Abstract
Both SM proteins (for Sec1/Munc18-like proteins) and SNARE proteins (for soluble NSF-attachment protein receptors) are essential for intracellular membrane fusion, but the general mechanism of coupling between their functions is unclear, in part because diverse SM protein/SNARE binding modes have been described. During synaptic vesicle exocytosis, the SM protein Munc18-1 is known to bind tightly to the SNARE protein syntaxin-1, but only when syntaxin-1 is in a closed conformation that is incompatible with SNARE complex formation. We now show that Munc18-1 also binds tightly to assembled SNARE complexes containing syntaxin-1. The newly discovered Munc18-1/SNARE complex interaction involves contacts of Munc18-1 with the N-terminal Habc domain of syntaxin-1 and the four-helical bundle of the assembled SNARE complex. Together with earlier studies, our results suggest that binding of Munc18-1 to closed syntaxin-1 is a specialization that evolved to meet the strict regulatory requirements of neuronal exocytosis, whereas binding of Munc18-1 to assembled SNARE complexes reflects a general function of SM proteins involved in executing membrane fusion.
Keywords: exocytosis, membrane fusion, neurotransmitter release, Sec1/Munc18-like proteins, synapse
Every eukaryotic cell relies on the precise and regulated trafficking of proteins, membranes, and other types of cargo between cellular compartments. The molecular machinery responsible for membrane traffic is partly conserved to provide for basic membrane fusion and fission reactions and is partly cell type- and compartment-specific to meet the unique requirements of a particular cellular locale (1). Neurons have arguably the most complex membrane trafficking of all cells, primarily because synaptic transmission requires continuous (but at the same time tightly regulated) synaptic membrane traffic. At a synapse, neurotransmitter release is effected by fusion of synaptic vesicles with the presynaptic plasma membrane. Synaptic vesicle fusion requires two conserved protein families that are universally involved in membrane fusion reactions: SNARE proteins, which are thought to pull membranes together by forming tight “SNARE complexes,” and Sec1/Munc18-like proteins (SM proteins), which perform an unknown but essential role in fusion and interact with SNARE proteins (reviewed in refs. 2–4).
Synaptic vesicle exocytosis involves one SM protein (Munc18-1) and three SNARE proteins (synaptobrevin/VAMP on synaptic vesicles, and SNAP-25 and syntaxin-1A/1B on the plasma membrane; syntaxin-1A and -1B are highly homologous and thought to have identical functions). All SNARE proteins contain a conserved 60- to 70-residue sequence, the SNARE motif, that assembles into a four-helical bundle during SNARE complex formation (5). Synaptobrevin/VAMP and SNAP-25 are “minimal SNARE proteins” that are composed of one (synaptobrevin) or two (SNAP-25) SNARE motifs and a membrane-anchor sequence. In syntaxin-1, however, the SNARE motif and transmembrane region occupy less than half of the sequence, with the remainder (the N-terminal ≈180 residues) consisting of a conserved but natively unstructured N-terminal sequence, an autonomously folded three-helical Habc domain (6), and a linker sequence (Fig. 1a). Interestingly, syntaxin-1 exists in two alternative conformations: a “closed” conformation in which the Habc domain folds back onto the SNARE motif, preventing its engagement into SNARE complexes, and an “open” conformation in which the SNARE motif is exposed to participate in SNARE complex formation (7).
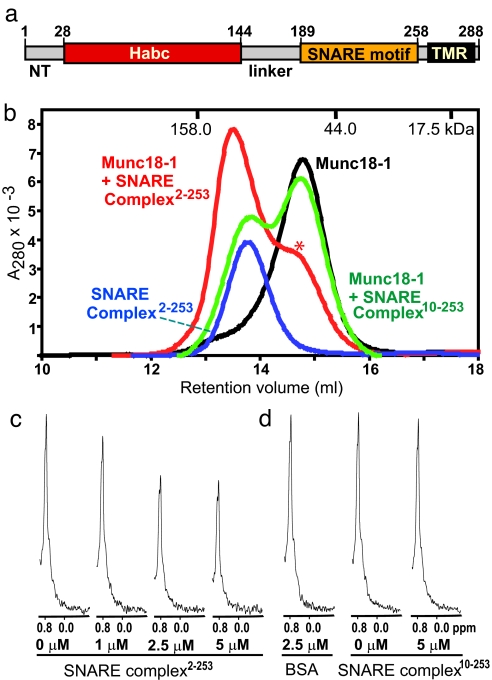
Fig. 1.
