Abstract
The presence of a solvent-exposed alanine residue stabilizes a helix by 0.4–2 kcal·mol−1 relative to glycine. Various factors have been suggested to account for the differences in helical propensity, from the higher conformational freedom of glycine sequences in the unfolded state to hydrophobic and van der Waals' stabilization of the alanine side chain in the helical state. We have performed all-atom molecular dynamics simulations with explicit solvent and exhaustive sampling of model peptides to address the backbone conformational entropy difference between Ala and Gly in the denatured state. The mutation of Ala to Gly leads to an increase in conformational entropy equivalent to ≈0.4 kcal·mol−1 in a fully flexible denatured, that is, unfolded, state. But, this energy is closely counterbalanced by the (measured) difference in free energy of transfer of the glycine and alanine side chains from the vapor phase to water so that the unfolded alanine- and glycine-containing peptides are approximately isoenergetic. The helix-stabilizing propensity of Ala relative to Gly thus mainly results from more favorable interactions of Ala in the folded helical structure. The small difference in energetics in the denatured states means that the Φ-values derived from Ala → Gly scanning of helices are a very good measure of the extent of formation of structure in proteins with little residual structure in the denatured state.
Keywords: folding, pathway, protein, stability, transition state
There have been many experimental and theoretical studies of the relative helix forming propensities of amino acids (1–9) since the structure of the α-helix was predicted by Pauling and coworkers in 1951 (10). Irrespective of the means of estimation, Ala consistently has a high propensity for helix formation, and Gly and Pro the lowest. The helix propensity of Ala relative to Gly does, however, vary significantly between different studies. The reported values include ≈0.9 kcal·mol−1 from studies where the mutations were made at solvent-exposed positions in a protein helix (3, 6), whereas the values range from ≈0.7 to 2.0 kcal·mol−1 in peptides (2, 7, 9). In proteins, the magnitude of the free energy change on mutation of Ala to Gly at an internal position of an α-helix typically ranges from 0.4 to 2.0 kcal/mol (4).
The origin of the difference in helical propensity between Ala and Gly is also a matter of some debate. Factors thought to be important include difference in backbone conformational entropy in the denatured state, burial of hydrophobic surfaces on folding, and disruption of hydrogen bonding between the protein and the solvent (4, 6, 11–19). The estimated relative importance of the contributing factors also changes depending on the method used or system in which they were measured. One of the earliest calculations by Leach and Scheraga estimated the difference in the backbone contribution to the entropy of unfolding between Ala and Gly to be −2.4 cal·mol−1·K−1, based on the excluded volume due to steric interactions (11). A very similar value was obtained by Freire and coworkers (14) in a mutational study using the leucine-zipper region of GCN4. In contrast, other studies have obtained a much smaller difference in backbone conformational entropy (17, 18).
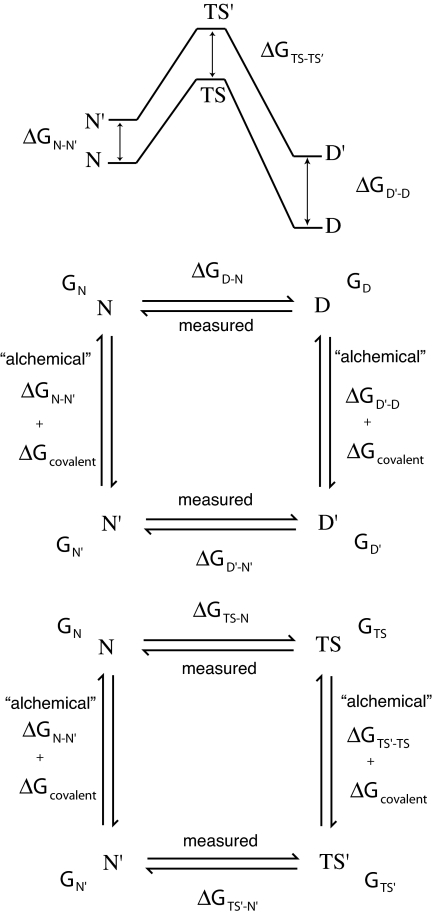
Here, we use all-atom molecular dynamics simulations with explicit solvent to estimate the backbone conformational entropy difference between Ala and Gly in the denatured state. We use a number of different models for the denatured state. Small peptides are used as models for a denatured state with minimal influence from the rest of the protein: AXA and GGXGG, where X is either Ala or Gly. We also use high-temperature unfolding simulations carried out as part of the dynameomics project (www.dynameomics.org) (20) to model denatured states of intact proteins. The results are of direct importance in the interpretation of Φ-values for protein folding derived from Ala → Gly scanning of helices whereby the changes in free energies of activation for folding on mutation of alanine to glycine are compared with the corresponding changes in free energy of denaturation to infer the extent of formation of structure in the transition state of folding (21, 22) (Fig. 1).
