Abstract
A subgroup of genes induced by IFN-γ requires both STAT1 and IRF1 for transcriptional activation. Using WT, stat1−/−, or irf1−/− cells, we analyzed the changes induced by IFN-γ in gbp2 promoter chromatin. STAT1 associated with the promoter independently of IRF1 and played an essential role in the ordered recruitment of the coactivator/histone acetyl transferase CREB-binding protein (CBP) and the histone deacetylase HDAC1. Hyperacetylation of histone 4 also required STAT1. Phosphorylation at S727 in the transactivating domain increased transcriptional activity of STAT1. In cells expressing a STAT1S727A-mutant CBP recruitment, histone 4 hyperacetylation and RNA polymerase II association with the gbp2 promoter were strongly reduced. IRF1 association with the gbp2 promoter followed that of STAT1, but STAT1 association with DNA or histone hyperacetylation were not necessary for IRF1 binding. RNA polymerase II association with the gbp2 promoter required both STAT1 and IRF1, suggesting that both proteins mediate essential steps in transcriptional activation. IRF1, but not STAT1, was found to coimmunoprecipitate with RNA polymerase II. Together, the data support the assumption that the main role of STAT1 in activating gbp2 transcription is to provide transcriptionally competent chromatin, whereas the function of IRF1 may lie in directly contacting RNA polymerase II-containing transcriptional complexes.
Keywords: chromatin, interferon, signal transduction, interferon regulatory factor
IFN-γ enhances cell-mediated immunity against both nonviral pathogens and viruses (1). STAT1, the central mediator of IFN-γ-induced gene expression is phosphorylated at Y701 by the IFN-γ receptor-associated Janus kinases Jak1 and Jak2, an essential prerequisite for dimerization and nuclear translocation (2). In addition, a serine/threonine kinase phosphorylates the STAT1 transactivating domain at S727 and increases transcriptional competence (3–5). Promoter sequences found in IFN response regions are the γ-IFN activated site (6) recognized by STAT1 dimers and the IFN-stimulated response element (ISRE) (7). ISRE sequences bind STAT complexes and also IFN regulatory factors (IRFs) (8, 9). In the context of the IFN-γ response, the ISRE mediates transcriptional effects of IRF1 and noncanonical STAT1 complexes, e.g., STAT1 dimers associated with IRF9 (10–12).
STAT1 has been linked predominantly to positive gene regulation, but some genes are repressed by STAT1 (13–15). Many genes stimulated by STAT1 in the context of an IFN-γ response require cooperative effects with other transcription factors, such as IRF-1, USF-1, SP1, or C/EBPβ (16). In most cases both STAT1 and the cooperating transcription factor bind to their cognate promoter sequences, although the cooperation with C/EBPβ is mediated by a sequence designated GATE, which binds C/EBPβ but not STAT1 (17).
The gbp1 and gbp2 genes are IFN-γ-inducible members of the p65 GTPase gene family with putative roles in the resistance to intracellular pathogens (18). gbp2 transcription in both humans and mice requires promoter binding sites for both STAT1 dimers and IRF transcription factors (6, 19). Guanylate-binding protein (GBP) expression in response to IFN-γ is virtually absent in cells from irf1 knockout mice (10, 11), The promoter of the irf1 gene contains a binding site for STAT1 dimers. Therefore, IRF1 accumulates in cells treated with IFN-γ (20).
Transcriptional activation of the gbp genes is accompanied by promoter acetylation (5). Consistently, STAT1 interacts with the coactivator/histone acetyl transferase (HAT) CREB-binding protein (CBP) that is required for STAT1-dependent transcription of chromatin templates in vitro (5, 21, 22). Moreover, microarray analysis of HDAC1-deficient cells identified gbp genes as belonging within a group of genes requiring histone deacetylase 1 (HDAC1) for IFN-γ-induced expression (23). STAT1 also directly binds a complex of MCM proteins that enhance gbp transcription most likely by providing helicase activity for strand separation in the initiation and elongation steps (24). BRG1, an ATPase subunit of the SWI/SNF chromatin remodeling complex, binds the human gbp promoter as a prerequisite for the association of STAT1 (25, 26).
Serving as a paradigm for the group of genes coregulated by STAT1 and IRF1, the gbp2 gene allows us to address two important open questions. (i) What is the relative importance of STAT1 for IFN-γ-induced gbp2 transcription as a transcriptional activator of the irf1 gene on the one hand and as a cognate binding factor of the gbp2 promoter on the other? (ii) What is the nature of the molecular mechanisms mediating cooperative stimulation of gbp transcription by STAT1 and IRF1? Our studies show that both STAT1 and IRF1 are required at the gbp2 promoter to recruit RNA polymerase II (RNA pol II) to the transcription initiation site. STAT1 and its phosphorylation at S727 are essential for CBP recruitment, and STAT1 also mediates the association of the promoter with HDAC1. By contrast, the binding of IRF1 to the gbp2 promoter occurs independently of STAT1 binding and histone acetylation, but it cooperates with STAT1's activities in recruiting RNA pol II.