Munc18-1 binds to the neuronal SNARE complex. (a) Domain structure of syntaxin-1A. NT, N-terminal sequence; Habc, Habc domain; TMR, trans-membrane regions. (b) Gel filtration profiles on Superdex S200 (10/300GL) of isolated Munc18-1 (black trace), purified SNARE complex2–253 (blue trace), and an equimolar mixture of Munc18-1 with purified SNARE complexes containing syntaxin-1A2–253 (red trace; ∗, shoulder due to free Munc18-1) or syntaxin-1A10–253 (green trace) are shown. The retention volumes of molecular mass standards are indicated at the top. (c and d) 1D 13C-edited 1H NMR spectra of 2 μM 13C-labeled Munc18-1 in the absence or presence of increasing concentrations of SNARE complex containing syntaxin-1A2–253 (c) and in the absence or presence of 2.5 μM BSA (BSA) or 5 μM SNARE complex containing syntaxin-1A10–253 (d). Concentrations of the control protein (BSA) or SNARE complexes used in the 1D NMR binding experiments are indicated below the spectra.
SM proteins are composed of a conserved ≈600-aa sequence that folds into an arch-shaped structure containing three domains (for Munc18-1; see refs. 8 and 9). In all membrane fusion reactions, SM proteins are as essential as SNARE proteins; in fact, at the vertebrate synapse, deletion of Munc18-1 has more severe consequences than deletion of either synaptobrevin or SNAP-25 (10–12). The first clue to the function of SM proteins was obtained from the observation that Munc18-1 directly binds to syntaxin-1A (13–15), but this function has remained enigmatic, in part because of the diversity of SM protein/SNARE interactions that have been described. Specifically, four types of interactions were observed: (i) Munc18-1 binds tightly to the closed conformation of syntaxin-1A that is incompatible with SNARE complex formation (7, 16); Munc18-1 was also reported to bind to the syntaxin-1/SNAP-25 heterodimer and to be released when synaptobrevin-binding completes SNARE complex formation (17). (ii) Sec1p binds to assembled SNARE complexes but not to the isolated syntaxin-1 homologues Sso1/2p (18, 19). (iii) Sly1, Vps45, and Munc18c bind to short N-terminal sequences in their cognate syntaxins, an interaction that is compatible with either a closed conformation of syntaxin outside of a SNARE complex or an open conformation within a SNARE complex (20–26). (iv) Vps33 is part of the large HOPS complex that interacts with assembled SNARE complexes, at least in part by binding to the N-terminal PX domain of the SNARE protein Vam7p (27, 28).
The different modes of SM protein/SNARE interactions may reflect different regulatory requirements of membrane traffic in different systems. However, the requirement for both protein families in intracellular membrane traffic suggests a common mechanism of SM protein/SNARE coupling that underlies the key function of SM proteins in membrane traffic. Previous studies suggested that, among SM proteins, only Munc18-1 cannot bind to assembled SNARE complexes (e.g., see ref. 16). However, membrane fusion may generally involve binding of SM proteins to assembled SNARE complexes (29), and such binding may have been missed for Munc18-1 in previous studies because it was masked by the extremely high affinity of the Munc18-1/syntaxin-1 binary interaction. We therefore have now reinvestigated whether Munc18-1 directly binds to SNARE complexes, and we show that Munc18-1 indeed interacts directly with the assembled neuronal SNARE complex in addition to isolated syntaxin-1 in the closed conformation. Together with increasing evidence for SM protein/SNARE complex interactions in other systems (18, 19, 24, 25, 30), our data strongly suggest that all SM proteins may be united by binding directly or indirectly to assembled SNARE complexes.
Results
Munc18-1 Coelutes with the Neuronal SNARE Complex in Gel Filtration.
To test the possibility that Munc18-1 might bind to the neuronal SNARE complex, we used methods that detect interactions in solution, do not require tags, and are not as prone to artifacts as GST pull-downs. Gel filtration of purified neuronal SNARE complexes containing the almost complete cytoplasmic region of syntaxin-1A (syntaxin-1A2–253) and the SNARE motifs of SNAP-25 and synaptobrevin revealed that the SNARE complex elutes considerably earlier than expected for its molecular mass (56.3 kDa) (Fig. 1b). This elution behavior is probably due to the extended conformation of the complex, where the N-terminal region of syntaxin-1A containing the Habc domain (5.5 nm long) (6) is flexibly linked to the elongated four-helix bundle formed by the SNARE motifs (10.5 nm long) (5) (see the NMR analysis below). Munc18-1, on the other hand, elutes close to its expected molecular weight, consistent with its compact structure (Fig. 1b). Strikingly, addition of stoichiometric amounts of the SNARE complex induced a marked shift in the elution volume of Munc18-1, showing that Munc18-1 interacts with the SNARE complex with a relatively high affinity. Note that these experiments used purified assembled SNARE complexes to exclude free syntaxin-1A2–253 and that the UV absorption was dominated by Munc18-1 because its extinction coefficient is considerably larger than that of the SNARE complex (Fig. 1b).