Fig. 1.
Thermodynamic cycles in mutagenesis linking measurements on wild-type and mutant proteins with calculated energies for changes within a structure on mutation (the “alchemical” terms). The experimentally measurable terms are ΔGD-N, the free energy for denaturation of wild-type protein, and ΔGD′-N′, that of mutant; ΔGTS-N, the free energy of activation of unfolding of wild-type protein, and ΔGTS′-N′, that of mutant; and ΔGTS-D, the free energy of activation of folding of wild-type protein, and ΔGTS′-D′, that of mutant. Changes in observables are related to changes in “alchemical” terms by ΔΔGD-N = ΔGD′-N′ − ΔGD-N = ΔGD′-D − ΔGN′-N; ΔΔGTS-N = ΔGTS′-N′ − ΔGTS-N = ΔGTS′-TS − ΔGN′-N; and ΔΔGTS-D = ΔGTS′-D′ − ΔGTS-D = ΔGTS′-TS − ΔGD′-D. The changes in covalent energies in the alchemistry cancel out. This figure is modified from ref. 33. Calculation of Φ-values from the free energies is given in Eqs. 1 and 2.
Results
Peptide Models of the Denatured State.
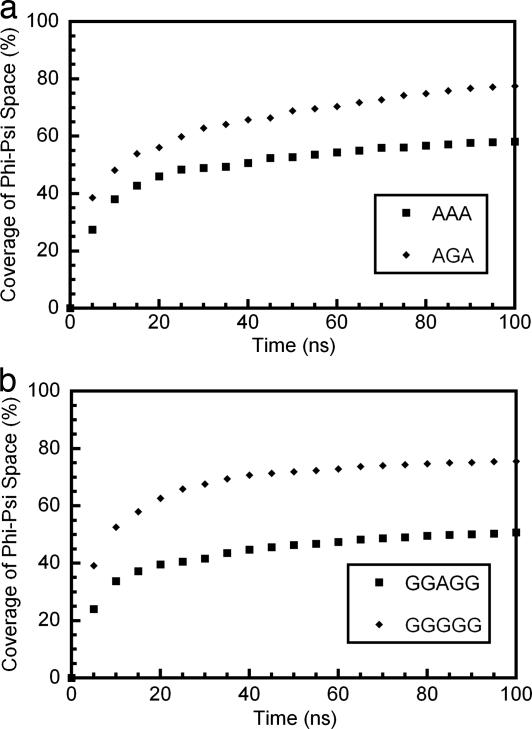
We used two peptide systems, AXA and GGXGG, to model a minimally hindered statistical-coil-like denatured state, where X is either Ala or Gly. One important question in this type of study is whether the simulations have been carried out long enough to achieve sufficient sampling of conformational space. To address this issue, we measured the coverage of conformational space at regular intervals throughout the 298 K simulations (Fig. 2). Coverage was assessed as the percentage of populated (Φ,Ψ) bins. After a rapid rise in coverage, an increasingly slow rise was seen, such that by 50 ns of simulation time ≈90% of the bins populated after 100 ns of simulation were already populated. At 75 ns, this percentage increased to ≈95%. These results suggest that we did have adequate sampling; extending the simulations would lead to little increase in coverage at the expense of a great deal of computation time.
Fig. 2.
Coverage of Φ-Ψ space converges over the simulation time course. (a) AXA peptides. (b) GGXGG peptides. The percentage coverage of Φ-Ψ space is shown at 5-ns intervals over the course of 100 ns of simulation. A 5° × 5° bin size was used in these calculations.
A further consideration is that our entropy calculation could depend on the size of the bins used in the subdivision of (Φ,Ψ) space. But, as shown for the 298-K GGXGG peptide simulations, entropy differences varied little with bin size (Table 1). In this case, the mean value of TΔS (A → G) at 298 K was −0.342 kcal·mol−1 with a standard deviation of 0.006 kcal·mol−1.
Table 1.
