig. 3D), and, in contrast to wild-type ASAP1, [R497K]ASAP1 increased the fraction of cells forming dorsal ruffles when treated with PDGF (Fig. 3D). The differences were unlikely caused by differences in levels of expression, because the fraction of cells transfected and the levels of overexpression (Fig. 3E) were nearly identical. Based on Western blotting and the fraction of cells transfected (17% for wild type and 14% for [R497K]ASAP1 in the experiment shown in Fig. 3E), 25- to 50-fold more ectopic than endogenous ASAP1 was expressed. [R497K]ASAP1 did not induce dorsal ruffles in the absence of PDGF. Taken together, these data support a role for the Arf GAP activity of ASAP1 in regulating the actin cytoskeleton.
Discussion
ASAP1 is localized to peripheral rather than central FAs and moves to the edge of the cell during cell spreading and to actin-rich dorsal ruffles in response to PDGF. In addition, overexpression of ASAP1 interferes with cell spreading and PDGF-induced FA turnover and dorsal ruffle formation. The Arf GAP activity of ASAP1 is necessary for these effects. These results implicate an Arf pathway as a regulator of cytoskeleton reorganization and a target of PDGF-mediated signaling. Given the interaction of ASAP1 with other regulators of the cytoskeleton, phosphoinositides and Src, the role of ASAP1 may be to integrate these signals, ensuring that membrane traffic and cytoskeletal changes are coordinated at each step of the cycle of events that result in cell movement.
Two lines of evidence support a role for ASAP1 as a regulatory rather than a structural component of FAs. First, although ASAP1 localizes to FAs and cycles with FA proteins, ASAP1's behavior could be discriminated from other FA proteins. ASAP1 seemed to dissociate from FAs earlier than and associate with FAs later than other FA proteins. ASAP1 also associated with peripheral FAs to a greater extent than more central FAs. Second, the effect of overexpressing ASAP1 suggests a regulatory role. Focal complexes, rather than FAs, are formed in cells overexpressing ASAP1. Overexpression of ASAP1 interfered with cell spreading on fibronectin and PDGF-induced ruffling.
Experiments comparing ASAP1 overexpression with a mutant ASAP1 lacking GAP activity, [R497K]ASAP1, suggest the effects of ASAP1 are consequent to interaction with Arf. Thus, [R497K]ASAP1, possibly functioning as a dominant negative to endogenous ASAP1 to prolong Arf activation, increased the fraction of cells forming dorsal ruffles after PDGF treatment. [R497K]ASAP1 had less of an effect on cell spreading than did wild-type ASAP1. [R497K]ASAP1 may have retained partial ability to inhibit spreading, because, in addition to inactivating Arf, ASAP1 may titrate a molecule such as Src or FAK. Which Arf is involved is unclear because ASAP1 can regulate Arf1, Arf5, and Arf6 in vitro. Of these Arfs, Arf6 has been implicated in plasma membrane events, and both Arf1 and Arf6 have been implicated as regulators of the cytoskeleton.
The effect of ASAP1 on the cytoskeleton and PDGF signaling could be related Arfs' impact on phospholipid metabolism. Arf proteins have been found to activate phospholipase D (46, 47) and phosphoinositide kinases (48–51) and to mediate PDGF-induced phospholipase D activation (52). In addition, Arf proteins are affected by phosphoinositides. Both Arf exchange factors (10, 53–56) and GAPs (27, 28, 42) contain PH domains and are activated by phosphoinositides. Thus, PtdInsP2 could function as a second messenger with effects on the cytoskeleton mediated by actin-binding proteins or Rho family proteins (39). A system of feedforward and feedback loops impinging on Arf would regulate PtdInsP2 production. ASAP1, a PtdInsP2- and phosphatidic acid-dependent Arf GAP, would limit the response (33), and overexpression of ASAP1 would increase the sensitivity of this feedback loop, consequently limiting PtdInsP2 production.
ASAP1 might also affect the activity of Rho family proteins with consequent changes in the cytoskeleton. Interaction between the activities of Arf and Rho family proteins has been noted previously (16, 57). Because both Arf and Rho regulate phospholipase D (58) and phosphatidylinositol 4-monophosphate (PtdIns4P) 5 kinase (51), the mechanisms described in the preceding paragraph might be involved. However, Arf and Rho family proteins could interact in other ways as well. For instance, Arf could affect the trafficking of Rac (16), or Rho and Arf could together affect the activity of a common protein, such as Partner of Rac (11, 59), PtdIns4P 5 kinase (60), or phospholipase D (58). Alternatively, Arf and Rho proteins could reciprocally affect each other's activation or inactivation. Several predicted proteins have been identified in sequencing databases that contain both Arf GAP and Rho GAP domains. Conceivably, Arf could affect Rho family proteins in a manner similar to the effect of heterotrimeric G proteins on this family through p115 (61).