Because the N-terminal sequence of various syntaxins has been previously implicated in interactions with SM proteins that are compatible with SNARE complex formation (20, 21), we examined the role of the syntaxin-1A N-terminal sequence in the Munc18-1/SNARE complex interaction. Remarkably, SNARE complexes with a syntaxin-1A fragment containing a short N-terminal truncation (syntaxin-1A10–253) did not produce a significant shift in the mobility of Munc18-1 in gel filtration, showing that the syntaxin-1A N-terminal sequence is critical for binding of Munc18-1 to the SNARE complex (Fig. 1b). In agreement with the previous notion that the N-terminal region of syntaxin-1A is not sufficient for Munc18-1 binding, we did not observe tight binding during gel filtration between Munc18-1 and a syntaxin-1A fragment containing its N-terminal region but not its SNARE motif (syntaxin-1A1–180; data not shown). These results indicate that both the N-terminal sequence of syntaxin-1A and the four-helical bundle formed by the SNARE motifs of syntaxin-1A, synaptobrevin, and SNAP-25 contribute to the Munc18-1/SNARE complex interaction. This notion correlates with the observation that the elution volume of the SNARE complex decreases only slightly upon Munc18-1 binding (Fig. 1b), suggesting that binding of both the syntaxin-1A N-terminal region and the SNARE four-helix bundle to Munc18-1 results in a compact structure with its longest dimension still dictated by the elongated SNARE four-helical bundle. This conclusion was further supported by dynamic light scattering experiments, which showed that the Munc18-1/SNARE complex “hypercomplex” is monodisperse and revealed only a slight increase in the apparent radius of the SNARE complex upon Munc18-1 binding (from 5.0 to 5.3 nm) [supporting information (SI) Fig. 5].
Confirmation of the Munc18-1/SNARE Complex Interaction by 1D NMR Spectroscopy.
1D 13C-edited 1H NMR spectroscopy measures binding by monitoring the intensity of the strongest methyl resonance of a 13C-labeled protein (SMRC), which can be observed with high sensitivity at low micromolar concentrations (31). Binding of an unlabeled protein causes a decrease in the SMRC intensity of the 13C-labeled protein because of the resonance broadening that results from the increase in molecular weight associated with complex formation; in contrast, no changes in SMRC intensity are expected if there is no binding. As shown in Fig. 1c, addition of increasing amounts of unlabeled purified SNARE complex containing syntaxin-1A2–253 to 13C-labeled Munc18-1 (2 μM) led to a progressive decrease of its SMRC intensity. In contrast, addition of BSA (66 kDa) used as a negative control had no effect (Fig. 1d). Direct SDS/PAGE analysis of the NMR sample containing 2.5 μM SNARE complex confirmed that the SNARE complex was fully assembled and that free syntaxin-1A was barely detectable (SI Fig. 6). Curve fitting of the decrease in the SMRC intensity of 13C-labeled Munc18-1 as a function of the SNARE complex concentration indicated a 1:1 stoichiometry with an estimated KD of 99 nM. Attempts to measure the affinity more accurately at lower 13C-labeled Munc18-1 concentrations yielded more noisy data, but the estimated KD values (100–300 nM) confirmed that Munc18-1 binds to the SNARE complex with a submicromolar affinity that is ≈10-fold lower than that of the Munc18-1/syntaxin-1 heterodimeric complex (data not shown). In additional experiments, addition of 5 μM unlabeled SNARE complex containing the N-terminally truncated syntaxin-1A fragment (syntaxin-1A10–253) to 2 μM 13C-labeled Munc18-1 barely decreased its SMRC intensity (Fig. 1d), confirming the importance of the syntaxin-1A N-terminal sequence for Munc18-1 binding.
Chemical Cross-Linking of Purified Munc18-1 to Assembled SNARE Complexes.
