Abstract
Molecular confinement offers new routes for arraying large DNA molecules, enabling single-molecule schemes aimed at the acquisition of sequence information. Such schemes can rapidly advance to become platforms capable of genome analysis if elements of a nascent system can be integrated at an early stage of development. Integrated strategies are needed for surmounting the stringent experimental requirements of nanoscale devices regarding fabrication, sample loading, biochemical labeling, and detection. We demonstrate that disposable devices featuring both micro- and nanoscale features can greatly elongate DNA molecules when buffer conditions are controlled to alter DNA stiffness. Furthermore, we present analytical calculations that describe this elongation. We also developed a complementary enzymatic labeling scheme that tags specific sequences on elongated molecules within described nanoslit devices that are imaged via fluorescence resonance energy transfer. Collectively, these developments enable scaleable molecular confinement approaches for genome analysis.
Keywords: DNA labeling, genomics, nanofabrication, polymer confinement
Approaches using single molecules are poised to radically alter the way most biological researchers conceive, perform, and analyze experiments. The use of single molecules as analytes represents absolute miniaturization and fosters new measurement schemes, allowing for a more comprehensive analysis. In comparison with traditional bulk measurement schemes, experiments using single molecules are direct and rapid, promising the generation of thousands of data points in a short time period. As such, the entry of single-molecule analytes into mainstream genomic investigation requires the development of high-throughput platforms, which in turn must effectively fuel new biological insights. To meet this challenge, integration of experimental or analytical components into working systems is a critical barrier because developments that simply identify new single-molecule phenomena or create new greatly miniaturized devices are merely the first few steps in this process. This implies that research efforts truly intent on impacting the genomic sciences will likely leverage single-molecule effects through the integration of robust devices with equally robust biochemical and detection schemes.
Almost paradoxically, many schemes for the analysis of single DNA molecules, such as sequencing, employ amplification steps (1, 2) using genomic DNA substrates. These steps, although obviating traditional clone libraries, also eliminate the principle advantage of single-molecule techniques for which measurements describe individual molecules, free from any consequences of an ensemble. Thus, new single-molecule approaches aimed at genomic analysis require a high level of precision and throughput before they become tools for research.
When large genomic DNA molecules are the primary analyte, how random coils are unraveled for high-resolution detection of markers (typically by fluorescence microscopy) as well as the presentation techniques employ a variety of schemes: devices that direct fluid flows for surface deposition of molecules (3, 4); agarose gel matrices (5, 6) or engineered nanopillar arrays (7, 8), which enable electrokinetically driven reptation or threading of DNA molecules; and, most recently, direct nanoconfinement (9, 10), where molecules are trapped in elongated configurations. During the last decade, nano- and microfabrication techniques have catalyzed these developments, allowing researchers to fabricate devices boasting complex geometries in both hard and soft materials. Traditionally, nano- and microscale fabrications for biological research have used silicon or quartz wafers as an adaptation from microelectromechanical systems (11), but there have been problems in integrating these materials with biological or genomic applications. Traditional device fabrication is technologically demanding and time-consuming, requiring expensive lithographic equipment (e.g., electron beam lithography for each nanoscale device) in a clean-room facility and later requiring complicated sealing approaches. Additionally, although electrokinetic control is very common for biomolecule manipulation, the semiconductor attributes of silicon wafers favor quartz fabrication due to its insulatory properties. However, toilsome and expensive device fabrication from quartz necessitates reuse, leading to contamination issues (12). Although nanoscale features are more rapidly rendered through molding techniques (13), the inability to produce a large number of disposable devices within a typical laboratory environment (14) has greatly hindered their use in high-throughput genomic applications.
To overcome the limitations of hard materials, soft lithography based on elastomer replicas (14, 15) has been developed, augmenting traditional lithography for biological applications using microfluidics (12, 16, 17). Soft lithography uses molds (masters) created from silicon wafers patterned with photoresist for producing disposable elastomeric replicas (14), most commonly from poly(dimethylsiloxane) (PDMS), ideally suited for high-throughput applications. Unfortunately, PDMS elasticity portends mechanical instability, especially for nanostructures of <100 nm, which often disappear by collapsing against flat substrates (18).
Given these concerns, our goal was to develop a high-throughput system for genome analysis by using disposable devices offering effective nanoscale geometries sufficient for the presentation of large elongated DNA molecules. The idea was to create the basis for a successor to the established optical mapping system (19, 20), which spans entire genomes through overlapping ordered restriction maps created from individual genomic DNA molecules. Our motivation for the work described here was advancement of whole-genome restriction mapping from a tool for discovery, or sequence validation, to a means for analysis of human populations and cancer genomes. We solved the conundrum of how to imbue features in disposable PDMS devices with nanoconfinement capabilities that closely parallel those found in devices with features of <100 nm and requiring traditional lithographic approaches.