Results
Organization of the Murine gbp1/gbp2 Promoters and Regulation of Their Activity by STAT1 and IRF1.
A cluster of five GBP genes maps to mouse chromosome 3 (www.ensembl.org). Previously, three groups described murine GBP promoters. The first report assigned the cloned promoter to the gbp1 gene (27). Two further groups subsequently cloned the same stretch of DNA and a further one differing by only a few base pairs but with an identical IFN response region (19, 28). Those two groups concurred in their interpretation that the cloned DNAs contained highly homologous promoters of the gbp1 and gbp2 genes. The ensembl database shows the gbp1 and gbp2 genes juxtaposed and in the same orientation, spaced by a short intergenic region of 1,466 bp. The previously described promoters are all highly homologous to the intergenic region containing the gbp2 promoter, but none shows significant homology to the region upstream of gbp1, suggesting that all of them represent allelic variations of the gbp2 promoter (see Fig. 2A). Quantitative real-time PCR analysis showed that the gbp1 and gbp2 genes are strictly coregulated, as would be expected from the use of common promoter elements (Fig. 1A). Both showed an identical requirement for the presence of STAT1 and IRF1, with the need for STAT1 being more stringent than that for IRF1. Inspection of the gbp1 upstream sequence revealed the presence of bona fide IFN-γ-activated site (GAS) and ISRE sequences at positions −253/−245 (TTCATAGAA) and −139/−127 (AATTTCACTTTCT), respectively. These gbp1 upstream sequences most likely constitute an IFN-γ response region, but functional analysis will be required to ascertain this assumption.
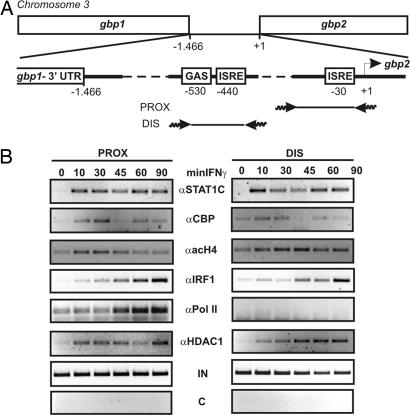
Fig. 2.
Recruitment of transcriptional regulators to the gbp2 promoter chromatin. (A) Graphic representation of the murine gbp2 promoter region. gbp1 and gbp2 are located on chromosome 3 and are separated by 1.466 bp of intergenic region. This region contains the regulatory GAS and ISRE elements. Primer pairs used for ChIP assays are depicted. (B) Recruitment of STAT1 (αS1C), IRF1 (αIRF1), RNA pol II (αPol II), CBP (αCBP), and HDAC1 (αHDAC1) to the distal (DIS) and proximal (PROX) regions of the gbp2 promoter, and hyperacetylation of histone 4 (αacH4) of the respective promoter regions, as analyzed by ChIP. Primary BMDM were treated with 10 ng/ml (240 units/ml) IFN-γ for the indicated time points (minIFN-γ), and formaldehyde-cross-linked chromatin was isolated and subjected to IP with the indicated Abs. The promoter elements were analyzed by amplification of the distal and proximal gbp2 promoter regions by PCR. The specificity for the IP was determined by using preimmune serum (C) as negative control and amplification of input DNA (IN) by PCR.
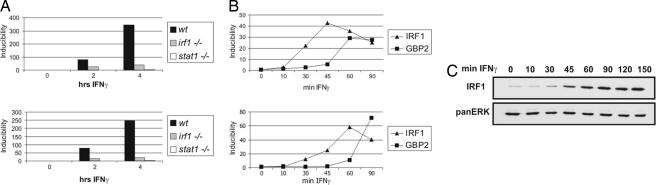
Fig. 1.
Regulation of the gbp1 and gbp2 genes by IFN-γ dependence on the presence of STAT1 and IRF1. (A) gbp1 (Upper) and gbp2 (Lower) expression in irf1- and stat1-deficient fibroblasts. Immortalized WT, irf1−/−, and stat1−/− fibroblasts were treated with 10 ng/ml (240 units/ml) IFN-γ for the indicated times and analyzed for expression of gbp1 and gbp2 by real-time PCR. Inducibility was calculated after normalizing to GAPDH mRNA levels. (B) Kinetics of irf1 and gbp2 expression. Primary BMDMs were treated with 10 ng/ml (240 units/ml) IFN-γ for the indicated times. Nuclear RNA (hnRNA), isolated from purified nuclei, and mRNA from whole-cell extracts were isolated and reverse transcribed. Inducibility of irf1 and gbp2 hnRNA (Upper) and mRNA (Lower) expression were analyzed by real-time PCR and normalized to endogenous GAPDH. (C) Western blot analysis of IRF1 protein expression was performed with lysates from WT BMDM treated with 10 ng/ml (240 units/ml) IFN-γ for the indicated time. Equal loading was determined by probing the membrane with anti-panERK Abs.
The IFN-response region of the gbp2 promoter is divided into an ISRE proximal to the cap site and more distal GAS and ISRE sequences (Fig. 2A). Our recent ChIP experiments showed that the proximal gbp2 promoter ISRE associates with a noncanonical, IFN-γ-activated STAT1 complex (5). The distal GAS element is a canonical, although imperfect, binding site for STAT1 dimers (19).