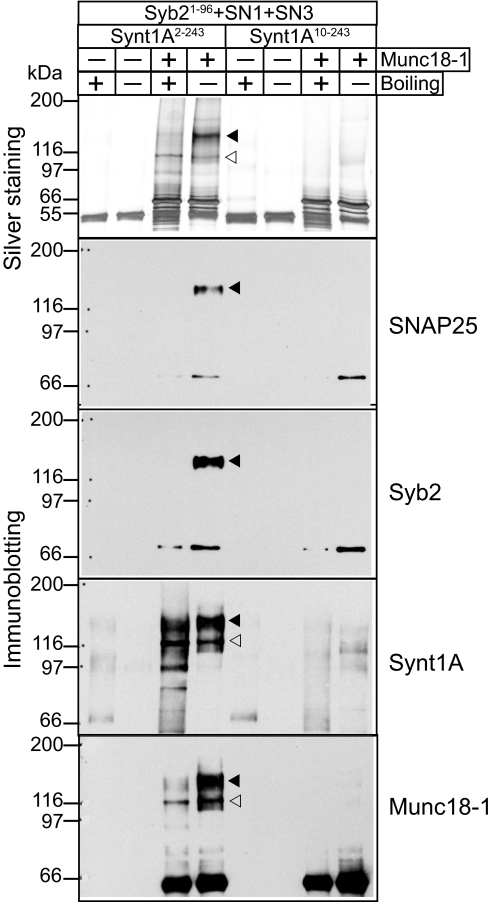
Chemical cross-linking of samples containing Munc18-1 and assembled SNARE complexes with the syntaxin-1A2–243 fragment revealed a major band containing SNAP-25 and synaptobrevin in addition to Munc18-1 and syntaxin-1A (Fig. 2). This band was sensitive to boiling, indicating that the SNARE proteins present are at least in part associated in a non-cross-linked manner. A smaller band (open arrowhead in Fig. 2) also contained Munc18-1 and syntaxin-1A, but not synaptobrevin and SNAP-25, and was insensitive to boiling, suggesting that it is composed of the syntaxin-1A/Munc18-1 complex, which cross-links very efficiently. Importantly, no cross-linking was observed in parallel experiments with a SNARE complex containing an N-terminally truncated syntaxin-1A10–243 fragment (Fig. 2).
Fig. 2.
Cross-linking analysis of the SNARE/Munc18-1 hypercomplex. Proteins were cross-linked by EDC and analyzed by SDS/PAGE with or without boiling. The top panel shows a silver-stained SDS/PAGE gel. The remaining panels show immunoblotting of the same samples with various Munc18-1 and SNARE protein antibodies. Filled arrowheads identify the positions of the SNARE/Munc18-1 hypercomplexes, and open arrowheads identify the positions of the Munc18-1/syntaxin-1A complexes. Syb2, synaptobrevin 2; synt1A, syntaxin-1A
Analysis of the Munc18-1/SNARE Complex Interaction by 2D NMR Spectroscopy.
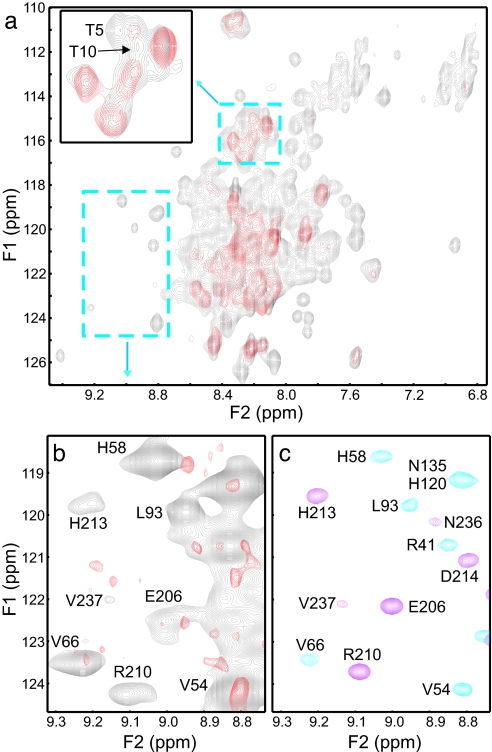
To further explore the Munc18-1/SNARE complex binding mode, we used 1H-15N heteronuclear single quantum correlation (HSQC) spectra enhanced by transverse relaxation optimized spectroscopy (TROSY). These experiments were performed with a 70 μM sample of a SNARE complex containing 2H,15N-labeled syntaxin-1A2–243 in the absence and presence of unlabeled Munc18-1 (Fig. 3). The spectrum of the isolated SNARE complex (Fig. 3a, gray contours) exhibits severe overlap in the center because it includes cross-peaks from the Habc domain and SNARE motifs of syntaxin-1A (which are highly helical and have few aromatic residues), from its flexible N-terminal sequence (residues 2–27), and from the likely flexible linker between the Habc domain and the SNARE motif (residues 145–188). Despite this overlap, cross-peaks corresponding to the Habc domain and the SNARE motif could be unambiguously assigned in well resolved regions of the spectrum (e.g., Fig. 3b, gray contours) by comparison with a 1H-15N HSQC spectrum of the syntaxin-1A N-terminal region (residues 1–180) (Fig. 3c, blue contours), and a 1H-15N TROSY-HSQC spectrum of a “minimal SNARE complex” corresponding to the four-helix bundle in which the syntaxin-1A SNARE motif was 2H,15N-labeled (Fig. 3c, magenta contours); both of these spectra were assigned previously (6, 32). Note that the cross-peaks from the Habc domain in the SNARE complex are much stronger than those from the SNARE motif (Fig. 3b, gray contours) because of the highly elongated nature of the four-helix bundle. These differential intensities show that the Habc domain and the four-helix bundle are indeed flexibly linked.