By considering DNA as a polyelectrolyte, its enlargement in terms of physical measures of size hinges on a polymer's persistence length, commonly ≈50 nm (21). Physical properties of DNA chains are derived from treatments modeling them as worm-like coils (22), where they exhibit both local rigidity and long-range flexibility. From the theoretical work of Odijk, Skolnick, and Fixman (23, 24), we know that ionic strength is an important factor governing intrachain electrostatic repulsion affecting the persistence length of worm-like polyelectrolyte coils, estimated from the Debye–Hückel screening length. This view was experimentally confirmed for large DNA molecules by Baumann et al. (21), who found that the persistence length of DNA molecules inversely varied with ionic strength, in good agreement with the theory of Odijk, Skolnick, and Fixman. Consequently, we reasoned that low-ionic-strength conditions would sufficiently increase DNA persistence length, allowing for larger channels to be used for polymer confinement. Thus, when comparable length scales are achieved (persistence length and channel dimensions), polymer confinement regimes shift, enabling extensive elongation of polymer chains (25) within channel dimensions readily supported by standard PDMS fabrication techniques. Accordingly, we report our findings showing that DNA molecules are stretched up to 60% of their polymer contour length in disposable PDMS devices having 100-nm × 1-μm channels under low-ionic-strength conditions; remarkably, these results are comparable with those previously obtained under standard buffer conditions using 30- × 40-nm channels (fused silica) employing nanoimprint or electron beam lithography (26).
Although low-ionic-strength buffers enable DNA elongation in larger nanoslits readily made of PDMS, the avoidance of biochemically meaningful salt concentrations causes problems for most DNA enzymes used for genome analysis. However, molecular confinement and DNA modification enzymes (e.g., restriction endonucleases) are not necessarily incompatible when used within standard <100-nm fabrications, as recently demonstrated by Riehn et al. (27). Instead, issues arise regarding the scalability of their device as a viable platform for genomic analysis due to the requirement that molecules must be continuously imaged for discernable biochemical events, greatly diminishing potential throughput. As such, we developed a single-molecule labeling scheme obviating these concerns while offering distinct advantages for robust detection and integration within a system for genome analysis.
Conventional hybridization techniques are not suited for marking discrete DNA molecules because presentation approaches require intact, double-stranded DNA molecules after processing for analysis. Because optical mapping successfully employs restriction enzymes for reliable placement of sequence-specific markers onto individual molecules, we reasoned that a new class of endonucleases nicking at specific sites (28) would also confidently mark molecules but without unwanted double-strand cleavage. Because nicks cannot be directly discerned by fluorescence microscopy, we label these sites by nick-translation of DNA molecules in bulk solution using fluorochrome-labeled nucleotides. We then counterstain DNA backbones with the bis-intercalator YOYO-1 before imaging. Specificity is enhanced by using ligase and dideoxynucleotide blocking steps, greatly diminishing labeling of preexisting random nicks. Finally, incorporated fluorochrome labels support FRET detection, which simplifies data acquisition by requiring one laser for excitation of DNA backbones (donor) and labels (acceptor).
Here, we report a series of interlocking developments and findings to potentiate a device design based on physical modification of large single DNA molecules through simple alterations of solution ionic strength. This developmental stance fosters creation of usable systems for advancing genome analysis. We demonstrate proof of principle with physical maps of BACs.
Results and Discussion
Multiscale Fabrication Facilitates DNA Loading and Elongation.
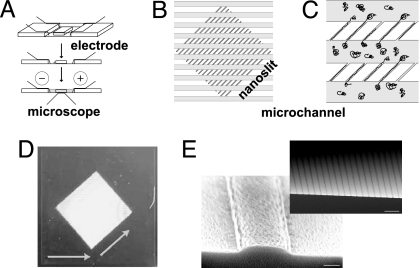
Previously, we used soft lithography techniques to fabricate a microchannel device designed to load and deposit large DNA molecules onto charged glass surfaces (3). Because this device has proven robust for routine optical mapping, we have adapted it here for capillary preloading of newly incorporated nanoslits (100 nm × 1 μm; Fig. 1). These PDMS devices are fabricated by following standard rapid prototyping procedures (12, 17) but with the inclusion of a dry-etch step, creating 100-nm-high features (Fig. 1), used because it persists through cycles of replica molding, fostering large-scale fabrication of devices. A photoresist pattern of microchannels is then overlaid, creating nanoslit and microchannel features on the same silicon wafer. The nanoslit geometry engenders nanoscale confinement conditions while employing simple relief-based fabrication techniques, and wider channel entrances offer simplified loading and less clogging. Because the relaxed dimensions of our molecules (λ and T4 bacteriophage DNA, Rg = 0.55 and 1.13 μm, respectively) are greater than the slit dimensions, they are excluded from entering (29) by simple diffusion after capillary loading; an electric potential (70 V) transports relaxed DNA coils to nanoslit entrances for subsequent elongation.