To determine the temporal sequence of IRF1 accumulation and gbp2 expression, we determined IFN-γ-induced accumulation of irf1 nuclear RNA (hnRNA), mRNA (Fig. 1B), and protein (Fig. 1C). Unspliced irf1 hnRNA was already 50% maximal 30 min after addition of cytokine. The accumulation of cytoplasmic mRNA was delayed by ≈15 min and protein synthesis by yet another 15 min. Nuclear gbp2 mRNA closely correlated with amounts of IRF1 protein, and the delay between hnRNA synthesis and cytoplasmic mRNA accumulation was similar as in the case of irf1. The data thus confirm that gbp2 mRNA transcription is mostly a secondary response to IFN-γ and that in the presence of STAT1 dimers, IRF1 availability limits the rate of gbp2 nuclear RNA synthesis.
Recruitment of Transcriptional Regulators to the gbp2 Promoter.
To address the changes of gbp2 promoter chromatin after IFN-γ stimulation, we performed antibody (Ab)-mediated ChIP. Consistent with our previous results (5), STAT1 was recruited rapidly to the proximal and distal response elements (Fig. 2B). Recruitment of the HAT CBP and histone 4 hyperacetylation closely paralleled STAT1 binding. Recruitment of HDAC1 also paralleled that of STAT1. This finding is consistent with earlier reports showing that STAT1 interacts with these proteins when coimmunoprecipitated from cell extracts or in pull-down assays (29). IRF1 also bound both the proximal and distal promoter regions (Fig. 2B). Promoter binding was delayed by ≈20 min compared with STAT1. The kinetics of RNA pol II recruitment were similar to those of IRF1 binding, which in turn closely paralleled the accumulation of IRF1 protein and nuclear gbp2 mRNA (Fig. 1).
Further investigation of the gbp2 promoter was performed in gene-disrupted fibroblasts. The kinetics of gbp2 expression and transcription factor recruitment in this cell type were not significantly different from those observed in macrophages (data not shown). IFN-γ-induced STAT1 binding was virtually unaffected by the absence of IRF1 (Fig. 3A). Slightly reduced association particularly with the proximal site reflects reduced STAT1 expression in irf1−/− cells (Fig. 3G), which is most likely due to the role of IRF1 in maintaining STAT1 expression through autocrine type I IFN production (30). IRF1 binding to the gbp2 promoter was completely abolished in stat1−/− cells (data not shown) because of the complete lack of IFN-γ-induced IRF1 synthesis (Fig. 3G).
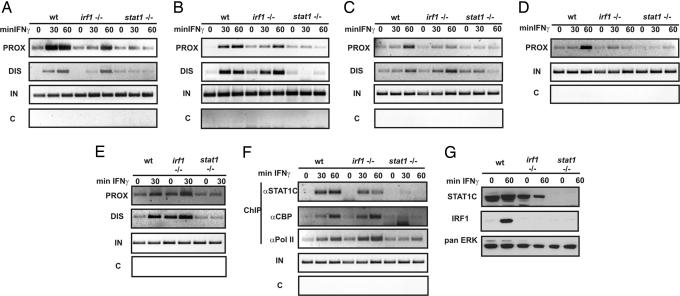
Fig. 3.
Impact of irf1 or stat1 deficiency on the IFN-γ-induced alterations of gbp2 promoter chromatin. (A–E) ChIP assays were performed with immortalized WT, stat1 (stat1−/−)-, and irf1 (irf1−/−)-deficient fibroblasts for binding of STAT1 (αS1C) (A), hyperacetylation of histone 4 (αacH4) (B), recruitment of CBP (αCBP) (C), RNA pol II (αPol II) (D), and HDAC1 (αHDAC1) (E) to the proximal (PROX) and distal (DIS) regions of the gbp2 promoter. The cells were treated for 30 or 60 min with 10 ng/ml (240 units/ml) IFN-γ, and formaldehyde-cross-linked chromatin was immunoprecipitated with the indicated Abs. (F) IFN-γ-dependent factor recruitment to the irf1 promoter. DNA isolated from A–E was subjected to PCR by using primers recognizing the irf1 promoter. (G) Western blot analysis of whole-cell extracts from WT, irf1-, and stat1-deficient fibroblast for protein expression of STAT1 and IRF1. The cells were treated for 60 min with 10 ng/ml (240 units/ml) IFN-γ or left untreated. The membrane was probed with a STAT1 C-terminal Ab, α-IRF1 Ab, and with anti-panERK Abs for equal loading.
STAT1 deficiency caused an almost complete absence of CBP and HDAC1 recruitment, histone 4 hyperacetylation, and RNA pol II binding (Fig. 3 B–F). By contrast, IRF1 deficiency had little impact on the association of HDAC1. CBP recruitment or the hyperacetylation of H4 particularly at the proximal promoter were reduced, but an IFN-γ-stimulated increase was clearly detectable. Strikingly, however, RNA pol II recruitment to the gbp2 cap site was highly dependent on the presence of IRF1. To demonstrate specificity of these findings, IFN-γ-dependent factor recruitment to the irf1 promoter was examined. Consistent with the lack of an IRF1-binding site in its IFN-γ-response region, CBP and, importantly, RNA pol II association were found to be unaffected by the absence of IRF1 protein (Fig. 3F). In contrast, neither protein was found to be associated with the irf1 promoter in absence of STAT1.