Fig. 3.
Heteronuclear NMR analysis of Munc18-1 binding to the SNARE complex. (a) Superposition of 1H-15N TROSY-HSQC spectra of 70 μM SNARE complex containing 2H,15N-labeled syntaxin-1A2–243 in the absence (gray contours) and presence (red contours) of stoichiometric amounts of unlabeled Munc18-1 at 28°C. (Inset) Expansion of the region denoted by the small dashed rectangle that includes cross-peaks corresponding to the two N-terminal threonine residues of syntaxin-1A (T5 and T10). (b) Superposition of the expansion from a well resolved region of the spectra denoted by the dashed rectangle on the lower left corner of a. Note that the spectra were plotted at different contour levels to emphasize distinct features (e.g., spectra in b were plotted at lower contour levels than in a to reveal all observable cross-peaks in this region). (c) Superposition of the same expansions of a 1H-15N HSQC spectrum of syntaxin2–180 (blue contours) at 28°C and a 1H-15N TROSY-HSQC spectrum of a minimal SNARE complex containing 2H,15N-labeled syntaxin SNARE motif (residues 191–253, magenta contours) at 32°C, which were used to assign the cross-peaks in a and b.
Addition of Munc18-1 led to severe broadening of most well resolved cross-peaks (Fig. 3 a and b, red contours). Although attempts to improve the quality of the spectra to precisely map the syntaxin-1A residues involved in binding by increasing the protein concentration were hampered by sample precipitation, our 1H-15N TROSY-HSQC data show that both the Habc domain and the four-helix bundle participate in Munc18-1 binding. Note that, because both structural elements are flexibly linked, Munc18-1 binding to only one of these elements is not expected to perturb the other element so severely. Furthermore, Munc18-1 binding also broadened beyond detection the cross-peaks from Thr-5 and Thr-10 of the syntaxin-1A N-terminal sequence (Fig. 3a Inset), confirming the involvement of this sequence in Munc18-1 binding to the SNARE complex. The picture that emerges from these results is that Munc18-1 binds to the SNARE complex in a multivalent interaction that involves the N-terminal sequence of syntaxin-1A, its Habc domain, and the SNARE four-helix bundle (see model in Fig. 4). This multivalency likely underlies the compact nature of the Munc18-1/SNARE complex hypercomplex indicated by our gel filtration and dynamic light scattering data.
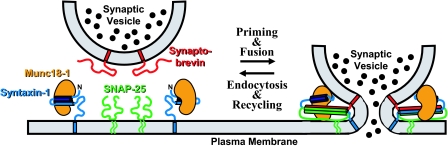
Fig. 4.
Model for the interactions of Munc18-1 with synaptic SNARE proteins. The diagram on the left exhibits the binding of Munc18-1 to the closed conformation of syntaxin-1 that is thought to operate before priming and fusion of synaptic vesicles, although this point has not been demonstrated. The diagram on the right displays the binding of Munc18-1 to assembled SNARE complexes during the fusion reaction, which may prevent diffusion of SNARE complexes into the space between the membranes or actively pull the SNARE complexes apart to open the fusion pore. Munc18-1 is shown contacting the membranes through two regions at opposite ends of the central cavity because these regions contains abundant basic residues, but diffusion of the SNARE four-helix bundle to the middle could also be hindered by steric and/or electrostatic repulsion with the membranes if Munc18-1 adopts a different orientation.