Fig. 1.
Microchannel–nanoslit device design and loading scheme. (A) Plexiglas slide (25.4 × 76.2 mm) with a rectangular opening to which a glass coverslip window (18 × 18 mm) is affixed with wax. The PDMS device is bonded to this window and immersed in buffer for electrokinetic loading via the indicated electrodes. Before buffer immersion, DNA is pipetted into microchannels for capillary loading. (B) Illustration (top view) shows nanoslits (diagonal, 100 nm high × 1 μm wide) overlaid with microchannels (horizontal, 3 μm × 100 μm wide). (C) Cartoon depicts relaxed and stretched DNA molecules occurring during electrokinetic loading within microchannels and nanoslits. (D) Photograph of silicon master mold bearing photoresist and etched features; arrows show the path of DNA molecules taken through the microchannel and nanoslit features. (E) Scanning electron micrograph of the silicon master shows a single nanoslit mold feature before the photoresist overlay, conferring microchannel features. (Scale bar, 300 nm.) The upper image shows many such nanoslit features spaced 4 μm apart (center-to-center). (Scale bar, 10 μm.)
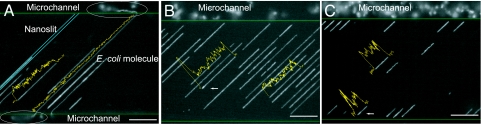
Fig. 2 shows images of electrokinetically loaded λ (48.5 kb), T4 (166 kb), and Escherichia coli genomic DNA molecules. For elongation, a molecule undergoes electrophoresis within a microchannel toward a nanoslit entrance, where it proceeds to enter then stretch. An E. coli genomic DNA molecule (Fig. 2A) shows a contour length exceeding that of a nanoslit so that its ends appear relaxed within the microchannels. Here, the largest E. coli genomic DNA fragment spans the 105-μm-long nanoslit and is qualitatively sized at 432 kb, assuming a stretch of 0.60 (see the next section); this stretch may be increased through tension exerted by the relaxed ends. Accordingly, the corresponding fluorescence intensity profile shows that this molecule is well stretched without detectable local folding back of DNA segments, also known as hairpins (30). The total size of this molecule, accounting for the relaxed portions, may exceed 1 megabase. Such stretching is contrasted with the molecules completely within the nanoslits that exhibit folding, as evidenced by heightened values in their fluorescence intensity profiles.
Fig. 2.
Gallery of fluorescence micrographs shows stretched and relaxed DNA molecules within the nanoslit device after electrokinetic loading; images were taken a few minutes after the electric field was shut off. (A) A large E. coli DNA molecule spans across the 105-μm-long nanoslit (0.01× TE buffer) showing relaxed ends (circled) within abutting microchannels. (B) T4 DNA (166 kb) molecules; 0.05× TE. (C) λ DNA (48.5 kb) molecules; 0.01× TE. Green lines demarcate nanoslit–microchannel interfaces; blue indicates a nanoslit, and yellow lines show integrated fluorescence intensity profiles revealing folded ends (B and C, arrows). Relaxed molecules within the microchannel regions appear as diffuse, partly out-of-focus, fluorescent balls, whereas stretched molecules present as long, linear objects. (Scale bars, 20 μm.)
Odijk (30) has put forward a theory of hairpins in various types of nanochannels (circular, square, and rectangular). The B-form of DNA remains intact so that the bending energy is purely elastic (see Eq. 3 below). The confines of a nanochannel cut off orientational and translational degrees of freedom, an entropic effect that tightens hairpins, forcing them toward the central axis of the channel. Because of this effect, which increases the free energy of the hairpin markedly, the distance between hairpins may reach tens or hundreds of micrometers, even for a nanoslit of the dimensions used here.
Low-Ionic-Strength Effects on DNA Elongation.
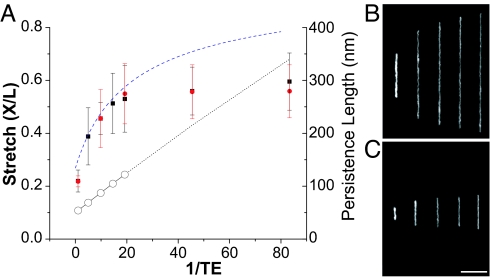
Diminished ionic strength conditions increase DNA intrachain electrostatic repulsion (23, 24), with a concomitant increase of the polymer persistence length, observable within a nanoslit as DNA stretching. We study these effects by using dilutions of the same working buffer, Tris–EDTA (1× TE; 10 mM Tris base/1 mM EDTA, pH 8.0, titrated with HCl; ionic strength, 8.4 mM), performed within the same slit geometry (100 nm × 1 μm) for all experiments presented here. In addition to the TE buffer system, sodium phosphate and diluted New England Biolabs buffer 4 (Materials and Methods) were evaluated, and each demonstrated DNA stretching within nanoslits at low apparent ionic strengths (data not shown). Fig. 2 B and C show typical images of T4 and λ DNA (polymer contour lengths, 74.5 and 21.8 μm, respectively) stretched within nanoslits under low-ionic-strength conditions (0.05× and 0.01× TE), with average apparent lengths of 40.9 ± 8.4 μm and 13.0 ± 2.4 μm, respectively. As a direct comparison, our value for the λ DNA is nearly equivalent to the findings of Reisner et al. (26), who used 30- × 40-nm channels and TBE buffer (0.5× TBE = 45 mM Tris base/1 mM EDTA/45 mM boric acid). Fig. 3 shows a plot of DNA elongation as a function of TE dilution (1/TE) reported as stretch, which is defined as the average apparent length, X, divided by the dye adjusted polymer contour length, L (e.g., L = 21.8 μm for λ DNA); a stretch of 1.0 indicates complete elongation.