Effect of Mutating the S727 Phosphorylation Site in the STAT1 Transactivating Domain.
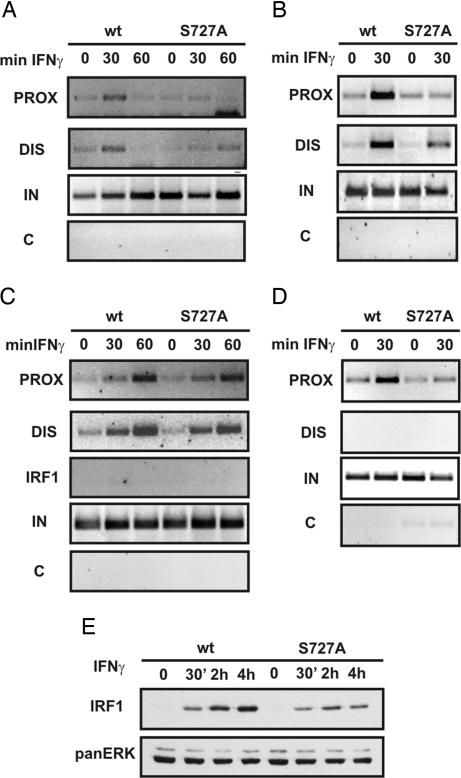
STAT1 S727 phosphorylation is essential for IFN-γ-induced, gbp2 promoter histone 4 hyperacetylation. Moreover, CBP does not efficiently bind STAT1S727A in IFN-γ-treated cells (5). Consistent with these earlier findings, the recruitment of CBP to gbp2 promoter chromatin was virtually absent in cells expressing a STAT1S727A phosphorylation site mutant, and absence of CBP coincided with strongly reduced H4 acetylation (Fig. 4 A and B). Accumulation of IRF1 protein in response to IFN-γ is reduced by >50% in cells expressing STAT1S727A (Fig. 4E). Despite this reduction and the lack of histone acetylation, particularly at the proximal gbp2 promoter, IRF1 association with the IFN response region was not very different from that found in cells expressing wild-type (WT) STAT1 (Fig. 4C). The specificity of IRF1 binding was confirmed by the lack of amplification of the irf1 promoter in the same ChIP DNA samples (Fig. 4C). The binding of RNA pol II was significantly decreased in IFN-γ-treated cells expressing STAT1S727A (Fig. 4D), which is in line with the strong effect of the STAT1S727A mutation on gbp transcription (5, 31).
Fig. 4.
Role of STAT1 S727 phosphorylation in the activation of the gbp2 promoter by IFN-γ. BMDM, obtained from WT and STAT1 S727A mice, were treated with 10 ng/ml (240 units/ml) IFN-γ for the indicated time points, and formaldehyde-cross-linked chromatin was isolated. (A–D) IP of sonicated fragments was performed overnight with polyclonal Abs against CBP (A), hyperacetylated histone 4 (acH4) (B), IRF1 (C), and RNA Pol II (D). (E) Western blot analysis of whole-cell extracts from WT and S727A macrophages. The cells were treated with IFN-γ and analyzed for IRF1 protein levels. Equal loading was determined by reprobing the membrane with anti-panERK Abs.
Analysis of gbp2 Expression and gbp2 Promoter Chromatin in Cells Expressing IRF1 in Absence of Stat1 Activity.
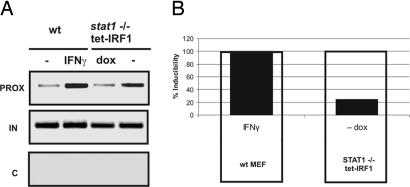
Irf1 being a STAT1-regulated gene, STAT1-independent effects of IRF1 on IFN-γ-regulated genes cannot be studied in stat1−/− cells. Therefore, we resorted to two different strategies to study IRF1 in the absence of STAT1 activity. First, a doxycyclin (dox)-repressed IRF1 gene was introduced into STAT1-deficient fibroblasts. ChIP analysis of IRF1 showed a strong increase of chromatin-associated IRF1 in transfected cells after dox withdrawal (Fig. 5A) and a concomitant expression of endogenous gbp2 mRNA. gbp2 expression caused by IRF1 alone was much lower than that noted in IFN-γ-treated WT cells (Fig. 5B). These results are consistent with previous reports (32) showing transcriptional effects of IRF1 overexpression. They demonstrate that IRF1 alone is able to bind gbp2 chromatin and stimulate target gene transcription but that expression is low compared with cytokine-treated cells. Therefore, the data also stress the important role of STAT1 dimer association with chromatin for gbp2 promoter activity. Similar conclusions could be drawn from experiments with a recently established line of murine fibroblasts expressing a fusion protein between IRF1 and the ligand-binding domain of the human estrogen receptor (IRF1-hER) (33, 34). In this cell line, IRF1 is constitutively expressed, but its transcription factor activity is strictly controlled by estrogen [see supporting information (SI) Fig. 7]. IRF1-ER bound the gbp2 promoter in absence of exogenous stimuli and binding was increased after treatment with estrogen, IFN-γ, or both. Induction of gbp2 expression by estrogen alone was low compared with that by IFN-γ.