ASAP1 has a number of domains, in addition to the Arf GAP domain, that mediate binding to signaling molecules, including phosphoinositides, Src, and FAK. Conceivably, ASAP1 could function as an Arf effector with Arf-GTP binding influencing the activity of other domains. Conversely, the GAP activity may be regulated through the interactions. In either case, ASAP1 could function to integrate these various signals, thereby ensuring that membrane traffic and cytoskeletal changes are coordinated.
Acknowledgments
We thank Drs. Douglas Lowy, Julie Donaldson, Trevor Jackson, Harish Radhakrishna, Edna Cukierman, J. Thomas Parsons, and Ken Yamada for discussions and advice; Dr. J. Thomas Parsons for sharing unpublished data; and Jenny Clark for expertise in nucleotide sequencing. The work was supported by the Division of Basic Sciences, National Cancer Institute (P.A.R.) and U.S. Public Health Service Grant CA41072 from the National Cancer Institute (J.A.C.).
Abbreviations
- PDGF
platelet-derived growth factor
- PH
pleckstrin homology
- FA
focal adhesion
- FAK
focal adhesion kinase
- GAP
GTPase-activating protein
- PtdInsP2
phosphatidylinositol 4,5-bisphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070552297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070552297
References
- 1.Bretscher M S. Cell. 1996;87:601–606. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- 2.Burridge K, Chrzanowska-Wodnicka M, Zhong C. Trends Cell Biol. 1997;7:342–347. doi: 10.1016/S0962-8924(97)01127-6. [DOI] [PubMed] [Google Scholar]
- 3.Gumbinar B M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 4.Lauffenburger D A, Horwitz A F. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison T J, Cramer L P. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 6.Sheetz M P, Felsenfeld D P, Galbraith C G. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson J G, Klausner R D. Curr Opin Cell Biol. 1994;6:527–532. doi: 10.1016/0955-0674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 8.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 9.Peters P J, Hsu V W, Ooi C E, Finazzi D, Teal S B, Oorschot V, Donaldson J G, Klausner R D. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franco M, Peters P J, Boretto J, Van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza-Schorey C, Boshoins R L, McDonough M, Stahl P D, Van Aelst L. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza-Schorey C, Van Donselaar E, Hsu V W, Yang C, Stahl P D, Peters P J. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank S R, Hatfield J C, Casanova J E. Mol Biol Cell. 1998;9:3133–3146. doi: 10.1091/mbc.9.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J, Khachikian Z, Radhakrishna H, Donaldson J G. J Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Cox D, Tseng C C, Donaldson J G, Greenberg S. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson J G. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 17.Donaldson J G, Cassel D, Kahn R A, Klausner R D. Proc Natl Acad Sci USA. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donaldson J G, Kahn R A, Lippincott-Schwartz J, Klausner R D. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- 19.Palmer D J, Helms J B, Beckers C J, Orci L, Rothman J E. J Biol Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- 20.Stamnes M A, Rothman J E. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- 21.Traub L M, Kornfeld S, Ungewickell E. J Biol Chem. 1995;270:4933–4942. doi: 10.1074/jbc.270.9.4933. [DOI] [PubMed] [Google Scholar]
- 22.Norman J C, Jones D, Barry S T, Holt M R, Cockcroft S, Critchley D R. J Cell Biol. 1998;143:1981–1995. doi: 10.1083/jcb.143.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Premont R T, Claing A, Vitale N, Freeman J L, Pitcher J A, Patton W A, Moss J, Vaughan M, Lefkowitz R J. Proc Natl Acad Sci USA. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagrodia S, Bailey D, Lenard Z, Hart M, Guan J L, Premont R T, Taylor S J, Cerione R A. J Biol Chem. 1999;274:22393–22400. doi: 10.1074/jbc.274.32.22393. [DOI] [PubMed] [Google Scholar]