Discussion
In eukaryotic cells, SNAREs and SM proteins are conserved components of the membrane fusion machinery. Previous studies suggested that SNAREs and SM proteins interact by diverse mechanisms: Munc18-1 (and likely Munc18-2) was shown to bind only to the closed conformation of syntaxin outside of the SNARE complex (7, 8, 16); Sly1, Vps45, and Munc18c were found to bind to the N-terminal sequences of their respective cognate syntaxins independent of whether they are in SNARE complexes (20, 21, 25, 26, 33); yeast Sec1p was observed to bind only to assembled SNARE complexes without participation of the N-terminal sequence of syntaxins Sso1/2p (18, 19); and Vps33p was found to bind to SNARE complexes by interacting with the PX domain of Vam7p (27, 28). Faced with this diversity of interactions and the fact that, among SM proteins, only Munc18-1 did not bind to assembled SNARE complexes, we have reinvestigated how Munc18-1 interacts with SNARE proteins.
Our results now show that, contrary to previous notions (7, 8, 16), Munc18-1 binds to fully assembled neuronal SNARE complexes in addition to syntaxin-1 alone. Binding of Munc18-1 to the SNARE complex is stable during gel filtration (Fig. 1a) and was independently confirmed by 1D and 2D NMR spectroscopy (Figs. 1 c and d and 3) and by chemical cross-linking (Fig. 2). Furthermore, we show that the Munc18-1/SNARE complex interaction involves the N-terminal sequence of syntaxin-1, its Habc domain, and the four-helix bundle of the SNARE complex (see model of Fig. 4). Hence, our data establish that Munc18-1 engages in two distinct interaction modes with SNARE proteins: the well characterized binding to the closed conformation of syntaxin-1 (7, 8) and the novel binding to assembled SNARE complexes. The latter interaction was probably overlooked in previous studies because it depends on a free but intact N terminus of syntaxin-1, and most studies used either truncated or N-terminally fused syntaxin-1 (e.g., see ref. 16). The dual Munc18-1/SNARE binding mode emerging from our results suggests that, during synaptic vesicle exocytosis, Munc18-1 first binds to closed syntaxin-1 in an interaction that may help organizing the assembly of SNARE complexes, but at the same time may impose an energy barrier to SNARE complex formation. SNARE complex assembly on Munc18-1 then leads to formation of the hypercomplex containing Munc18-1 and the SNARE complex by a mechanism that requires opening of syntaxin-1 and is likely assisted by other factors such as Munc13 (34–36).
General Features of the SM Protein Interaction with SNARE Proteins.
Because Munc18-1 was the only SM protein that was thought to be unable to bind to SNARE complexes, the current finding that Munc18-1 in fact does bind to such complexes suggests that SM proteins are united by a common interaction, directly or indirectly, with SNARE complexes. However, only for Sec1p in yeast (18) and for Munc18-1 (Fig. 3) was there any demonstrated contact between the SM protein and the four-helix bundle of the assembled SNARE complex, and it is thus unclear whether such a binding mode is general. Among all SM proteins, Munc18-1 (and possibly Munc18-2) is unique in that it appears to interact with SNARE proteins in two distinct modes (see model in Fig. 4): the previously described binding to the closed conformation of syntaxin-1 (7, 8) and the binding to assembled SNARE complexes described here. Of these binding modes, the first mode appears to be an evolutionary specialization that may have emerged to accommodate the exquisite spatial and temporal requirements of neuronal exocytosis. Specifically, the extremely high levels of SNARE proteins in nerve terminals may necessitate an additional control of SNARE complex assembly, a control that could be provided by the binding of Munc18-1 to the closed conformation of syntaxin-1. Future studies are needed to examine whether this interaction is in fact functionally important and to test this hypothesis. In contrast, the second mode appears to be a general mechanism that mediates fusion and is equivalent to many other SM protein/SNARE interactions.
Function of Munc18-1 and Other SM Proteins in Fusion.