Fig. 3.
DNA stretch varies with TE concentration. (A) Ionic strength varied through successive 5-, 10-, 15-, 20-, 50-, and 100-fold dilutions of 1× TE buffer (ionic strength, 8.4 mM). The reciprocal TE buffer concentrations vs. the stretch of λ (black square) and T4 (red circle) DNA are plotted along with the calculated persistence length (Eq. 1) using ionic strengths determined from dilutions (solid line with open circles). A dotted line continues the calculated persistence length for ionic strengths at <0.5 mM, accounting for uncertainties associated with very low ionic strength. A dashed blue line represents a fitted curve using Eq. 12. The stretch is defined by apparent length (X) divided by the polymer contour length (L) of YOYO-1-stained DNA. Each data point represents measurements from 50 to 300 molecules; error bars show standard deviations on these means. (B and C) Fluorescence images (a combination of five separate experiments) show T4 DNA (B) and λ DNA (C) at five different TE dilutions, 1.002× (0.9980), 0.102× (9.80), 0.0520× (19.2), 0.0220× (45.5), and 0.0120× (83.3), with the corresponding dilution factors shown in parentheses. (Scale bar, 10 μm.)
This plot (Fig. 3) shows that DNA stretch is size-independent and inversely related to salt concentration or ionic strength according to persistence length calculations using Eq. 1. The persistence length (P) is related to the ionic strength (I) by an expression derived by Odijk, Skolnick, and Fixman (23, 24) and interpreted by Baumann et al. (21):
where Po is the nonelectrostatic intrinsic persistence length due to base stacking, Pel is the electrostatic persistence length due to intrachain repulsion, κ−1 is the Debye–Hückel screening length, and lB is the Bjerrum length (≈7 Å in water). Although the value of Po varies according to environmental conditions (21), here we take Po to be 50 nm. Accordingly, we see that stretch plateaus at a 20× dilution (effective ionic strength, 0.45 mM), with a corresponding persistence length of 122 nm, a value greater than our nanoslit height (100 nm). Although we interpret observed DNA stretching to be strictly a function of persistence length determined at a given salt concentration, such lengths may not be accurate due to additional low-ionic-strength effects that we are not taking into account here, including local DNA melting, particularly at AT-rich regions, or experimental limitations, for example, carbonate dissolution or contamination, which alters ionic strength at very low salt concentrations. A dotted line is used for describing alterations of persistence length at <0.05× TE (20× dilution) in Fig. 3.
Polymer Elongation Regimes Under Confinement.
DNA elongation is greatly enhanced when the scale of confinement geometry approaches a value comparable to the radius of gyration (29, 31, 32) or persistence length (24, 26). Previously, Brochard and de Gennes (29) developed a scaling argument explaining polymer stretching under confinement within a channel, where a self-avoiding chain is considered as a series of blobs provided that channel dimensions are much larger than the persistence length (D ≫ P) and the polymer contour length is greater than D (L ≫ D). In their treatment, the stretch (X/L) of a polymer (persistence length P and molecule width w) in a channel (width D1 and height D2) is given by X/L ≅ (wP/D1D2)1/3 (10, 29, 32, 33); however, this argument is not valid for highly stretched polymers (e.g., X/L > 0.5). Instead, for highly stretched polymer chains under nanoconfinement, a different scaling argument (25) treats polymer stretching within a nanochannel as a long chain deflected by the walls of a nanochannel with the stretch given by
Here, we wish to estimate the numerical coefficient. We introduce an analytical treatment based on a rescaling of the harmonic potential to the hard wall repulsions experienced by a worm-like chain confined in a slit geometry, followed by comparison with our experimental observations (Fig. 3). The regime of strong elongation is complicated because of the formation of hairpins (30). Because of this, the crossover between the two regimes is nontrivial and not understood at present.
Calculation of Stretch Within a Slit Regime.