Backbone conformational entropy for Ala and Gly
| Model* | −Σpilnpi Ala | −Σpilnpi Gly | ΔSback Ala → Gly, cal·mol−1·K−1 | TΔSback Ala → Gly 298 K, kcal·mol−1 | Φ-Ψ coverage Ala, % | Φ-Ψ coverage Gly, % |
|---|---|---|---|---|---|---|
| AXA | ||||||
| 298 K | 6.5 | 7.0 | −0.9 | −0.3 | 58 | 77 |
| GGXGG | ||||||
| 298 K 2° × 2° bins | 8.8 | 9.3 | −1.1 | −0.3 | 37 | 60 |
| 298 K 3° × 3° bins | 8.0 | 8.6 | −1.1 | −0.3 | 43 | 70 |
| 298 K 5° × 5° bins | 7.0 | 7.6 | −1.2 | −0.4 | 51 | 76 |
| 298 K 10° × 10° bins | 5.7 | 6.3 | −1.2 | −0.4 | 60 | 81 |
| 498 K | 7.5 | 8.1 | −1.2 | −0.4 | 68 | 82 |
| Dynameomics | ||||||
| 298 K sequence matched† | 4.5 | 5.0 | −0.9 | −0.3 | 28 | 49 |
| 498 K sequence matched† | 7.1 | 7.8 | −1.4 | −0.4 | 88 | 91 |
| 498 K 0–20% helix‡ | 7.5 | 8.1 | −1.1 | −0.3 | 89 | 91 |
| 498 K 20–40% helix‡ | 7.2 | — | −1.7§ | −0.5§ | 88 | — |
| 498 K 40–60% helix‡ | 6.7 | — | −2.7§ | −0.8§ | 86 | — |
| 498 K 60–100% helix‡ | 5.6 | — | −4.9§ | −1.5§ | 77 | — |
*A 5° by 5° bin size is used unless otherwise stated.
†More Ala than Gly residues have a helical conformation, thus the number of Ala residues analyzed must be reduced. In this case, for each Gly residue, an Ala with the same flanking (i− 1 and i+ 1) amino acids is selected.
‡Ala and Gly residues were classified based on the percentage of time helical structure was adopted in the denatured state. A residue was considered helical if it was one of three contiguous residues with helical (Φ,Ψ) angles.
§The value for Gly with 0–20% helical structure in the denatured state is used as a reference.
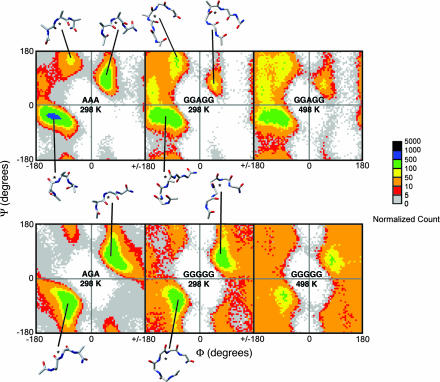
The backbone conformational entropy difference between Ala and Gly in both of the peptide models is shown in Table 1. The (Φ,Ψ) distributions for GGAGG and AAA, and GGGGG and AGA peptides were very similar (Fig. 3), as reflected by the similar backbone conformational entropy differences at 298 K. The value for the GGXGG peptides was ≈1.2 times that of the AGA peptides with both values being <0.5 kcal·mol−1. We used a high-temperature model of the unfolded state to estimate the backbone conformational entropy difference between Ala and Gly in helices. It is useful, therefore, to determine how high temperature changes the (Φ,Ψ) distributions of the smaller peptides. In the 498-K GGXGG simulations, both Ala and Gly had a greater coverage of (Φ,Ψ) space than did the simulations at 298 K. GGAGG populated 68% of the bins at 498 K and 51% at 298 K, whereas GGGGG populated 82% and 76% of the bins at 498 K and 298 K, respectively. The (Φ,Ψ) distribution also changed to some extent with temperature (Fig. 3). The change was more marked for the GGGGG peptide, where the populations of the upper right and lower left quadrants were reduced and that of the upper left and lower right quadrants increased at high temperature. This change in distribution resulted in an increase in the conformational entropy (S = −Σpilnpi) for both Ala and Gly. However, Δ(−Σpilnpi) (A → G) was similar in both cases, being 0.6 at both 298 and 498 K. If the distribution seen at 498 K was appropriate for use as a model for the unfolded state at 298 K, this would give a value for TΔS (A → G) at 298 K of −0.4 kcal·mol−1, similar to that seen for the GGXGG peptides at 298 K (−0.3 kcal·mol−1).