Fig. 5.
Analysis of gbp2 promoter chromatin and of gbp2 in Stat1−/− cells. (A) STAT1-deficient fibroblasts were pretreated with dox for 6 h and then transiently transfected with pRETRO-tet-OFF-FLAG-IRF1. Twenty-four hours after transfection, dox was removed from the cells (−) or left on the cells (dox) for an additional 24 h. Control cells were treated with 10 ng/ml (240 units/ml) IFN-γ for 1 h. Formaldehyde-cross-linked chromatin was isolated and subjected to IP with IRF1 Abs. The proximal gbp2 promoter was analyzed by PCR. The specificity for the IP was determined by using preimmune serum (C) as negative control, and amplification of input DNA (IN) by PCR. (B) RNA was isolated, reverse-transcribed, and analyzed for endogenous gbp2 expression by real-time PCR.
IRF1, but Not STAT1, Is Found in Complexes with RNA Pol II in IFN-γ-Treated Cells.
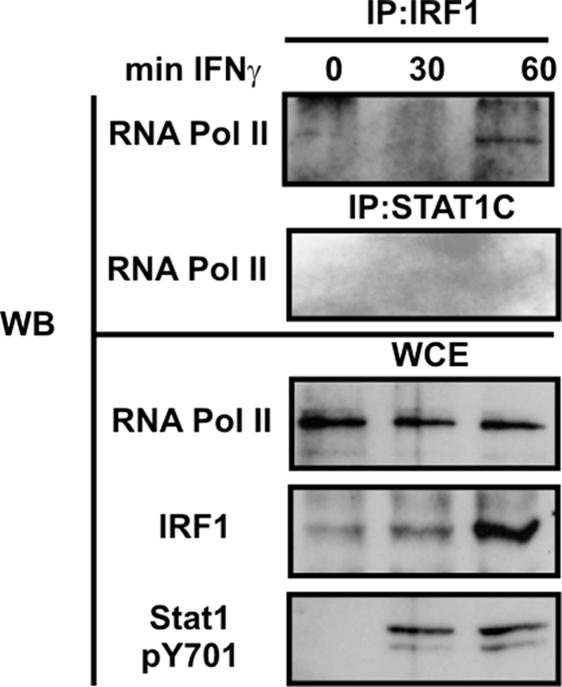
STAT1 being the major player in gbp2 promoter histone hyperacetylation, we tested whether the role of IRF1 might be to directly contact protein complexes containing RNA pol II. Extracts from IFN-γ-treated macrophages were precipitated with Abs to either IRF1 or STAT1, and the precipitates were analyzed by Western blot for the presence of RNA pol II. Fig. 6 shows that RNA pol II was associated with IRF1 60 min after IFN-γ treatment. By contrast, association of RNA pol II with STAT1 was not detected at this or earlier time points. Control blots demonstrated the expected increase in STAT1 tyrosine phosphorylation and IRF1 protein expression.
Fig. 6.
IRF1 associates with RNA pol II complexes in IFN-γ-treated cells. BMDM obtained from WT mice were treated with 10 ng/ml (240 units/ml) IFN-γ for 30 or 60 min. Nuclei were isolated, and IP with the indicated Abs was performed. Western blot membranes were probed with an Ab to RNA pol II. Aliquots from the lysates were recovered before the IP (WCE) and analyzed by Western blot for the input of RNA pol II, Y701-phosphorylated STAT1 (pY701), and IRF1.
Discussion
The STAT and IRF protein families contribute in numerous ways to the development and regulation of innate and adaptive immune responses. As an example, STATs and IRFs interact functionally both in the synthesis of and response to type I IFN (35, 36). For the subgroup of genes represented by gbp2, the functional interaction does not appear to require tight association or cooperative binding of the two proteins, but rather results from a requirement for both STAT1 and IRF1 in the process of transcriptional activation. The main goal of this study was to gain insight into the IFN-γ-induced chromatin changes requiring, or resulting from, the STAT1–IRF1 interaction.
Both the distal and proximal IFN-γ response regions of the gbp2 promoter contain a canonical IRF binding site. Our data as well as those from previous studies (11, 19) show that these sites are occupied by IRF1 during an IFN-γ response. Residual gbp2 transcription still occurred in absence of IRF1 and may reflect exclusive STAT1 action or result from the activity of a different IRF family member. Distinguishing these possibilities requires further investigation. STAT1 binds the distal part of the IFN-γ response region using an imperfect GAS, whereas the proximal site represents one of the rather rare cases (15) where STAT1 binds a different promoter sequence as a consequence of IFN-γ signaling. In an analogous situation, the human 9/27 gene is rendered IFN-γ-inducible by a STAT1/IRF9 complex associating with an ISRE (12). The presence of IRF9 at the gbp2 promoter in IFN-γ-treated cells has not been tested.