A rigorous analytical calculation is impossible because of the hard wall boundary conditions previously mentioned in ref. 25. Nevertheless, an exact computation is feasible for a worm-like chain confined in a harmonic potential, V(x) (34). The Hamiltonian H in one dimension may be written as
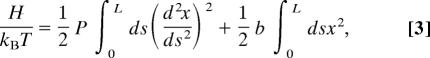 |
where the first term is the scaled bending energy of the chain and the second is V[x(s)], which simulates the confinement. The position of the chain is x(s) at contour distance s from one end. Statistical mechanics applied to Eq. 3 may be shown to lead to a Gaussian distribution for the worm (as L → ∞)
In this way, we may eliminate the dummy variable b. From the orientation– translation distribution derived by Burkhardt (35), one may obtain (in one dimension)
and
One has to subtract the energy from the external field to retain only the configurational free energy Fconf, which is entropic in origin [Fconf = (3/4)Ftot; for a detailed discussion, see appendix 1 in ref. 36]. After eliminating b, we obtain
Burkhardt (34) also computed Fconf for a worm in a hard slit, but numerically,
where Arec = 1.1036.
Rescaling.
If we now assume that a hard wall may be simulated by a Gaussian, then we must identify d via the respective free energies (Eqs. 8 and 9) in two dimensions x, y:dx, dy. For instance, in the x direction, we have
Hence, we then insist on
Accordingly, we arrive at the following approximate expression for the relative elongation:
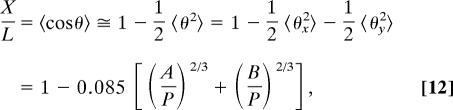 |
where A = 1,000 nm and B = 100 nm in our experimental condition. Of course, as 〈cosθ〉 diverges further away from unity, it becomes more approximate because we assume there is no back-folding. We note that the numerical coefficient in Eq. 12 is remarkably low. It implies that the formation of hairpins must be difficult even when A and B are of the order of the persistence length. This corroborates the computations of the high hairpin free energies under the same conditions (30). A fit of Eq. 12 shows good agreement with the experimental observations in Fig. 3, discounting TE buffer dilutions below 1/TE = 20, where measured stretch values plateau, as attributed to the previously stated experimental conditions associated with very low ionic strength effects. We also point out that the effect of the electrostatic interaction between the DNA and the negatively charged walls of the nanoslit in our expansion for the stretch Eq. 12 is quite negligible. Although the Debye scaling length at 1/TE = 20 is a substantial 15 nm long and should result in decreasing the height B to an effective value of ≈70 nm, the width A is altered little, if at all. The stretch is then predicted to increase by a few more percentage points because of electrostatics, which is well within the experimental margin of error displayed in Fig. 3A.
Equilibration of DNA Stretch.
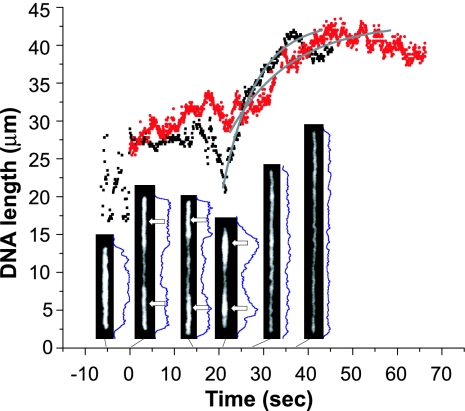
DNA molecules electrokinetically move through the microchannels as relaxed coils until approaching a nanoslit entrance. There, as one end of a molecule enters the nanoslit, it transiently elongates, adopting a conformation we describe as a single-ended dumbbell. The dumbbell progressively disappears as it threads into the nanoslit, adopting a nascent, fully confined conformation while approaching equilibrium after the field is shut-off. Fig. 4 shows a plot of length vs. time for DNA molecules reaching such equilibrium, where the inset images show dynamic shrinking and unfolding of T4 DNA molecules. These experiments used New England Biolabs buffer 4 at a 1,000× dilution, resulting in a stretch of 0.58 (42.8 ± 3.6 μm; 81 molecules) (data not shown). As seen in Fig. 4, DNA molecules expectedly enter nanoslits with different conformations and reach equilibrated forms via different processes. For example, the apparent equilibration time constants for these molecules are 7.4 and 13.7 s. These findings bear interesting semblance to a recent report in which Mannion et al. (37) investigated DNA equilibration within nanochannels under relatively high salt buffer conditions (5× TBE; 445 mM Tris borate/10 mM EDTA). In their 100- × 100-nm nanochannels, T4 DNA molecules are initially stretched up to 40 μm and exponentially contract down to 26.4 μm (X/L = 0.23) with a relaxation time constant of 9.3 s, whereas in our 100-nm × 1-μm nanoslits, DNA molecules under low salt conditions start at ≈25 μm and then stretch up to ≈40 μm (X/L = 0.55).
Fig. 4.