Fig. 3.
Ramachandran plots for peptide models of a random-coil-like denatured state: AAA, AGA, GGAGG, and GGGGG at 298 K, and GGAGG and GGGGG at 498 K. The Ramachandran plot is divided into 5° × 5° bins and colored according to the population each bin. A total of 100,000 conformations from 100 ns of simulation were counted in total. The backbone conformations of representative structures are shown alongside the 298 K plots. The central residue (for which Φ and Ψ angles were calculated) is indicated by an asterisk.
Dynameomics: Full-Length Protein Models of the Denatured State.
Small peptides are appropriate models for random coil regions of denatured states. We initially attempted to model the effects of A → G mutations in structured regions of denatured states using isolated helices from barnase and the engrailed homeodomain. However, 300 ns of simulation time resulted in insufficient sampling. To circumvent inadequate sampling, we used data from our dynameomics database, in which native and unfolding trajectories for representatives of all protein folds are being simulated from a list of 1,130 nonredundant folds as initial targets (www.dynameomics.org) (20). Of the currently analyzed 145 proteins, 114 have left the native and first intermediate clusters (if present) by 10 ns of simulation time at 498 K (see Methods). The database of 114 proteins contains a wealth of data that can be compared with our small peptide models.
We calculated the backbone conformational entropy difference between Ala and Gly in helices from the dynameomics data. Ala and Gly residues that occupied the central position in a three-residue stretch of helical structure in the starting structure of the 114 target proteins were first identified. By this definition, 386 Ala and 110 Gly residues adopted a helical conformation in the starting structure. It was necessary to reduce the number of Ala residues sampled to be the same as the number of glycine residues: firstly, residues were selected based on the sequence of the flanking amino acids; and in the second, residues were categorized by the amount of time spent in contiguous helical structure in the denatured state and by estimating the effect of residual structure on the entropy difference.
Entropy calculated from sequence matching.
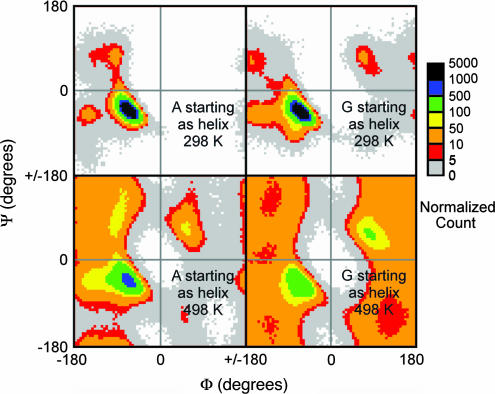
For each of the 110 Gly residues that were helical in the native state, an Ala with the same flanking (i − 1 and i + 1) amino acids was picked out of the data set of 386 Ala residues. Selecting residues in this way both ensured that the same number of samples was compared and also compensated to some extent for the sequence differences between proteins. A match could not be found for all Gly residues, leading to a sample size of 78 residues. The Ramachandran plots for the selected residues at both 298 and 498 K are shown in Fig. 4. As expected, at 298 K both Ala and Gly residues originating in helical structure remained in predominantly helical conformations, with low population outside the most favored helical region. At 498 K the (Φ,Ψ) distributions become much more like that of the corresponding GGXGG peptide (Figs. 3 and 4). However, for Ala in particular, the dynameomics data had a higher population of the helical region than did GGXGG at the same temperature. There was a marked difference between the values of −Σpilnpi in the 298 and 498 K data sets (Table 1). For example, in the native state (298 K trajectories), the value of −Σpilnpi for Ala was 4.5, whereas in the high-temperature denatured state, it was 7.1. However, the values of Δ(−Σpilnpi) (A → G) were similar: −0.5 in the native state, and −0.7 in the high-temperature denatured state. For the native state this gave a backbone conformational entropy difference between Ala and Gly of approximately −0.3 kcal·mol−1. Making the assumption that the conformations seen at 498 K are representative of the denatured ensemble at 298 K would give a value for TΔS (A → G) of approximately −0.4 kcal·mol−1. We would expect that the value calculated by using this assumption to underestimate the value calculated with a true 298 K denatured ensemble.
Fig. 4.