Consistent with recently reported immunoprecipitation (IP) experiments (5, 29), our study demonstrates that CBP and HDAC1 are recruited to promoter chromatin in a STAT1-dependent manner. The almost identical kinetics of recruitment after IFN-γ treatment suggest that complexes of STAT1 dimers with CBP and/or HDAC1 are either formed before binding to gbp2 chromatin or that their assembly at the promoter is extremely rapid. Both the HAT function of CBP and HDAC1 activity are needed to efficiently activate gbp2 transcription (5, 23). CBP function closely correlates with promoter histone hyperacetylation, but the target of HDAC1 is still unknown. The concomitant recruitment of CBP and HDAC1 favors the assumption that their target proteins are different. Alternatively, they might be regulated to act on the same targets in different phases of the transcriptional cycle. The highly transient nature of histone 4 hyperacetylation at the gbp2 promoter is in good agreement with the assumption that deacetylase activity must closely follow that of the HAT.
Absence of IRF1 reduced, but did not abrogate CBP recruitment and gbp2 promoter hyperacetylation. The rather small decrease was closely correlated with, and most likely due to, the reduced association of the promoter with STAT1 dimers in irf1−/− cells. Reduced levels of STAT1 in gene-targeted cells result from IRF1's role in regulating constitutive STAT1 expression downstream of autocrine type I IFN activity (30). This IRF1 activity and its delayed binding to gbp2 promoter chromatin with respect to that of STAT1 binding argue against a helper function of IRF1 for Stat1 association. More likely the reduction of CBP recruitment and histone acetylation in irf1-deficient cells results from a combination of low STAT1 amounts and low affinity of the protein for the gbp2 IFN response region.
Using cells expressing IRF1 in absence of STAT1 dimers, we were able to show that IRF1 associates with the gbp2 promoter in the absence of prebound STAT1. Hence, STAT1 must neither directly contact IRF1 nor modify chromatin as a prerequisite for IRF1 binding. Furthermore, IRF1 association with the gbp2 promoter chromatin was unaffected by the absence of STAT1 TAD phosphorylation at S727 and the concomitant decrease in CBP association to STAT1 dimers and histone 4 hyperacetylation (5). Unperturbed association of IRF1 with the gbp2 promoter under these conditions shows that the reduction of IFN-γ-induced gbp2 expression in cells expressing STAT1S727A is correlated with an absence of histone hyperacetylation, not a lack of IRF1 binding. Our findings also suggest a very limited potential of IRF1 to recruit HATs to the gbp2 promoter. Otherwise, defective histone acetylation in STAT1S727A cells should be rescued by IRF1. gbp2 belongs with a group of IFN-γ-induced genes that strongly require STAT1 serine phosphorylation. It is tempting to speculate that IFN-γ-induced genes that are less dependent on the STAT1 serine phosphorylation require STAT1 interaction with transcriptional proteins that either reduce the need for histone hyperacetylation, or that, unlike IRF1, significantly contribute to HAT recruitment.
RNA pol II binding shows a virtually complete dependence on IRF1. Gain-of-function analysis in cells expressing IRF1 in absence of active STAT1 shows its limited intrinsic ability to stimulate gene expression, and IPs demonstrate that IRF1 and RNA pol II are parts of the same transcriptional complex. Therefore, our data are consistent with a division of labor between STAT1 and IRF1 in stimulating gbp2 expression. IRF1 plays an essential role in directing RNA pol II to the CAP site through its ability to contact either the enzyme itself or associated proteins, one of which might be TFIIB (37). The important role of IRF1 in RNA pol II recruitment is suggested not only by the coimmunoprecipitation experiment, but also by the nearly identical kinetics of IRF1 and RNA pol II association with gbp2 chromatin and the onset of nuclear gbp2 RNA accumulation. For STAT1, our findings support (at least) two different tasks in activating gbp2 gene transcription. First, it must stimulate irf1 mRNA transcription. Second, it must directly contribute to gbp2 promoter activation by creating a more permissive chromatin environment for RNA pol II through the recruitment of CBP and possibly other HATs. STAT1 is also essentially required for HDAC1 association with the gbp2 promoter chromatin, and HDAC1 is important for gbp2 expression (23). Identifying the relevant targets for this enzyme and determining whether they are identical to those associated with type I IFN-induced transcription (29, 38, 39) will be an important future task. Besides CBP STAT1 is instrumental in directing MCM proteins and possibly also other chromatin remodeling factors to target promoters (24). The question of whether IRF1 additionally contributes to the remodeling of promoter chromatin and structure will need to be answered. Several events appear to be parallel between the gbp2 promoter and the pIV promoter of the cIIta gene, encoding the master regulator of MHC II genes. Both promoters recruit STAT1 and IRF1 with similar kinetics and mediate a secondary, delayed response to IFN-γ (this work and refs. 26 and 40). Both employ the HAT activity of P300/CBP and the chromatin remodeling activity of the SWI/SNF subunit BRG1 (25, 26, 41). On the other hand, HDAC1 has so far not been implicated in CIITA regulation, and, conversely, the E box binding protein USF1, which is needed for cIIta gene stimulation (42), has not been linked to gbp2 mRNA expression. It will be interesting to see whether these differences really distinguish the two promoters or whether they reflect incomplete knowledge of their regulation. In the latter situation, the identical mechanisms of induction may represent a molecular paradigm for secondary response promoters stimulated by IFN-γ.