T4 DNA relaxation within nanoslits after electrokinetic loading. Plots show the relaxation kinetics (0.4 mM ionic strength, New England Biolabs buffer 4) (see Materials and Methods) of two DNA molecules [the first (black) enters nanoslits in a folded state, and the second (red) is not significantly folded] gauged by apparent length measurements as a function of time; 0 s corresponds to the electric field being shut off. Six images of molecule 1 show evolution of fluorescence intensity profiles (blue traces) echoing conformational and length changes obtained at −5, 0, 14, 21, 27, and 37 s, with arrows indicating putatively folded regions flagged by increased fluorescence intensities. We interpret this analysis as indicating that molecule 1 enters a nanoslit highly folded, with both ends tucked in. After the field is shut off, folded arms appear, signaled by the fluorescence intensity profiles, and continue unfolding up to ≈40 s, until the entire molecule appears fully equilibrated at 50 s. Interestingly, at 21 s, the molecule shrinks (also at −5 s), then proceeds to relax in an exponential fashion; curve fits (gray lines) of molecules 1 and 2 show time constants of 7.4 and 13.7 s, respectively. For further details, see SI Movie 1.
DNA Barcoding via Fluorochrome Labeling of Specific Nick Sites.
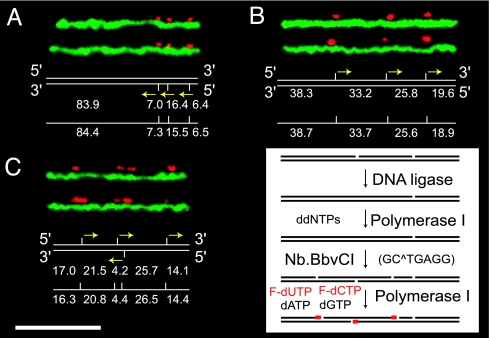
Fig. 5 describes a molecular barcoding approach compatible with low salt conditions producing sequence-specific landmarks detectable by fluorescence microscopy. Importantly, all steps are performed in “test tubes” and then loaded into the nanoslit device after adjustment of ionic strength. Here, a nicking enzyme, Nb.BbvCI, cleaves only cognate sites on single strands of double-stranded molecules, made detectable by nick translation using fluorochrome-labeled nucleotides. Because preexisting DNA nicks would produce spurious signals, such sites are repaired or disabled by using T4 ligase and polymerase incorporation of dideoxyribonucleotides (ddNTPs) before labeling. Because nick translation efficiently incorporates fluorochrome-labeled nucleotides, crossing of mobilized nick sites on complementary DNA strands occurs and produces double-strand breaks. We attenuate this action by the continued presence of ddNTPs in the labeling reaction mix, thus limiting the number of nucleotides incorporated per nick site through chain termination. This step also controls the size of fluorescent punctates, which would otherwise expand into each other if nucleotide incorporation were unchecked, thus diminishing the number of discrete markers.
Fig. 5.
Molecular barcoding scheme and maps of nicked, fluorochrome-labeled BAC molecules imaged by fluorescence microscopy. (Inset) First, DNA ligation and nick translation with ddNTPs obviate inherent nicks before barcoding; second, Nb.BbvCI places site-specific nicks on these molecules; and last, E. coli DNA polymerase I incorporates fluorochrome-labeled deoxyribonucleotides F-dCTP and F-dUTP (Alexa Fluor 647-aha-dCTP, dUTP) into nick sites. Fluorescence images of labeled DNA molecules (pseudocolored DNA backbones are green and FRET imaged punctates red) compared with expected labeling patterns (measured in kilobases) from sequence, and unity-based maps constructed from analyzed molecules (Materials and Methods) are shown for BAC79 (113.7 kb) (A), BAC150 (116.8 kb) (B), and BAC614 (82.5 kb) (C). Yellow arrows orient nick translation on DNA strands. DNA molecules were stretched in 0.01× TE buffer within described nanoslits. (Scale bar, 10 μm.)
FRET Imaging of Barcoded Molecules in Nanoslits.
Because labeled DNA molecules are stained with the intercalating dye YOYO-1, FRET operates with incorporated fluorochrome labels (Alexa Fluor 647; FRET acceptor). Fig. 5 shows FRET detection of barcode features, mapping their spacing (measured in kilobase pairs) by using integrated fluorescence intensity measurements of intervening YOYO-1 signals (intervals). Averaging of such barcodes from multiple molecules produces “unity-based maps” (4) that are compared against in silico barcodes constructed from sequence information. There is good agreement with the in silico references constructed from sequence information [supporting information (SI) Fig. 6]. Comparisons show an average absolute relative error of 2.79% and a pooled standard deviation of 3.34 kb, demonstrating development of a DNA barcoding system (for additional analysis, see SI Text).
Conclusion.