Ramachandran plots for Ala and Gly residues beginning in a helical conformation in the dynameomics data set. (Upper Left) Ala at 298 K. (Upper Right) Gly at 298 K. (Lower Left) Ala at 498 K. (Lower Right) Gly at 498 K. The Ramachandran plot is divided into 5° × 5° bins and colored according to the number of counts in each bin. 1,716,000 samples were taken from a total of 78 residues in each case. The data were reduced to 100,000 samples in this figure to facilitate comparison with the small peptides (Fig. 3). Because Ala is better represented than Gly in the database, not all Ala residues were used for this calculation. Instead, for each Gly residue an Ala with the same flanking (i − 1 and i + 1) amino acids was chosen out of the larger data set.
Residues categorized by the amount of helical structure in the denatured state.
The percentage of time a helical conformation was adopted in the denatured state was calculated for each of the 110 Gly and 386 Ala residues. A residue was considered helical if it was one of three contiguous residues with helical (Φ,Ψ) angles. The calculation was carried out between 10 and 21 ns in the 498 K trajectories (see Methods). The residues were then split into four categories according to the percentage of time helical structure was adopted (0–20%, 20–40%, 40–60%, and 60–100% of the time in a helical conformation). The proportion of residues adopting helical conformations for <20% of the denatured state was higher for Gly (61% of residues) than for Ala (38% of residues). The value of −Σpilnpi was calculated for each category for Ala and for residues with 0–20% helical structure for Gly (too few Gly residues were present in the other categories). The sample size used was 65 residues in each case, corresponding to the number of Gly residues with 0–20% helical structure (Table 1). These data can be used to estimate the variation of the difference in entropy between Ala and Gly in a helix with the amount of structure in the denatured state. Where both Ala and Gly residues have 0–20% helical structure, Δ(−Σpilnpi) (A → G) was −0.6, which was very similar to the value of −0.7 seen when residues were selected by sequence matching. At 298 K this would give a value for TΔS (A → G) of approximately −0.3 kcal·mol−1.
In estimating the effect of residual helical structure on the conformational entropy difference, we compared each Ala data set with the Gly residues having 0–20% helical structure in the denatured state. This represents the case where mutation of Ala to Gly leads to significant reduction in residual helical structure and thus gives an upper limit on the size of the conformational entropy difference between Ala and Gly for each category. As expected, the conformational entropy difference between Ala and Gly residues increased significantly as the percentage of time the Ala residues adopt helical structure in the denatured state increases (Table 1). The largest value of Δ(−Σpilnpi) (A → G) is −2.5 at 298 K, which gives a value of TΔS (A → G) of −1.5 kcal·mol−1.
Experimental Data for Changes of Free Energy on Ala → Gly Mutations.
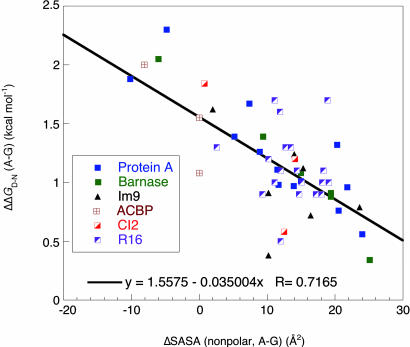
We modeled the mutation of Ala to Gly at sites in the middle of helices that are exposed to solvent and do not involve the participation of unpaired hydrogen bond donors or acceptors for representative proteins. We plotted (Fig. 5) the experimentally measured changes in free energy of denaturation against the calculated changes in solvent-accessible nonpolar surface area for the B domain of protein A (22), barnase (4, 5), Im9 (23), ACBP (24), CI2 (25), and R16 (26). The data paralleled those of the earlier studies on barnase with a good correlation between the changes in free energy in stability and the surface area of the native state, with a slope of ≈35 cal Å−2, showing that burial of solvent-accessible surface area of the alanine methyl group in the native state is a dominant factor in the overall energetics.
Fig. 5.
Plot of change in free energy of denaturation of representative proteins upon mutation of A to G against change in solvent-accessible nonpolar surface area for solvent-exposed sites in the middle of helices.
Discussion
We calculated that the presence of glycine in simple unfolded peptides contributed a favorable conformational entropy equivalent of approximately −0.3–0.4 kcal·mol−1 to the free energy at 298 K (= −TΔS), relative to the presence of alanine. We estimated the same contribution for regions of proteins that lack significant residual structure in the denatured state. This energy difference is small and clearly does not account for the observed stabilization of helices by Ala relative to Gly of up to 2 kcal·mol−1. Indeed, the correlation between the stabilization energy introduced by mutation of Gly to Ala and the change in solvent-accessible surface of the native helix (4, 5) (Fig. 5) strongly suggest that interactions of alanine in the folded helix play the dominant role in stabilization of helices.