The shared binding, directly or indirectly, of all SM proteins to assembled SNARE complexes is likely associated with a common function in membrane fusion. Munc18-1 was initially proposed to have an active role in fusion as an essential part of a hypothetical fusion machinery (13, 37), a notion that is attractive considering the large size and evolutionary conservation of SM proteins. Conversely, overexpression experiments suggested that Munc18-1 may act as an inhibitor of exocytosis (38), but a role merely as an inhibitor was ruled out by the total abrogation of neurotransmitter release observed in Munc18-1-deficient flies and mice (10, 39). Another hypothesis posits that Munc18-1 and other SM proteins serve as platforms for the assembly of cognate SNARE complexes (reviewed in ref. 37). Because SNARE complexes can assemble in both parallel and antiparallel fashions (40), such a role could ensure the proper orientation and specificity in SNARE complex formation. However, a role for SM proteins in providing SNARE complex specificity is difficult to reconcile with the relatively small number of SM proteins in yeast (four) and vertebrates (seven). A specific hypothesis for how Munc18-1 may play an active role in fusion (a hypothesis that motivated in part the present study) suggests that binding of the bulky Munc18-1 to assembling SNARE complexes may prevent diffusion of the thin, elongated, four-helix bundle to the center of the space between the membranes, where they could inhibit fusion (29). However, the precise geometry and dynamics of assembling SNARE complexes in membranes that are about to fuse remain to be established. Regardless of these possibilities, it seems likely that the active role of Munc18-1 in release is associated with its interaction with the SNARE complex uncovered here.
Materials and Methods
Plasmids and Recombinant Proteins.
Constructs for bacterial expression of synaptobrevin and SNAP-25 recombinant fragments were described previously (32). Constructs for bacterial expression of syntaxin-1A fragments were generated by standard PCR-based procedures and subcloned into pGEX-KT (41) vector. Recombinant proteins were expressed as GST fusions in bacteria and purified essentially as described (7). After thrombin cleavage to release the recombinant proteins from the GST moiety, all syntaxin-1A recombinant fragments have two additional N-terminal residues (Gly-Ser) derived from the vector. Recombinant full-length rat Munc18-1 was expressed in bacteria as a GST fusion protein, affinity-purified on glutathione-Sepharose beads, cleaved on beads with thrombin, eluted in a buffer containing 50 mM Tris·HCl (pH 8.0)/200 mM NaCl/2 mM DTT/1 mM EDTA/1 mM EGTA/1 mM AEBSF/Sigma Protease Inhibitors mixture/0.3% CHAPS, and further purified by size-exclusion chromatography on a S200 Superdex column (Amersham Biosciences, Pittsburgh, PA) in a buffer suitable for the following gel filtration or NMR experiment.
Assembly and Purification of SNARE Complexes.
SNARE complexes were assembled from highly purified recombinant fragments of rat synaptobrevin 2 (residues 1–96), rat syntaxin-1A (residues 2–253 or 10–253), and human SNAP-25 (residues 11–82 and 96–111) by overnight incubation at 32°C in 50 mM Tris·HCl (pH 7.4)/150 mM NaCl/2 mM TCEP/1 mM EDTA/1 mM EGTA/1 mM AEBSF/Sigma Protease Inhibitor mixture using ≈40–50 μM syntaxin and an ≈1.5 molar excess of all other SNAREs. The efficiency of SNARE complex formation was ≈85–90% as judged by nonreducing SDS/PAGE. The assembled SNARE complexes were purified from unassembled fragments by size-exclusion chromatography on S200 Superdex column (Amersham Biosciences).
Gel Filtration Experiments.
Purified SNARE complexes and Munc18-1 were incubated singly or together in a 200-μl volume at 4 μM in 40 mM Tris·HCl (pH 7.3)/100 mM NaCl/2 mM TCEP. Samples were cleared by centrifugation at 16,000 × g at 4°C and applied to a Superdex S200 (10/300GL) column (Amersham Biosciences).
Cross-Linking Experiments.
Cross-linking was performed with highly purified recombinant proteins at low micromolar concentrations at approximately equimolar ratios in 150 mM NaCl/25 mM Hepes (pH 7.6)/1 mM EDTA/protease inhibitors. All samples were treated with 5 mM EDC (Pierce, Rockford, IL) for 2 h on ice. Excess of cross-linker was quenched by adding 2-mercaptoethanol. Samples were analyzed by SDS/PAGE and immunoblotting that were performed by using standard protocols.
NMR Spectroscopy.
Proteins were uniformly labeled by growing bacteria in M9 minimal medium containing 15NH4Cl (CIL) as the sole nitrogen source (for 15N-labeling) or 13C6-glucose as the sole carbon source (for 13C-labeling) with H2O or D2O as a solvent. For the experiments with full-length Munc18-1, syntaxin-1A2–243/Munc18-1 complexes were initially assembled at low protein concentration (<10 μM) and concentrated to a 0.2 mM final concentration by using 30,000 MNWL concentrator (Millipore, Billerica, MA). 1D 13C-edited 1H NMR spectra were measured on a Varian INOVA600 spectrometer at 25°C in 20 mM sodium phosphate (pH 7.1)/150 mM NaCl/2 mM DTT, acquiring the first trace of a 1H-13C HSQC spectrum (1,500 scans; 30-min total acquisition time). 2D 1H-15N HSQC spectra of SNARE complexes were acquired at 28°C on a Varian INOVA600 spectrometer equipped with a cold probe (total acquisition times 2–24 h) except for the spectrum of the minimal SNARE complex that was described previously (32). All data were processed with NMRPipe (42) and analyzed with NMRView (43).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant NS37200 (to J.R.).