We have shown that DNA stretch is possible in relatively large-scale nanoslits by using low-ionic-strength buffers. Such polymer behavior was considered by using an analytical treatment explicitly dealing with slit geometries that engender molecular nanoconfinement. These insights were leveraged through the synergetic effects of low-ionic-strength buffers and the design of disposable PDMS devices bearing nanoslits for DNA analysis through codevelopment of a compatible labeling scheme for barcoding single molecules.
The correlation of increasing DNA elongation with decreasing ionic strength is explained by the electrostatic repulsive force within a persistence length resulting from the Debye screening length. By minimizing the constraints of confinement dimensions required for elongating DNA molecules and instead focusing on salt conditions, PDMS nanostructures become useful tools for future genomic and polymer physics studies. For genomic applications, a stretch of 0.60 is capable of providing valuable data fueling biological investigations, as was previously obtained with the optical mapping system (3). As such, future efforts will leverage fully automated imaging, processing, and barcode construction (preliminary system created; data not shown) for the comprehensive assessment of errors, allowing for full integration with new algorithms created for assembling maps that span entire genomes.
Materials and Methods
Nanoslit Preparation.
Fabrication of PDMS devices used standard rapid prototyping procedures (12). A master wafer of nanoslits (100 nm high, 1 μm wide, and 5 mm long) was dry-etched and overlaid with microchannels (3 μm high, 100 μm wide, and 10 mm long) made of negative photoresist; replica devices were produced by curing PDMS onto such master wafers (for details, see SI Text).
DNA Sample Preparation and Loading.
DNA samples [1 ng/μl λ DNA (New England Biolabs, Ipswich, MA), 0.78 ng/μl T4 DNA (Waco Chemicals, Waco, TX), and E. coli DNA (38)], contained 0.25 μM YOYO-1, from 1× to 0.01× TE buffer, 4% (vol/vol) 2-mercaptoethanol, and 0.1% (wt/vol) POP6 (Applied Biosystems, Foster City, CA) to suppress electroendosmosis (10). In addition to TE buffer, sodium phosphate buffer (pH 7.9; a 10:93 mixture of 10 mM NaH2PO4/10 mM Na2HPO4) and New England Biolabs buffer 4 with 20 mM EDTA were also used. YOYO-1-stained DNA molecules were loaded into the microchannels via capillary action and then entered the nanoslits by using an applied electrical field (70 V) (Fig. 1) with platinum electrodes inserted into the reservoirs.
Microscopy and Image Processing.
The microscopy setup for single color imaging and imaging flattening processes for shading correction is as reported in ref. 3 (for details, see SI Text).
DNA Barcoding.
The circular DNA molecules BAC79, BAC150, and BAC614 were linearized with FseI or SpeI (New England Biolabs). Preexisting nicks were repaired by using 2 units of T4 DNA ligase (1 mM ATP) at 16°C for 2 h, with a total volume of 17.5 μl of New England Biolabs buffer 4 or New England Biolabs buffer 2. The mix was then heat-inactivated at 65°C for 10 min. Endonuclease-free-grade E. coli DNA polymerase I (10 units) (Roche Applied Sciences, Indianapolis, IN) and added ddNTPs at 0.2 μM each (Amersham Biosciences, Piscataway, NJ) blocked remaining nicks at 37°C for 30 min (total volume, 40 μl). The labeling reaction mix [3 units of Nb.BbvCI (New England Biolabs)/2 μM Alexa Fluor 647-aha-dCTP/2 μM Alexa Fluor 647-aha-dUTP (Invitrogen, Carlsbad, CA)/20 μM dATP/20 μM dGTP/1 μM dCTP/1 μM dTTP] was then added, and the reaction was incubated for 30 min at 37°C. Enzymes were digested with 100 ng/μl proteinase K in 0.1% (wt/vol) N-lauroylsarcosine for 3 h at 50°C. Samples were diluted 4,000-fold, or buffer conditions were adjusted by dialysis against 2 liters of 0.01× TE buffer solution overnight with a microdispodialyzer (Spectrum Laboratories, Rancho Dominguez, CA) at 4°C.
Supplementary Material
Acknowledgments
We thank S. Zhou, K. Potomousis, A. Ramme, K. Kounovsky, G. Ananiev, T. Durfee, D. Frisch, N. Hermersmann, G. Plunkett III, R. Roberts, and S. Y. Xu for assistance. This work was supported by National Institutes of Health Grant 5R01HG000225 and National Science Foundation Grant NSEC DMR-0425880.
Abbreviations
- PDMS
poly(dimethylsiloxane)
- ddNTP
dideoxyribonucleotide
- TE
Tris–EDTA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611151104/DC1.