Materials and Methods
Antibodies.
Antiserum to the STAT1 C terminus used for Western blot analysis and ChIP assays was as described (43). STAT1 phospho-Y701 Ab was purchased from New England Biolabs (Beverly, MA). Monoclonal panERK Abs were purchased from Transduction Laboratories (Lexington, KY). Abs for IRF1 (M-20) and RNA Pol II (N-20) and a polyclonal antiserum to the N terminus of CBP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Ab against acetylated H4 was purchased from Upstate Biotechnology (Lake Placid, NY). Affinity-purified rabbit Abs were used to analyze HDAC1 by ChIP (23).
Cytokines and Reagents.
Recombinant mouse IFN-γ was used at a final concentration of 10 ng/ml (240 units/ml). Dox was purchased from Sigma–Aldrich (St. Louis, MO) and used at a final concentration of 1 μg/ml.
Cells.
Bone marrow-derived macrophages (BMDM) were obtained by culture of bone marrow in L-cell derived CSF-1 as described (44). Immortalized fibroblasts from WT, stat1−/− (45), and irf1−/− mice (46) (kindly provided by J. Pavlovic, University of Zurich, Zurich, Switzerland) were cultured in DMEM containing 10% FCS.
Plasmids and Transfections.
irf1 cDNA was generated by using RNA from IFN-γ-treated macrophages and inserted into pRetro-Off (Clontech, Palo Alto, CA). STAT1-deficient cells were pretreated with dox for 6 h before transfection by using ExGen (Fermentas) reagent according to the manufacturer's instructions. Cells were analyzed 48 h after transfection. Expression of IRF1 from the transfected plasmid was induced by removal of dox for 24 h.
ChIP.
A total of 107 cells (primary macrophages or fibroblasts with identical results) were used per time point. ChIP assays were performed as recently described (23) with the following Abs: αSTAT1-C (1:100), αIRF1 (3 μg), αRNA Pol II (3 μg), αCBP (3 μg), αacH4 (4 μg), and αHDAC1 (1:100). Primers used for the analysis of the proximal (PROX) and distal (DIS) gbp2 promoter were as described (5). For the irf1 promoter the following primers were used: forward, 5′-AGCACAGCTGCCTTGTACTTCC-3′, and reverse, 5′-CTTAGACTGTGAAAGCACGTCC-3′. All ChIP data presented in this work represent a minimum of three independent experiments.
RNA Preparation, cDNA Synthesis, and Quantitative RT-PCR.
Total RNA was isolated from 1 × 106 macrophages or fibroblasts by using the NucleoSpin RNA II kit (Macherey & Nagel, Düren, Germany) as described (47). Nuclear RNA was isolated from nuclear extracts, prepared as described (48). The cDNAs were reverse-transcribed from 5 μg of total or nuclear RNA. Real-time PCR experiments were normalized to the GAPDH housekeeping gene. Primers for real-time PCR of the GAPDH, gbp2, and irf1 genes were used as described (31, 47). Unspliced hnRNA was analyzed by real-time PCR using primer pairs recognizing exon–intron borders. hn-irf1: forward, 5′-ACATCGATGGCAAGGGATAC-3′, and reverse, 5′-GCATGCTGGGATGCTTTAAT-3′; hn-gbp2: forward, 5′-TCCAAGGCAGATGTTGTT-3′, and reverse, 5′-CTCCACAAC TGAGGACTCCA-3′.
Coimmunoprecipitation.
A total of 107 cells were harvested in PBS and resuspended in 300 μl of sucrose buffer (0.32 M sucrose/10 mM Tris·HCl, pH 8/3 mM CaCl2/2 mM MgOAc/0.1 mM EDTA/0.5% Nonidet P-40/1 mM DTT/0.5 M PMSF). The nuclei were pelleted and washed twice in sucrose buffer (without Nonidet P-40) and resuspended in 100 μl of low-salt buffer (20 mM Hepes/1.5 mM MgCl2/20 mM KCl/0.2 mM EDTA/25% glycerol/0.5 mM DTT/0.5 mM PMSF). Then, 100 μl of high-salt buffer (20 mM Hepes/1.5 mM MgCl2/800 mM KCl/0.2 mM EDTA/25% glycerol/1% Nonidet P-40/0.5 mM DTT/0.5 mM PMSF) was added slowly to the suspension. The lysates were incubated for 45 min at 4°C on a rotating wheel and centrifuged for 15 min at 14,000 × g. One-tenth of the supernatant was used as IP-input control and boiled with Laemmli buffer for 10 min. Binding of STAT1 C-terminal Ab (1:100 dilution) or with 3 μg of IRF1 Ab was performed overnight at 4°C. Protein A beads were added for 2 h, and the immunocomplexes were washed three times in low-salt buffer. The beads were boiled in 50 μl of Laemmli buffer and subjected to SDS gel electrophoresis.