In regions of proteins that have residual structure in the denatured state, the conformational entropy of glycine versus alanine contributed up to 1.5 kcal·mol−1 of free energy. This was because alanine induced structure. In terms of the overall free energy of the denatured state, therefore, the interactions of the side chain of alanine with other side chains in the structured denatured regions must contribute an enthalpy term that counterbalances the loss of conformation free energy.
The results above are of interest in themselves, but they are highly pertinent to Φ-value analysis. In protein folding studies, Ala → Gly scanning is an important tool for probing the structure of the transition state of folding (TS) (21, 22). In particular, this method is commonly used to determine the extent of structure formation in solvent-exposed positions in α-helices. The degree of structure in the TS is quantified by the Φ-value, calculated from measured equilibrium and kinetic data for denaturation of wild-type and mutant proteins (Fig. 1, where the mutant is denoted by a prime) (27–30). The Φ-value for folding is defined by
In terms of the other changes in the cycles in Fig. 1,
Where there is complete formation of local structure in the transition state, ΔGTS′-TS = ΔGN′-N, and Φ = 1. Where local structure in the transition state is as denatured as in the denatured state, ΔGTS′-TS = ΔGD′-D, and Φ = 0. The ΔGD′-D term in Eq. 2 contributes to the nonlinear dependence of Φ on the extent of formation of structure between the extreme values of 0 and 1. Accordingly, Φ-values are different from the α of classical rate equilibrium–free energy relationships (31).
The value of ΔGD′-D can be considered as the sum of two components ΔGreorg(D′-D) and ΔGsolv(D′-D) (28). ΔGreorg(D′-D) is the change in energy of the denatured state as it undergoes reorganization of structure upon mutation, and ΔGsolv(D′-D) is the corresponding change in solvation free energy. In some circumstances, such as the mutation of larger to smaller aliphatic side chains, when there is no residual structure in the denatured state, ΔGD′-D is close to zero, as measured from the change in free energy of transfer from the gas phase to water (30). The magnitude of ΔGsolv(D′-D) is typically <0.2 kcal·mol−1 for these residues (32). In such cases, Φ ∼ ΔGTS′-TS/ΔGN′-N, which in turn can be a good measure of the number of hydrophobic interactions made in the TS relative to N. For this reason, the larger to smaller hydrophobic mutations have been the mutations of choice for Φ-value analysis (30, 33). This relationship is also the basis for the widely used calculation of Φ-values from simulations (34–36). For Ala to Gly, the ΔGsolv(D′-D) is 0.45 kcal·mol−1 (32), the more favorable solvation energy of the alanine side chain almost exactly cancels the lower conformational free energy in the fully unfolded state. Thus, ΔGD′-D is approximately zero for Ala → Gly mutations where there is no residual structure in the denatured state. In this case, Φ ∼ ΔGTS′-TS/ΔGN′-N, as for hydrophobic deletion mutations. Ala → Gly scanning is thus an exceptionally good Φ-value probe.
Methods
Peptide Simulations.
AXA and GGXGG trajectories were simulated by using in lucem molecular mechanics (ilmm) (37) with an 8-Å force-shifted nonbonded cutoff and a nonbonded update cycle of 3. The peptides were acetylated at the N terminus and amidated at the C terminus. All atoms of the solute and the surrounding water were explicitly present, and the force fields of Levitt et al. (38, 39) were used. Simulations were carried out for 101 ns at 298 K (AXA and GGXGG) and 498 K (GGXGG only). Structures were saved at 1-ps intervals. The first 1 ns of each simulation was excluded from the analysis to allow for thermal equilibration of the system.
Dynameomics Protocol.
The dynameomics project (www.dynameomics.org) aims to simulate representative members of all protein folds under native and denaturing conditions (20). Native trajectories were simulated at 298 K for 21 ns in ilmm by using a 10-Å force-shifted nonbonded cutoff and a nonbonded update cycle of 3; structures were saved for analysis at 1-ps intervals. Duplicate unfolding trajectories were simulated at 498 K for 21 ns each, also using ilmm. An 8-Å force-shifted nonbonded cutoff and a nonbonded update cycle of 3 were used, and structures were saved for analysis at 1-ps intervals.