References
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, Porreca GJ, Reppas NB, Lin XX, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Dimalanta ET, Lim A, Runnheim R, Lamers C, Churas C, Forrest DK, de Pablo JJ, Graham MD, Coppersmith SN, Goldstein S, et al. Anal Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 4.Jing J, Reed J, Huang J, Hu X, Clarke V, Edington J, Housman D, Anantharaman TS, Huff EJ, Mishra B, et al. Proc Natl Acad Sci USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz DC, Koval M. Nature. 1989;338:520–522. doi: 10.1038/338520a0. [DOI] [PubMed] [Google Scholar]
- 6.Guo XH, Huff EJ, Schwartz DC. Nature. 1992;359:783–784. doi: 10.1038/359783a0. [DOI] [PubMed] [Google Scholar]
- 7.Volkmuth WD, Austin RH. Nature. 1992;358:600–602. doi: 10.1038/358600a0. [DOI] [PubMed] [Google Scholar]
- 8.Chan EY, Goncalves NM, Haeusler RA, Hatch AJ, Larson JW, Maletta AM, Yantz GR, Carstea ED, Fuchs M, Wong GG, et al. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Yu ZN, Wang J, Tegenfeldt JO, Austin RH, Chen E, Wu W, Chou SY. Appl Phys Lett. 2002;81:174–176. [Google Scholar]
- 10.Tegenfeldt JO, Prinz C, Cao H, Chou S, Reisner WW, Riehn R, Wang YM, Cox EC, Sturm JC, Silberzan P, et al. Proc Natl Acad Sci USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manz A, Miyahara Y, Miura J, Watanabe Y, Miyagi H, Sato K. Sens Actuators B. 1990;1:249–255. [Google Scholar]
- 12.Effenhauser CS, Bruin GJM, Paulus A, Ehrat M. Anal Chem. 1997;69:3451–3457. doi: 10.1021/ac9703919. [DOI] [PubMed] [Google Scholar]
- 13.Chou SY, Krauss PR, Renstrom PJ. Appl Phys Lett. 1995;67:3114–3116. [Google Scholar]
- 14.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Biebuyck HA, Abbott NL, Whitesides GM. J Am Chem Soc. 1992;114:9188–9189. [Google Scholar]
- 16.Delamarche E, Bernard A, Schmid H, Michel B, Biebuyck H. Science. 1997;276:779–781. doi: 10.1126/science.276.5313.779. [DOI] [PubMed] [Google Scholar]
- 17.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 18.Odom TW, Thalladi VR, Love JC, Whitesides GM. J Am Chem Soc. 2002;124:12112–12113. doi: 10.1021/ja0209464. [DOI] [PubMed] [Google Scholar]
- 19.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou SG, Allen AE, Apt KE, Bechner M, et al. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 20.Zhou S, Herschleb J, Schwartz DC. New Methods for DNA Sequencing. Amsterdam: Elsevier; 2007. [Google Scholar]
- 21.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc Natl Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schellman JA. Biopolymers. 1974;13:217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- 23.Skolnick J, Fixman M. Macromolecules. 1977;10:944–948. [Google Scholar]
- 24.Odijk T. J Polym Sci B Polym Phys. 1977;15:477–483. [Google Scholar]
- 25.Odijk T. Macromolecules. 1983;16:1340–1344. [Google Scholar]
- 26.Reisner W, Morton KJ, Riehn R, Wang YM, Yu ZN, Rosen M, Sturm JC, Chou SY, Frey E, Austin RH. Phys Rev Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 27.Riehn R, Lu MC, Wang YM, Lim SF, Cox EC, Austin RH. Proc Natl Acad Sci USA. 2005;102:10012–10016. doi: 10.1073/pnas.0503809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heiter DF, Lunnen KD, Wilson GG. J Mol Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 29.Brochard F, de Gennes PG. J Chem Phys. 1977;67:52–56. [Google Scholar]
- 30.Odijk T. J Chem Phys. 2006;125:1–8. doi: 10.1063/1.2400227. [DOI] [PubMed] [Google Scholar]
- 31.Jendrejack RM, Schwartz DC, Graham MD, de Pablo JJ. J Chem Phys. 2003;119:1165–1173. doi: 10.1063/1.1637331. [DOI] [PubMed] [Google Scholar]
- 32.Turban L. J Phys (Paris) 1984;45:347–353. [Google Scholar]
- 33.Odijk T. Biopolymers. 1979;18:3111–3113. doi: 10.1002/bip.1979.360181215. [DOI] [PubMed] [Google Scholar]
- 34.Burkhardt TW. J Phys A Math Gen. 1997;30:L167–L172. [Google Scholar]
- 35.Burkhardt TW. J Phys A Math Gen. 1995;28:L629–L635. [Google Scholar]
- 36.Ubbink J, Odijk T. Biophys J. 1999;76:2502–2519. doi: 10.1016/S0006-3495(99)77405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mannion JT, Reccius CH, Cross JD, Craighead HG. Biophys J. 2006;90:4538–4545. doi: 10.1529/biophysj.105.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J, Qi R, Aston C, Jing J, Anantharaman TS, Mishra B, White O, Daly MJ, Minton KW, Venter JC, et al. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.