Western Blot.
A protocol for this procedure was recently described (31).
Supplementary Material
Acknowledgments
We thank Manuela Baccarini for critical reading of our manuscript. This work was supported by Austrian Research Foundation Grant SFB F28-B13, Subproject 2803 (to T.D.). G.Z. and C.S. were supported by the Austrian Federal Ministry for Education, Science, and Culture (GEN-AU Project “Epigenetic Plasticity of the Mammalian Genome”) and Austrian Research Foundation Project P16443 (to C.S.).
Abbreviations
- BMDM
bone marrow-derived macrophage
- CBP
CREB-binding protein
- dox
doxycyclin
- IP
immunoprecipitation
- GAS
IFN-γ-activated site
- GBP
guanylate-binding protein
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- IRF
IFN regulatory factor
- ISRE
IFN-stimulated response element.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610944104/DC1.
References
- 1.Schroder K, Hertzog PJ, Ravasi T, Hume DA. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Wen Z, Zhong Z, Darnell JE., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 4.Decker T, Kovarik P. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 5.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. Immunity. 2003;19:793–802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 6.Lew DJ, Decker T, Strehlow I, Darnell JE. Mol Cell Biol. 1991;11:182–191. doi: 10.1128/mcb.11.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 8.Pine R, Decker T, Kessler DS, Levy DE, Darnell JE., Jr Mol Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 10.Kimura T, Nakayama K, Penninger JM, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, Taniguchi T. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 11.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, et al. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 12.Bluyssen HA, Muzaffar R, Vlieststra RJ, van der Made AC, Leung S, Stark GR, Kerr IM, Trapman J, Levy DE. Proc Natl Acad Sci USA. 1995;92:5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramana CV, Grammatikakis N, Chernov M, Nguyen H, Goh KC, Williams BR, Stark GR. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Proc Natl Acad Sci USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decker T, Kovarik P. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weihua X, Kolla V, Kalvakolanu DV. Proc Natl Acad Sci USA. 1997;94:103–108. doi: 10.1073/pnas.94.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacMicking JD. Trends Immunol. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Briken V, Ruffner H, Schultz U, Schwarz A, Reis LF, Strehlow I, Decker T, Staeheli P. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pine R, Canova A, Schindler C. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JJ, Vinkemeyer U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakharova N, Lymar ES, Yang E, Malik S, Zhang JJ, Roeder RG, Darnell JE., Jr J Biol Chem. 2003;278:43067–43073. doi: 10.1074/jbc.M308166200. [DOI] [PubMed] [Google Scholar]
- 23.Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder M, He W, Zhang JJ. Proc Natl Acad Sci USA. 2005;102:14539–14544. doi: 10.1073/pnas.0507479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattenden SG, Klose R, Karaskov E, Bremner R. EMBO J. 2002;21:1978–1986. doi: 10.1093/emboj/21.8.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni Z, Karaskov E, Yu T, Callaghan SM, Der S, Park DS, Xu Z, Pattenden SG, Bremner R. Proc Natl Acad Sci USA. 2005;102:14611–14616. doi: 10.1073/pnas.0503070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolet CM, Paulnock DM. J Immunol. 1994;152:153–162. [PubMed] [Google Scholar]
- 28.Anderson SL, Carton JM, Zhang X, Rubin BY. J Interferon Cytokine Res. 1999;19:487–494. doi: 10.1089/107999099313938. [DOI] [PubMed] [Google Scholar]
- 29.Nusinzon I, Horvath CM. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi T, Takaoka A. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 31.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy DE, Muller M, Decker T. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pine R. J Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhoff S, Schaper F, Hauser H. Nucleic Acids Res. 1993;21:2881–2889. doi: 10.1093/nar/21.12.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroger A, Dallugge A, Kirchhoff S, Hauser H. Oncogene. 2003;22:1045–1056. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- 35.Levy DE, Marie I, Prakash A. Curr Opin Immunol. 2003;15:52–58. doi: 10.1016/s0952-7915(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 36.Honda K, Yanai H, Takaoka A, Taniguchi T. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 37.Wang IM, Blanco JC, Tsai SY, Tsai MJ, Ozato K. Mol Cell Biol. 1996;16:6313–6324. doi: 10.1128/mcb.16.11.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I, Levy DE. Proc Natl Acad Sci USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakamoto S, Potla R, Larner AC. J Biol Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 40.Morris AC, Beresford GW, Mooney MR, Boss JM. Mol Cell Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 43.Kovarik P, Stoiber D, Novy M, Decker T. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baccarini M, Bistoni F, Lohmann Matthes ML. J Immunol. 1985;134:2658–2665. [PubMed] [Google Scholar]
- 45.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 46.Reis LF, Ruffner H, Stark G, Aguet M, Weissmann C. EMBO J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, et al. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 48.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.