Twenty nanoseconds of each 298-K dynameomics trajectory was analyzed. Only the last 10 ns of the 498-K dynameomics trajectories was analyzed, giving the same number of samples at both 298 and 498 K. A total of 145 targets were considered here. The 20-ns simulation time was insufficient for a number of the larger proteins to unfold to any great extent. Comparison with earlier, well characterized and experimentally validated, simulations from our laboratory suggests that proteins with a CONGENEAL structural dissimilarity (40) score of >0.5 had left both the native and first intermediate (if present) clusters (41–44). Proteins where the CONGENEAL score remained <0.5 were therefore excluded. The CONGENEAL dissimilarity score, introduced by Yee and Dill (40), is defined for comparison of two structures with the same number of residues as
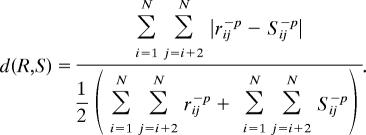 |
For two structures R and S, rij is the distance between atoms i and j in structure R, and sij is the distance in structure S. These distances are raised to the power −p, where p = 2 in this case. The CONGENEAL score is a measure of dissimilarity where more emphasis is placed on close neighbors than residues far apart in sequence. When compared with the Cα root mean square deviation the CONGENEAL score is less dominated by rigid body motion.
Entropy Calculations.
The entropy was calculated for backbone conformations from the distribution of (Φ,Ψ) angles as S = −RΣ[piln(pi)], where pi is the fractional population of bin i. To test whether our results depended on the division of (Φ,Ψ) space, the entropy was calculated by using 2 × 2, 3 × 3, 5 × 5, and 10 × 10° bins for the GGAGG and GGGGG peptides. The entropy difference was shown to be independent of bin size, and a 5 × 5° bin size was used for all other systems.
Calculation of Changes in Solvent-Accessible Surface Area on Mutation of Ala to Gly in Proteins.
Mutations of Ala to Gly were modeled by using Swiss-PdbViewer (45) for representative proteins [the B domain of protein A (PDB entry 1SS1), barnase (1A2P), Im9 (1IMQ), ACBP (1NT1), C12 (2CI2), and R16 (1CUN)]. The changes in solvent-accessible surface area were calculated by using the program NACCESS (S. J. Hubbard and J. M. Thornton, University College London).
Acknowledgments
The computational work was supported by National Institutes of Health Grant GM 50789 (to V.D.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Padmanabhan S, Marqusee S, Ridgeway T, Laue TM, Baldwin RL. Nature. 1990;344:268–270. doi: 10.1038/344268a0. [DOI] [PubMed] [Google Scholar]
- 2.O'Neil KT, DeGrado WF. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 3.Horovitz A, Matthews JM, Fersht AR. J Mol Biol. 1992;227:560–568. doi: 10.1016/0022-2836(92)90907-2. [DOI] [PubMed] [Google Scholar]
- 4.Serrano L, Neira JL, Sancho J, Fersht AR. Nature. 1992;356:453–455. doi: 10.1038/356453a0. [DOI] [PubMed] [Google Scholar]
- 5.Serrano L, Sancho J, Hirshberg M, Fersht AR. J Mol Biol. 1992;227:544–559. doi: 10.1016/0022-2836(92)90906-z. [DOI] [PubMed] [Google Scholar]
- 6.Blaber M, Zhang XJ, Matthews BW. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabartty A, Kortemme T, Baldwin RL. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creamer TP, Rose GD. Proteins Struct Funct Genet. 1994;19:85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Spek EJ, Gong Y, Zhou H, Kallenbach NR. Protein Sci. 1997;6:1264–1272. doi: 10.1002/pro.5560060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauling L, Corey RB, Branson HR. Proc Natl Acad Sci USA. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemethy G, Leach SG, Scherage HA. J Phys Chem. 1966;70:998–1004. [Google Scholar]
- 12.Matthews BW, Nicholson H, Becktel WJ. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang AS, Honig B. J Mol Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- 14.D'Aquino JA, Gomez J, Hilser VJ, Lee KH, Amzel LM, Freire E. Proteins Struct Funct Genet. 1996;25:143–156. doi: 10.1002/(SICI)1097-0134(199606)25:2<143::AID-PROT1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Brady GP, Sharp KA. Curr Opin Struct Biol. 1997;7:215–221. doi: 10.1016/s0959-440x(97)80028-0. [DOI] [PubMed] [Google Scholar]