- 25.Blader I J, Cope M J, Jackson T R, Profit A A, Greenwood A F, Drubin D G, Prestwich G D, Theibert A B. Mol Biol Cell. 1999;10:581–596. doi: 10.1091/mbc.10.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner C E, Brown M C, Perrotta J A, Riedy M C, Nikolopoulos S N, McDonald A R, Bagrodia S, Thomas S, Leventhal P S. J Cell Biol. 1999;145:851–863. doi: 10.1083/jcb.145.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M T, Andrade J, Radhakrishna H, Donaldson J G, Cooper J A, Randazzo P A. Mol Cell Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreev J, Simon J P, Sabatini D, Kam J, Plowman G, Randazzo P A, Schlessinger J. Mol Cell Biol. 1999;19:2338–2350. doi: 10.1128/mcb.19.3.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown M T, Cooper J A. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 30.Parsons J T, Parsons S J. Curr Opin Cell Biol. 1997;9:187–192. doi: 10.1016/s0955-0674(97)80062-2. [DOI] [PubMed] [Google Scholar]
- 31.Thomas S M, Brugge J S. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 32.Fincham V J, Frame M C. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kam, J. L., Miura, K., Jackson, T. R., Gruschus, J., Roller, P., Stauffer, S., Clark, J., Aneja, R. & Randazzo, P. A. (2000) J. Biol. Chem.275, in press. [DOI] [PubMed]
- 34.Breuer D, Wagener C. Exp Cell Res. 1989;182:659–663. doi: 10.1016/0014-4827(89)90268-1. [DOI] [PubMed] [Google Scholar]
- 35.Wennstrom S, Hawkins P, Cooke F, Hara K, Yonezawa K, Kasuga M, Jackson T, Claesson-Welch L, Stephens L. Curr Biol. 1994;4:385–393. doi: 10.1016/s0960-9822(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 36.Sakisaka T, Itoh T, Miura K, Takenawa T. Mol Cell Biol. 1997;17:3841–3849. doi: 10.1128/mcb.17.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng F, Lowell C A. EMBO J. 1998;17:4391–4403. doi: 10.1093/emboj/17.15.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddhanta U, McIlroy J, Shah A, Zhang Y, Backer J M. J Cell Biol. 1998;143:1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toker A. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- 40.Radhakrishna H, Klausner R D, Donaldson J G. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randazzo P A, Weiss O, Kahn R A. Methods Enzymol. 1995;257:128–135. doi: 10.1016/s0076-6879(95)57018-7. [DOI] [PubMed] [Google Scholar]
- 42.Randazzo P A, Kahn R A. J Biol Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- 43.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 44.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 45.Scheffzek K, Ahmadian M R, Wittinghofer A. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 46.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 47.Cockcroft S, Thomas G M H, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Truong O, Hsuan J J. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 48.Godi A, Pertile P, Meyers R, Marra P, DiTullio G, Iurisci C, Luini A, Corda D, DeMatteis M A. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 49.Fensome A, Cunningham I, Prosser S, Tan S K, Swigart P, Thomas G, Hsuan J, Cockcroft S. Curr Biol. 1996;6:730–738. doi: 10.1016/s0960-9822(09)00454-0. [DOI] [PubMed] [Google Scholar]
- 50.Martin A, Brown F D, Hodgkin M N, Bradwell A J, Cook S J, Hart M, Wakelam M J O. J Biol Chem. 1996;271:17397–17903. doi: 10.1074/jbc.271.29.17397. [DOI] [PubMed] [Google Scholar]
- 51.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris A J, Frohman M A, Kanaho Y. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 52.Shome K, Nie Y, Romero G. J Biol Chem. 1998;273:30836–30841. doi: 10.1074/jbc.273.46.30836. [DOI] [PubMed] [Google Scholar]
- 53.Klarlund J K, Guilherme A, Holik J J, Virbasius J V, Chawla A, Czech M P. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 54.Klarlund J K, Rameh L E, Cantley L C, Buxton J M, Holik J J, Sakelis C, Patki V, Corvera S, Czech M P. J Biol Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- 55.Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 56.Franco M, Boretto J, Robineau S, Monier S, Goud B, Chardin P, Chavrier P. Proc Natl Acad Sci USA. 1998;95:9926–9931. doi: 10.1073/pnas.95.17.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Calafat J, Janssen H, Greenberg S. Mol Cell Biol. 1999;19:8158–8168. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Exton J H. Biochim Biophys Acta. 1999;1439:121–133. doi: 10.1016/s1388-1981(99)00089-x. [DOI] [PubMed] [Google Scholar]
- 59.Kanoh H, Williger B-T, Exton J H. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- 60.Aspenström P. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 61.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]