Abbreviations
- HSQC
heteronuclear single quantum correlation
- TROSY
transverse relaxation optimized spectroscopy.
Note Added in Proof.
After this paper was accepted for publication, Shen et al. (44) also reported findings that Munc18-1 binds to the neuronal SNARE complex.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611318104/DC1.
References
- 1.Palade G. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 2.Brunger AT. Q Rev Biophys. 2005:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R, Scheller RH. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Südhof TC. Nat Rev Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 5.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 7.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Südhof TC, Rizo J. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misura KM, Scheller RH, Weis WI. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 9.Bracher A, Weissenhorn W. J Mol Biol. 2001;306:7–13. doi: 10.1006/jmbi.2000.4347. [DOI] [PubMed] [Google Scholar]
- 10.Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, et al. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 11.Schoch S, Deak F, Konigstorfer A, Mozhayeva M, Sara Y, Südhof TC, Kavalali ET. Science. 2001;294:1117–1122. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- 12.Washbourne P, Thompson PM, Carta M, Costa ET, Mathews JR, Lopez-Bendito G, Molnar Z, Becher MW, Valenzuela CF, Partridge LD, et al. Nat Neurosci. 2002;5:19–26. doi: 10.1038/nn783. [DOI] [PubMed] [Google Scholar]
- 13.Hata Y, Slaughter CA, Südhof TC. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 14.Garcia EP, Gatti E, Butler M, Burton J, De Camilli P. Proc Natl Acad Sci USA. 1994;91:2003–2007. doi: 10.1073/pnas.91.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pevsner J, Hsu SC, Scheller RH. Proc Natl Acad Sci USA. 1994;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B, Steegmaier M, Gonzalez LC, Jr, Scheller RH. J Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilly FE, Sorensen JB, Jahn R, Lang T. PLoS Biol. 2006;4:e330. doi: 10.1371/journal.pbio.0040330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Proc Natl Acad Sci USA. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Südhof TC. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 21.Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Südhof TC, Rizo J. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols BJ, Holthuis JC, Pelham HR. Eur J Cell Biol. 1998;77:263–268. doi: 10.1016/s0171-9335(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 23.Kosodo Y, Noda Y, Yoda K. Biochem Biophys Res Commun. 1998;250:212–216. doi: 10.1006/bbrc.1998.9288. [DOI] [PubMed] [Google Scholar]
- 24.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latham CF, Lopez JA, Hu SH, Gee CL, Westbury E, Blair DH, Armishaw CJ, Alewood PF, Bryant NJ, James DE, et al. Traffic. 2006;7:1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- 26.Dulubova I, Yamaguchi T, Arac D, Li H, Huryeva I, Min SW, Rizo J, Sudhof TC. Proc Natl Acad Sci USA. 2003;100:32–37. doi: 10.1073/pnas.232701299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins KM, Thorngren NL, Fratti RA, Wickner WT. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroupe C, Collins KM, Fratti RA, Wickner W. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizo J, Chen X, Arac D. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Peng R, Gallwitz D. J Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arac D, Murphy T, Rizo J. Biochemistry. 2003;42:2774–2780. doi: 10.1021/bi0272050. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Südhof TC, Rizo J. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 33.Bracher A, Weissenhorn W. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augustin I, Rosenmund C, Südhof TC, Brose N. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 35.Richmond JE, Weimer RM, Jorgensen EM. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn R, Lang T, Südhof TC. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 38.Wu MN, Littleton JT, Bhat MA, Prokop A, Bellen HJ. EMBO J. 1998;17:127–139. doi: 10.1093/emboj/17.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison SD, Broadie K, van de Goor J, Rubin GM. Neuron. 1994;13:555–566. doi: 10.1016/0896-6273(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 40.Weninger K, Bowen ME, Chu S, Brunger AT. Proc Natl Acad Sci USA. 2003;100:14800–14805. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hakes DJ, Dixon JE. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA, Blevins RA. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 44.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.