- 16.Luo PZ, Baldwin RL. Proc Natl Acad Sci USA. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal D, Chakrabarti P. Proteins Struct Funct Genet. 1999;36:332–339. [PubMed] [Google Scholar]
- 18.Zaman MH, Shen MY, Berry RS, Freed KF, Sosnick TR. J Mol Biol. 2003;331:693–711. doi: 10.1016/s0022-2836(03)00765-4. [DOI] [PubMed] [Google Scholar]
- 19.Richardson JM, Lopez MM, Makhatadze GI. Proc Natl Acad Sci USA. 2005;102:1413–1418. doi: 10.1073/pnas.0408004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day R, Beck DAC, Armen RS, Daggett V. Protein Sci. 2003;12:2150–2160. doi: 10.1110/ps.0306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews JM, Fersht AR. Biochemistry. 1995;34:6805–6814. doi: 10.1021/bi00020a027. [DOI] [PubMed] [Google Scholar]
- 22.Sato S, Religa TL, Daggett V, Fersht AR. Proc Natl Acad Sci USA. 2004;101:6952–6956. doi: 10.1073/pnas.0401396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friel CT, Capaldi AP, Radford SE. J Mol Biol. 2003;326:293–305. doi: 10.1016/s0022-2836(02)01249-4. [DOI] [PubMed] [Google Scholar]
- 24.Kragelund BB, Osmark P, Neergaard TB, Schiodt J, Kristiansen K, Knudsen J, Poulsen FM. Nat Struct Biol. 1999;6:594–601. doi: 10.1038/9384. [DOI] [PubMed] [Google Scholar]
- 25.Itzhaki LS, Otzen DE, Fersht AR. J Mol Biol. 1995;254:260–288. doi: 10.1006/jmbi.1995.0616. [DOI] [PubMed] [Google Scholar]
- 26.Scott KA, Randles LG, Clarke J. J Mol Biol. 2004;344:207–221. doi: 10.1016/j.jmb.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Serrano L, Matouschek A, Fersht AR. J Mol Biol. 1992;224:805–818. doi: 10.1016/0022-2836(92)90563-y. [DOI] [PubMed] [Google Scholar]
- 28.Fersht AR, Matouschek A, Serrano L. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 29.Matouschek A, Kellis J, Jr, Serrano L, Bycroft M, Fersht AR. Nature. 1990;346:440–445. doi: 10.1038/346440a0. [DOI] [PubMed] [Google Scholar]
- 30.Matouschek A, Kellis JT, Serrano L, Fersht AR. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 31.Fersht AR. Proc Natl Acad Sci USA. 2004;101:14338–14342. doi: 10.1073/pnas.0406091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfenden R, Andersson L, Cullis PM, Southgate CCB. Biochemistry. 1981;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 33.Fersht AR, Sato S. Proc Natl Acad Sci USA. 2004;101:7976–7981. doi: 10.1073/pnas.0402684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li A, Daggett V. Proc Natl Acad Sci USA. 1994;91:10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li A, Daggett V. J Mol Biol. 1996;257:412–429. doi: 10.1006/jmbi.1996.0172. [DOI] [PubMed] [Google Scholar]
- 36.Daggett V, Li AJ, Itzhaki LS, Otzen DE, Fersht AR. J Mol Biol. 1996;257:430–440. doi: 10.1006/jmbi.1996.0173. [DOI] [PubMed] [Google Scholar]
- 37.Beck DAC, Daggett V. Methods. 2004;34:112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. J Phys Chem. 1997;101:5051–5061. [Google Scholar]
- 39.Levitt M, Hirshberg M, Sharon R, Daggett V. Comp Phys Commun. 1995;91:215–231. [Google Scholar]
- 40.Yee DP, Dill KA. Protein Sci. 1993;2:884–899. doi: 10.1002/pro.5560020603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor U, Johnson CM, Daggett V, Fersht AR. Proc Natl Acad Sci USA. 2000;97:13518–13522. doi: 10.1073/pnas.250473497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bond CJ, Wong KB, Clarke J, Fersht AR, Daggett V. Proc Natl Acad Sci USA. 1997;94:13409–13413. doi: 10.1073/pnas.94.25.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daggett V. Acc Chem Res. 2002;35:422–429. doi: 10.1021/ar0100834. [DOI] [PubMed] [Google Scholar]
- 44.Day R, Bennion BJ, Ham S, Daggett V. J Mol Biol. 2002;322:189–203. doi: 10.1016/s0022-2836(02)00672-1. [DOI] [PubMed] [Google Scholar]
- 45.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]